1. Introduction
Although years have passed since the description of the first cases of SARS-CoV-2 infection in Wuhan, China, COVID-19 remains active but without considerable morbidity. SARS-CoV-2 infected individuals in general develop a severe acute respiratory syndrome, however, neurological [
1], cardiovascular, cutaneous, and renal manifestations [
2,
3] and multiple organ systems disorders [
4] are also observed. Most individuals with COVID-19 develop a mild respiratory illness, such as a dry cough, fever, dyspnea, and asymptomatic cases. However, the severity of COVID-19 clinical symptoms seems to be related to different factors, such as the period of virus exposure and consequent viral load, host immune capacity, population immunization rate, age, and characteristics of the circulant SARS-CoV-2 variant [
5].
In Brazil, so far, three epidemic waves have been observed driven by different variants of SARS-CoV-2. The first wave occurred in early April 2020 and was triggered by the strains B.1.1.28 and B.1.1.33 [
6]. The second and most devastating epidemic wave occurred between December 2020 and March 2021 and was marked by thousands of fatal cases due to the introduction of the P.1 (Gamma) strain [
7,
8,
9]. In December 2021, the number of positive cases for SARS-CoV-2 increased again, due to the rapid emergence of the Omicron variant.
The Omicron variant emergence brought a worldwide concern related to the number of positive cases of SARS-CoV-2 [
10,
11]. According to the literature, the rapid spread of Omicron is attributed to the presence of multiple mutations in the spike protein that may confer greater binding affinity to the host cell receptor and increased ability to evade immunity induced by vaccination and previous infection.
Although Omicron causes milder symptoms and less clinical severity in vaccinated individuals compared to previous SARS-CoV-2 variants, there are public health concerns due to the large number of cases, uncertainty regarding the effectiveness of the vaccines, and especially because in some countries children were not eligible for vaccination or boosters during the Omicron wave period [
12]. Since, even a milder infection can lead to an uncontrolled number of infections, which, in turn, will cause an overload of healthcare systems. Furthermore, observations regarding the reduced severity of clinical disease induced by Omicron may not accurately reflect the virulence of this variant, as significant mortality due to Omicron has been observed in elderly and under-vaccinated populations [
13]. Omicron also was associated with an increased risk of maternal morbidity and severe complications mostly among symptomatic and unvaccinated women [
14]. In this sense, to understand the epidemiological characteristics during the Omicron variant wave, we investigated, through genomic surveillance, the frequency, introduction, viral load in children, young people, and elderly people infected by SARS-CoV-2 in the state of São Paulo.
2. Materials and Methods
2.1. Sample collection
Nasopharyngeal swabs were collected from individuals presenting acute respiratory symptoms (fever, cough, sore throat, rhinorrhea, myalgia, headaches, anosmia, ageusia, and fatigue) in the Hospital das Clínicas of the Medical School of Ribeirão Preto/USP and State Health Departments. After that, the samples were received and processed by the COVID-RP Laboratory of the Blood Center of Ribeirão Preto to perform the molecular SARS-COV-2 diagnostic.
The samples were collected during the first 10 epidemiological weeks of 2022 (01.02.2022 to 03.12.2022). A total of 43.206 individuals from the region of Southeast Brazil were submitted to molecular diagnostics of SARS-CoV-2 and incidence of SARS-CoV-2 positive samples were monitored month by month, including Ct value and demographic profile of the patients. During this period, 17.695 (40.95%) samples were SARS-CoV-2 positive, including 779 (4.40%) from children of up to 10 years of age, 1.633 (9.23%) from adolescent population (11-19 years of age), 13.244 (74.85%) from adult population aged (20 to 59 years of age) and 2.039 (11.52%) from individuals >60 years old. In addition, we performed the genome analysis in 3.099 samples. The mean age of sequenced SARS-CoV-2 positive individuals was 37 ±18 years, 1.804 (58%) female and 1.295 (42%) male.
2.2. RNA extraction and SARS-CoV-2 infection diagnosis
For RNA extraction we used Extracta kit fast - DNA and RNA (Loccus) in automatic extractor Extracta 32 (Loccus) according to manufacturer’s instructions. The SARS-CoV-2 infection diagnosis was performed using the COVID19 Plus RealAmp Gene FinderTM kit (OSANG Healthcare Co. Ltd.). This assay allows for the molecular detection of the RdRp (RNA-dependent RNA polymerase), E and N genes of the SARS-CoV-2 virus and an internal control by reverse transcription and real-time multiplex PCR. The amplification reaction was performed on an ABI Prism 7500 Sequence Detection System (Thermo Fisher Scientific). Data analysis, including threshold values (cycle threshold (Ct)), baseline start, final values and interpretation of results, was defined according to the manufacturer’s instructions. All samples with cycle threshold (Ct) amplification values ≤35 for the internal control were considered for the interpretation of the results. Samples with amplification for the RdRp, E, and N genes of SARS-CoV-2 exhibiting Ct ≤40 were considered positive, except for the E gene alone. All samples with Ct ≤ 35 for at least two viral genes were selected for the SARS-CoV-2 genomic sequencing.
2.3. SARS-CoV-2 genomic sequencing
The SARS-CoV-2 genomic libraries were generated using the COVIDSeq kit (Illumina, San Diego, CA) [
15] and Oxford Nanopore Sequencing Kit (SQL-LSK 109) [
16,
17] following the manufacturer’s specifications. All samples with Ct ≤ 35 for at least two viral genes were selected for the SARS-CoV-2 genomic sequencing.
The PCR reaction products were combined for the tagmentation step of the genomic library. The tagged amplicon was purified and then amplified using indexes. After this point, the reactions were purified with the aid of magnetic beads, and the product from the libraries was grouped forming a single genomic library. We proceeded with library purification using 1x AMPure XP Beads following the manufacturer’s protocol. At the end of the process, the library was eluted in a specific buffer and the genetic material was quantified using Qubit® dsDNA HS DNA Quantitation Kits (Invitrogen), following the manufacturer’s instructions. Paired-end libraries were sequenced on Illumina’s MiSeq (V2 kit, 2150 cycles) or NextSeq. 2000 (P2 kit, 2100 cycles) platform.
For Nanopore technology, amplicons were purified using 1x AMPure XP beads, and cleaned-up PCR product concentrations were measured using the Qubit dsDNA HS assay kit. DNA library preparation was carried out using the ligation sequencing kit and the native barcoding kit (NBD104 and NBD114, Oxford Nanopore Technologies, Oxford, UK) [
17,
18]. Sequencing libraries were loaded into a (FLO-MIN106) flow cell (Oxford Nanopore Technologies). In each sequencing run, we used negative controls to prevent and check for possible contamination with less than 2% mean coverage.
2.4. Generation of consensus sequences
Illumina raw sequences were submitted to quality control analysis using the FastaQC software version 0.11.8. Cleaning was performed using Trimmomatic version 0.3.9. to select the best quality sequence score (>30). We mapped the quality-filtered sequences against the SARS- CoV-2 reference (Genbank RefSeq NC_045512.2) using BWA and used SAMtools for indexing the mapping results.
The mapped files were submitted for improvement using Pilon to correct possible deletions and insertions caused by the mapping process. The quality-filtered sequences were subjected to a remapping against the genome improved by Pilon. Finally, we use bcftools for variant calling and seqtk for the assembly of the consensus SARS-CoV-2 genomes. Positions covered by fewer than 10 reads (DP < 10) and bases with a quality score lower than 30 were considered as an assembly gap and thus converted into Ns. Coverage values for each genome were calculated using SAMtools v1.12. We assessed the consensus genome sequence quality using Nextclade v0.8.1.11.
Nanopore raw files were basecalled using Guppy, and barcode demultiplexing was performed using qcat. Consensus sequences were generated by de novo assembling using Genome Detective (
https://www.genomedetective.com/).
2.5. Phylogenetic analysis
For phylogenetic analysis, we analyzed 471 newly obtained sequences against a dataset containing 4660 SARS-CoV-2 genomes which were obtained from GISAID (
https://www.gisaid.org/). For virus taxonomy, we followed the steps of an already established canonical pipeline as implied by Nextstrain v.3.0.3. [
18]. In brief viral genomes were aligned using MAFFT v.7 and IQtree v.3 was used for phylogenetic tree reconstruction. The maximum likelihood (ML) phylogenetic trees were visualized using FigTree v. 1.4.4 and additionally edited by the R ggtree package (R package for the visualization of tree and annotation package).
2.6. Statistical analysis
The statistical analysis was performed in GraphPad Prism version 5.0 (La Jolla). We applied Kruskal-Wallis and Dunn tests for multiple comparisons. We considered α= 0.05 and *p<0.05, ** p<0.01 and *** p<0.001 were considered statistically significant.
3. Results
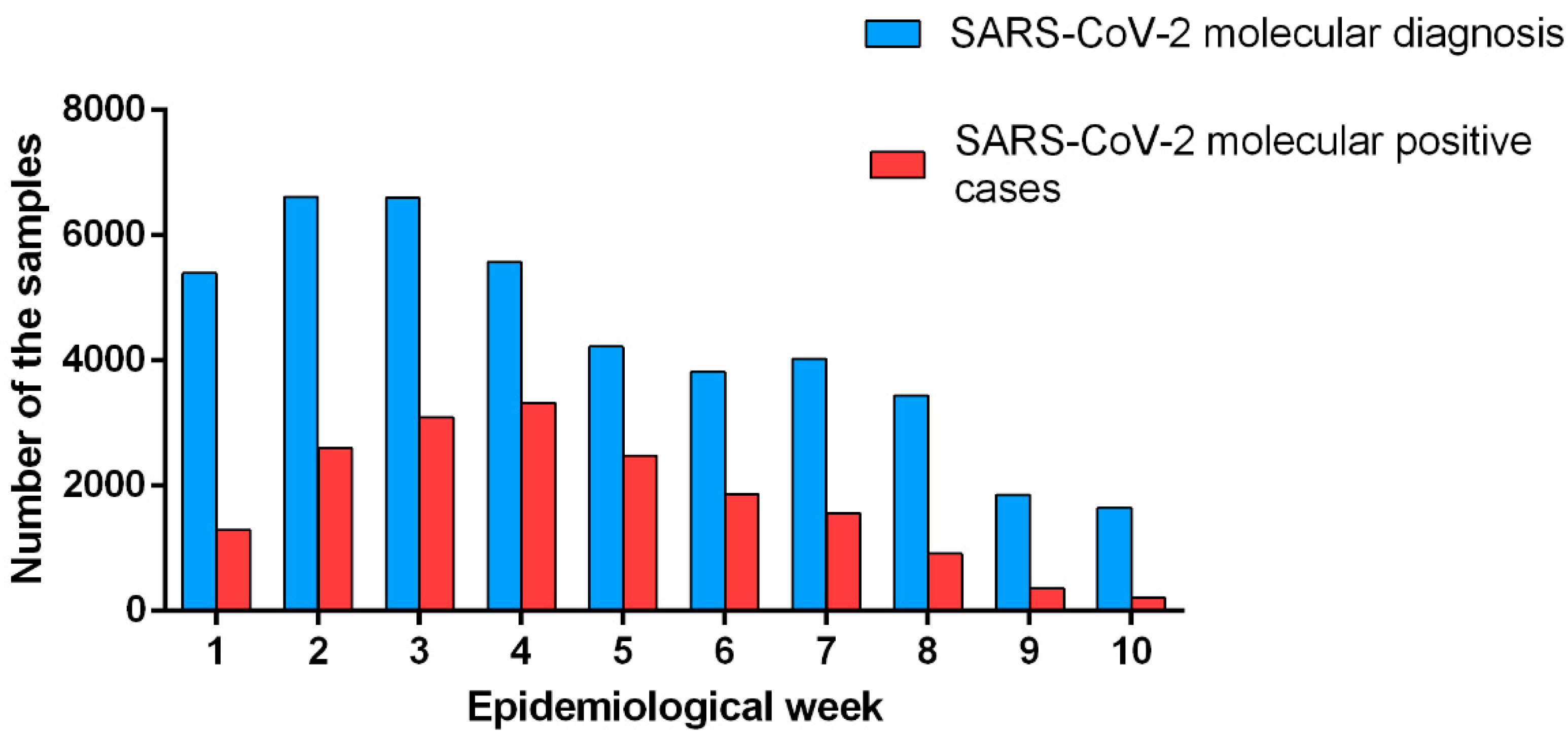
Initially, we performed an analysis to investigate the incidence of SARS-CoV-2 positive cases in our region in the first 10 epidemiological weeks of 2022. During this period, the positivity rate for SARS-CoV-2 reached 41%, gradually increasing month by month. According to
Figure 1, there was an increase of 1.2-fold in the number of tested SARS-CoV-2 samples during the 1st (01.02.2022 to 01.08.2022) to the 3rd epidemiological week (01.16.2022 to 01.22.2022). The increasing number of molecular tests was accompanied by an increase in the SARS-CoV-2 positivity rising from 24% to 47% respectively. From the 4th epidemiological week (01.23.2022 to 01.29.2022), the number of SARS-CoV-2 tests decreased, however, the number of positive cases remained stable and reached high reaching levels of 60% of the total number of infected individuals. From the 5th epidemiological week (01.30.2022 to 02.05.2022) the number of SARS-CoV-2 molecular exams continued to fall and the number of SARS-CoV-2 positive cases began gradually decreasing over the epidemiological weeks, reaching 13% of positivity rate for SARS-CoV-2 in the 10th epidemiological week (
Figure 1).
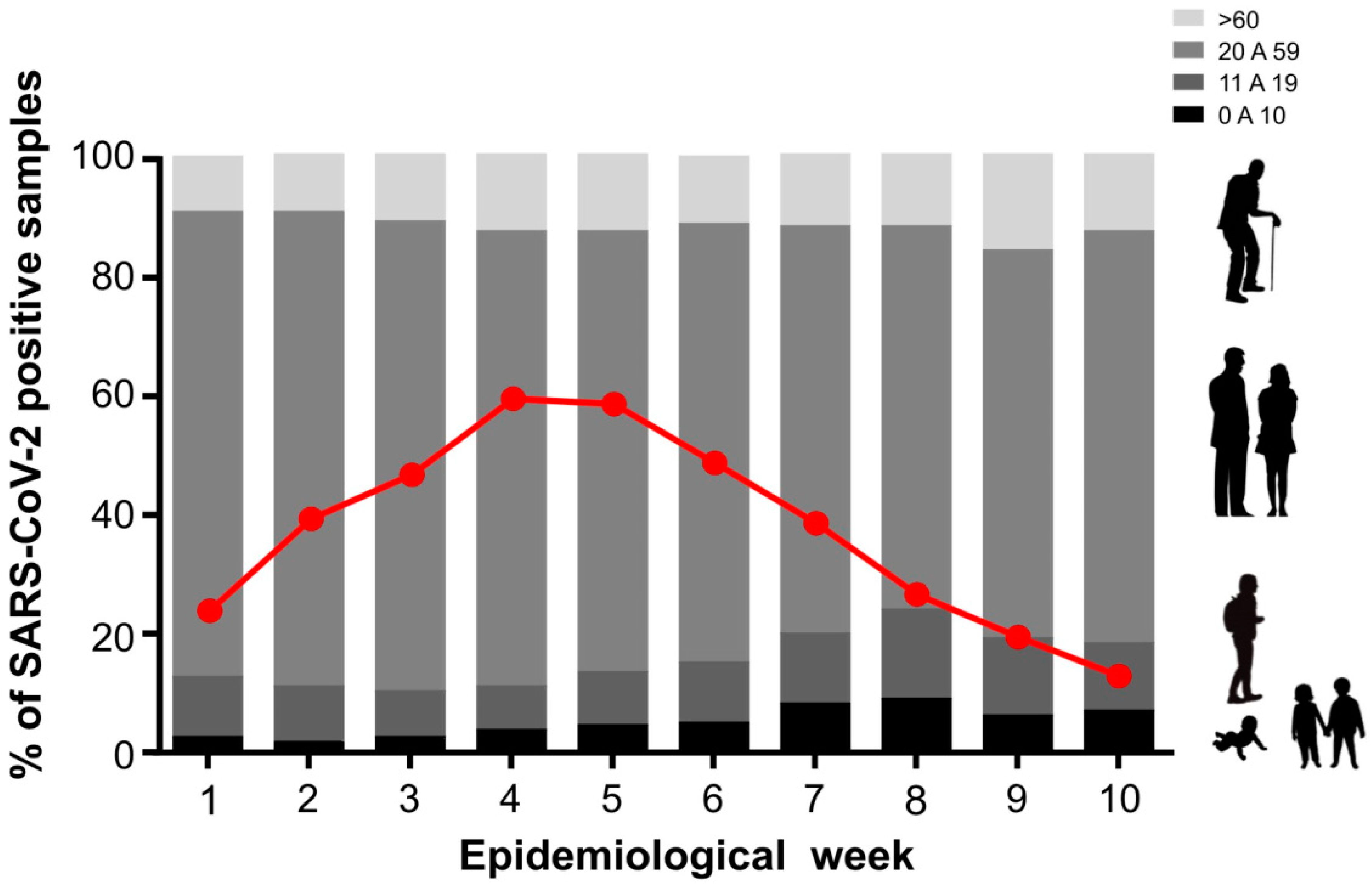
Due to the increase in SARS-CoV-2 cases observed in the first months of the year 2022, we examined the demographic profile of the SARS-CoV-2-positive cases. Our main question was to understand whether SARS-CoV-2 was more prevalent according to age since we consider that at the time of this work, the majority of the adult population, that is, those over 20 years old, would be immunized with both vaccine doses and also the booster dose. As shown in
Figure 2A, the majority of SARS-CoV-2 positive cases occurred between the epidemiological weeks 4-5, and we also observed an increase in children of up to 10 years of age (from 2.8% in the 1st epi week to 9.2% in the 8th epi week) (
Figure 2B). The average age of this group of children was 6 ± 3.2 years, 48% female and 52% male. When we evaluated the same period for the adolescent population (11-19 years of age), we also observed an increase in the number of SARS-CoV-2 positive cases, rising from 9.8% to 14.8% in the 8th epidemiological week (
Figure 2B). In the adolescent population, the mean age was 17 ± 2.6 years, with 54% female and 46% male. The frequency of SARS-CoV-2 positive cases for the adult population (20 to 59 years of age) remained unchanged. This was also regarding adults over 60 years old, we noted no significant difference in the number of SARS-CoV-2 infections over the epidemiological weeks (
Figure 2). The increase in the number of pediatric SARS-CoV-2 infected individuals may be related to the emergence of a new variant or also due to the availability of vaccines, considering the immunization scheme adopted in Brazil for different ages (
Figure 3).
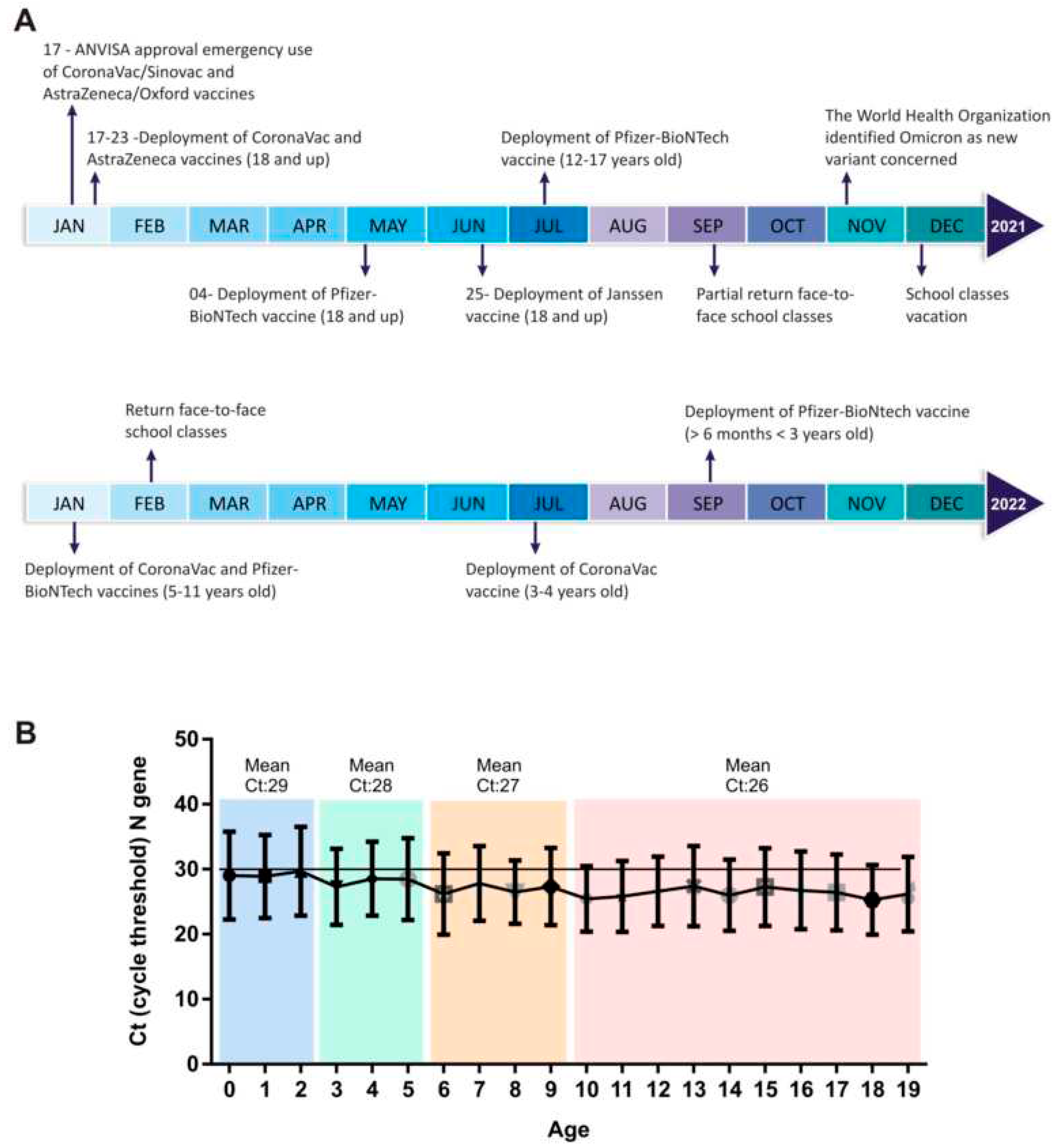
To understand the relationship between the burden of SARS-CoV-2 and age group, we compared the amplification Cts of the SARS-CoV-2 N gene obtained from individuals with different age groups. In the majority of the epidemiological weeks, we observed a significant difference in Ct of samples belonging to younger patients compared to other age groups. The Ct of samples from children aged 0-10 years were higher than the Ct observed in individuals >60 years old (Supplementary material). According to our results, the group of early age presents a higher Ct value for the viral SARS-CoV-2 N gene, suggesting a lower load in this group of patients (
Figure 3B). Interestingly, when the pediatric group was divided by age into four groups, it was possible to observe a gradual decrease in the Ct value (higher viral load). Children aged up to 2 years have average Ct= 29 and the age group from 3 to 5 years has an average Ct=28. Along with the increase in age, the Ct value decreased, once children aged 6 to 9 years old had a mean Ct=27 and from 10 years old onwards the mean Ct was 26 (
Figure 3B).
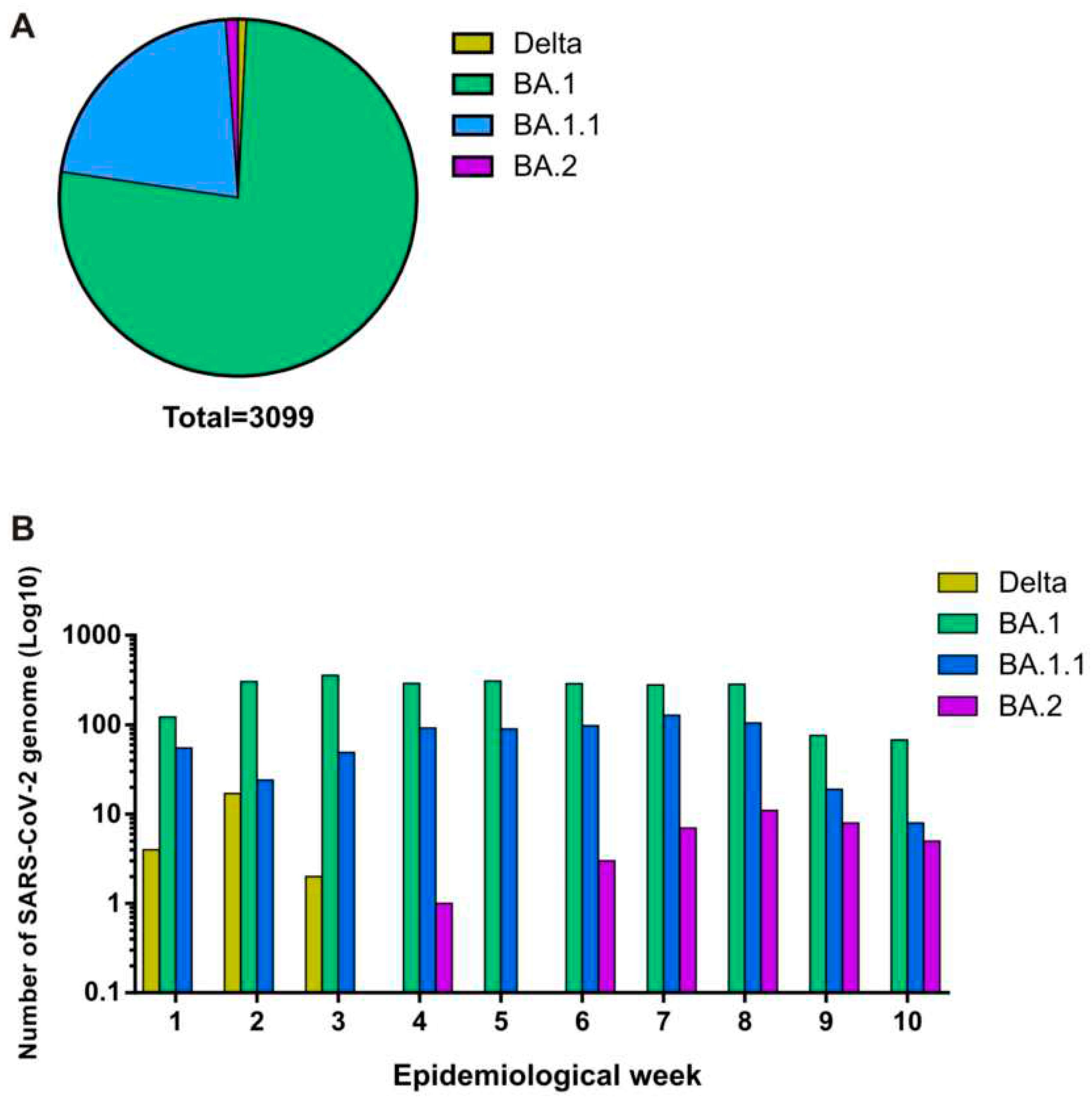
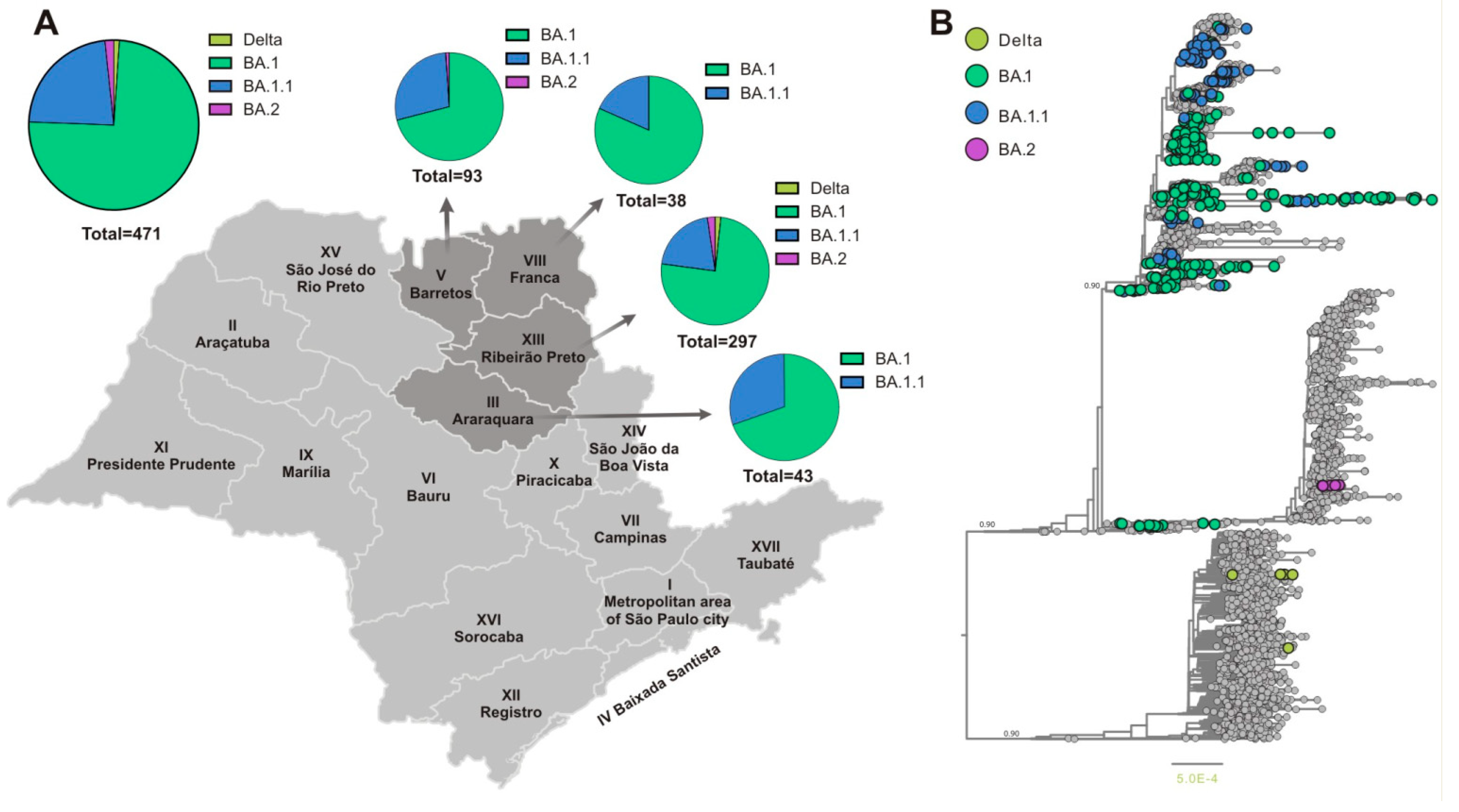
Subsequently, we investigated the circulating SARS-CoV-2 variants from a region of Southeast Brazil, composed of 90 counties, including Araraquara (24), Barretos (18), Franca (22), and Ribeirão Preto (26) and together they reached a population coverage of 3.307.320 habitats. A total of 3,099 whole genomes were generated which were collected between 02 January and 12 March 2022 and data analysis revealed that SARS-CoV-2 sequencing achieved an average coverage of 98% of the gene and an average depth of 3.414 (±1941) times per sample. In accordance with the epidemiological situation, SARS-CoV-2 variants BA.1(76.5 %), BA.1.1(21.5 %), BA.2 (1.3%), and Delta (0.75%) were identified (
Figure 4A). Our analysis revealed the predominance of the Omicron lineage during the first 10 epidemiological weeks of the year 2022. The complete lineage substitution of Delta to Omicron lineage was observed after epidemiological week 3 (
Figure 4B). The first case of BA.2 was also identified in epidemiological week 4 and this variant was frequently detected until the final of the evaluated period (
Figure 4B).
Considering the increased number of SARS-CoV-2 infections in younger patients and that during the period of this study, they were partially immunized becoming an important source of viral dissemination, we investigated the distribution of the SARS-CoV-2 variants in this group. Demographic characteristics showed that the mean age of younger patients (0 to 19 years) was 14 ± 5.6 years, with 236 (50.1%) females and 235 (49.9%) males. Similar to adult (20 to 59 years of age) and older (>60 years old) groups in younger patients the lineage Omicron (BA.1) was the most prevalent (74.5%), followed by BA.1.1 (23%), BA.2 (1.7%) and Delta (1%) (Figura 5A). Then we investigate the distribution of SARS-CoV-2 variants in these younger patients (0 to 19 years of age) considering four different Department of Public Health (DPH) from São Paulo State (Araraquara, Barretos, Franca, and Ribeirão Preto). Our analysis revealed that in this group 100% of the SARS-CoV-2 genomes from Araraquara (III) and Franca (VIII) DPH were classified as Omicron. A predominance of Omicron BA.1 was also observed in Barretos (V) (71%) and Ribeirão Preto (XIII) (75%) DPH (
Figure 5B). The variant BA.1.1 appeared as the second variant well represented in our dataset, reaching 23% of all genomes. However, in Barretos, 1% of the sequence was classified as BA.2 and Ribeirão Preto showed the presence of BA.2 (2.3%) and Delta (1,7%) (
Figure 5B).
The performed phylogenetic analysis (
Figure 5B) showed that almost all analyzed sequences (n=458/471) belonged to the Omicron VOC and a small number (n=5/471) to Delta VOC and the BA.2 sublineage (n=8/471). The small number of Delta VOC sequences corresponded to the gradual replacement of the Delta VOC in Brazil by the Omicron VOC. The Omicron VOC sequences BA.1. and BA.1.1 that were obtained from younger patients formed several small monophyletic clusters which were randomly interspersed along the whole Omicron VOCs circulating worldwide. This clusterization might be related to several independent BA.1 and BA1.1. introductions in this part of the São Paulo state which corresponded to the epidemiological situation and the rapid dissemination of the Omicron VOC in the state of São Paulo by that time. We also were able to identify a small number of BA.2 sequences that were clustered together, probably indicating sustained transmission of this Omicron VOC sublineage by that time. However, due to the very initial steps of BA.2 introduction we suggest that this clustering reflects the initial dissemination steps of this VOC in this part of the State, rather than specific genomic features of the detected BA.2 sequences.
4. Discussion
Genomic surveillance of SARS-CoV-2 has allowed the monitoring of the infection, as well as the knowledge to follow the evolution of the SARS-CoV-2 variants worldwide. In this work, we monitored the SARS-CoV-2 infection in people living in southeastern Brazil during the first 10 weeks of the year of 2022. We observed a growing increase in the number of SARS-CoV-2 positive cases until the 3rd epidemiological week. According to our results, this increase in infected individuals is directly related to the introduction of the Omicron variant in our region, since, the phylogenetic analysis revealed that 76% and 22% of the SARS-CoV-2 genomes evaluated in this period were classified as BA.1 e BA.1.1 respectively. Other studies corroborate our finding, showing that the arrival of Omicron VOC caused an important change in the epidemiological scenario, culminating in the replacement of the Delta variant by Omicron in approximately 3.5 weeks, and reached a dominant plateau in different Brazilian capitals (Silva et al., 2023). Furthermore, one of the relevant features of the Omicron variant is its ability to cause reinfection. This aspect has been reinforced by research that demonstrated low cross-neutralization against the Omicron variant from previous non-Omicron viral infection or two-dose mRNA vaccination [
19].
When we consider the age of the SARS-CoV-2 positive sample, it was observed that the number of children and adolescents infected had increased over the epidemiological weeks. This observation was also reported by studies that evaluated the pandemic caused by SARS-CoV-2 in other countries and raised the hypothesis that this increase was due to the greater potential for transmission of the Omicron variant, less frequent use of face masks among children than adults, and low vaccination rate in the pediatric population, with only children aged 12 years and older being eligible for vaccination at the time of the study [
20]. Another issue that we investigated was the relationship between age and SARS-CoV-2 viral load by analyzing the Ct (threshold cycle) of the nucleocapsid viral region (N gene). Significant differences were found related to the Ct value and age, suggesting that early age presents a higher Ct value for the viral SARS-CoV-2 N gene, and probably the lower proviral load should be observed in this group of patients. Three previous studies have described those viral loads in children with asymptomatic SARS-CoV-2 infection or mild illness are comparable [
21] or slightly lower than viral loads in adults with SARS-CoV-2 infection [
22,
23]. These findings are reinforced by Jia 2021 which showed that the cytokine profiles of children with mild symptoms of COVID-19 resemble those of healthy children, reflecting a low level of inflammation [
24].
It is important to note that according to our data, the number of children and adolescents with COVID-19 is increasing and this group may be responsible for the spread of new variants of SARS-CoV-2. During the period of this study, this particular group of pediatric individuals was not eligible or had not completed the anti-SARS-CoV-2 immunization (
Figure 3A)[
20]. In addition, schools returned to face-to-face classes and many Brazilian states eased security measures by allowing large events, and the mandatory use of masks is no longer in force. Our analysis found that in the pediatric group, 1.7% of the samples were classified as BA.2 sublineage. The emergence of Omicron lineage and subvariants BA.1, BA.1.1, BA.2, BA.3, BA.4, BA.5, and BA.2.12.2 had been reported in different continents [
25,
26]. The Omicron variant presents milder disease but it is significantly more contagious and has caused more hospitalizations, especially in unvaccinated children younger than 5 years and unvaccinated or incompletely vaccinated adults [
27,
28].
All this data reinforces the considerable positive effect of vaccination against COVID-19. Our study compared the epidemiological data during the Omicron wave in the unvaccinated (under 5 years), partially vaccinated (5-11 years) and completely vaccinated (over 12 years). Even though we do not have data related to the symptoms of evaluated individuals, we consider that the high number of SARS-CoV-2- infected did not increase the number of deaths, suggesting that vaccines offered low effectiveness against Omicron infection, but significant protection against disease severity, especially after the booster dose. According to Araujo da Silva, 2022, the distribution age of patients admitted to a hospital in Rio de Janeiro during the Omicron season, showed more children between 0 and 2 years and fewer children older than 12 years compared with the previous two years [
29]. Furthermore, it was observed that only 2 (3.3%) of hospitalized patients had received the full available COVID-19 vaccine, and the majority of admitted cases of the disease occurred in children where the vaccine was not available for their respective ages or did not receive two doses [
29].
In conclusion, we should be aware of the variation in the number of SARS-CoV-2-infected individuals, which may represent an evolutionary pressure for immune escape and encourage the emergence of new variants. Even if there remains some uncertainty about the future directions of SARS-CoV-2 infection, constant genomic surveillance for SARS-CoV-2 strains remains essential to determine the role of viral evolution and the spread of variants in different Brazilian regions.
Supplementary Materials
Real-time PCR analysis showing the CT values for the N gene of SARS-CoV-2. Each graphic corresponds to one specific epidemiological week (EW) (1 to 10). The different shades of gray represent the different age groups evaluated.
Acknowledgements
We thank all contributors from GISAID. We also thank Sandra Navarro Bresciani for the artwork and Lilian Beatriz Moreira and Ana Carla Medeiros for their help with the SARS-CoV-2 molecular diagnosis. This study was supported by the Fundação Butantan, Fundação de Amparo à Pesquisa do Estado de São Paulo (Grant Numbers: 2020/10127- 1; 2020/06441-2), Genomic surveillance of SARS-CoV-2 in collaboration with health service partners of SUS founded by PAHO/WHO and Fundação Hemocentro Ribeirão Preto.
Ethics Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee of the Faculty of Medicine of Ribeirão Preto, University of São Paulo (CAAE: 50367721.7.1001.5440).
References
- Liguori, C. , Pierantozzi, M., Spanetta, M., Sarmati, L., Cesta, N., Iannetta M., et al. (2020) Subjective neurological symptoms frequently occur in patients with SARS-CoV2 infection. Brain Behav. Immun. 88:11-16.
- Parker, ER. , Fitzpatrick, A. (2022) A case report of COVID-19-associated erythema nodosum: a classic presentation with a new trigger. Fam. Pract. 25:cmab177. [CrossRef]
- Armaly, Z. , Kinaneh, S., Skorecki, K. (2021) Renal Manifestations of Covid-19: Physiology and Pathophysiology. J. Clin. Med.10(6):1216. [CrossRef]
- Raman, B. , Bluemke, D.A., Lüscher, T.F., Neubauer, S. (2022) Long COVID: post-acute sequelae of COVID-19 with a cardiovascular focus. Eur. Heart. J. 14;43(11):1157-1172. [CrossRef]
- Long B, Carius BM, Chavez S, Liang SY, Brady WJ, Koyfman A, Gottlieb M. Clinical update on COVID-19 for the emergency clinician: Presentation and evaluation. Am J Emerg Med. 2022 Apr;54:46-57. [CrossRef]
- Alcantara, L.C.J. , Nogueira, E., Shuab, G., Tosta, S., Fristch, H., Pimentel, V., et al. (2022) SARS-CoV-2 epidemic in Brazil: how variants displacement have driven distinct epidemic waves. Virus Res. 315:198785. [CrossRef]
- Naveca, F.G. , Nascimento, V., de Souza, V.C., Corado, A.L., Nascimento F., Silva G., et al. (2021) COVID-19 in Amazonas., Brazil., was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 27(7):1230-1238. [CrossRef]
- Giovanetti M, Slavov SN, Fonseca V, Wilkinson E, Tegally H, Patané JSL, et al. Genomic epidemiology of the SARS-CoV-2 epidemic in Brazil. Nat Microbiol. 2022 Sep;7(9):1490-1500. [CrossRef]
- Alcantara, L.C.J. , Nogueira, E., Shuab, G., Tosta, S., Fristch, H., Pimentel, V., et al. (2022) SARS-CoV-2 epidemic in Brazil: how variants displacement have driven distinct epidemic waves. Virus Res. 315:198785. [CrossRef]
- Viana, R. , Moyo, S., Amoako, DG., Tegally, H., Scheepers, C., Althaus, C.L., et al. (2022) Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature.603(7902):679-686. [CrossRef]
- Callaway, E. Heavily mutated Omicron variant puts scientists onalert. Nature 2021; 600:21.
- Hyams C, Challen R, Marlow R, Nguyen J, Begier E, Southern J, King J, Morley A, Kinney J, Clout M, Oliver J, Gray S, Ellsbury G, Maskell N, Jodar L, Gessner B, McLaughlin J, Danon L, Finn A; AvonCAP Research Group. Severity of Omicron (B.1.1.529) and Delta (B.1.617.2) SARS-CoV-2 infection among hospitalised adults: A prospective cohort study in Bristol, United Kingdom. Lancet Reg Health Eur. 2023 Feb;25:100556. [CrossRef]
- Taylor, L. Covid-19: Hong Kong reports world’s highest death rate as zero covid strategy fails. BMJ. 2022 Mar 17;376:o707. [CrossRef] [PubMed]
- Villar J, Soto Conti CP, Gunier RB, Ariff S, Craik R, Cavoretto PI, Rauch S, Gandino S, Nieto R, Winsey A, Menis C, Rodriguez GB, Savasi V, Tug N, Deantoni S, Fabre M, Martinez de Tejada B, Rodriguez-Sibaja MJ, Livio S, Napolitano R, Maiz N, Sobrero H, Peterson A, Deruelle P, Giudice C, Teji JS, Casale RA, Salomon LJ, Prefumo F, Cheikh Ismail L, Gravett MG, Vale M, Hernández V, Sentilhes L, Easter SR, Capelli C, Marler E, Cáceres DM, Albornoz Crespo G, Ernawati E, Lipschuetz M, Takahashi K, Vecchiarelli C, Hubka T, Ikenoue S, Tavchioska G, Bako B, Ayede AI, Eskenazi B, Thornton JG, Bhutta ZA, Kennedy SH, Papageorghiou AT; INTERCOVID-2022 International Consortium. Pregnancy outcomes and vaccine effectiveness during the period of omicron as the variant of concern, INTERCOVID-2022: a multinational, observational study. Lancet. 2023 Feb 11;401(10375):447-457. [CrossRef]
- DNA Pipelines R&D., Farr, B., Rajan, D., Betteridge, E., Shirley, L., Quail, M., et al.., (2020) COVID-19 ARTIC v3 Illumina library construction and sequencing protocol. ProtocolsIo. 5:1-16. [CrossRef]
- Quick, J. , Grubaugh, N.D., Pullan, S.T., Claro, I.M., Smith, A.D., Gangavarapu, K., et al. (2017) Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 12:1261–1276. https:// doi.org/10.1038/nprot.2017.066.
- Quick, J. (2020) nCoV-2019 sequencing protocol. protocols.
- Lima, A.R.J. , Ribeiro, G., Viala, V.L., de Lima, L.P.O., Martins, A.J., Barros, C.R.D.S., et al. (2022) SARS-COV-2 genomic monitoring in the state of São Paulo unveils two emerging AY.43 sublineages. J Med Virol. 1:10.1002/jmv.27674. [CrossRef]
- Zou J, Xia H, Xie X, Kurhade C, Machado RRG, Weaver SC, Ren P, Shi PY. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat Commun. 2022 Feb 9;13(1):852. [CrossRef]
- National Plan for the Operationalization of Vaccination Against Covid-19. Coordenação-Geral de Documentação e Informação – Editora MS – OS 2022/0441. 2 Edition. Ministério da Saúde:bvsms.saude.gov.br.
- Jones, TC. , Biele, G., Mühlemann, B., Veith, T., Schneider, J., Beheim-Schwarzbach, J., et al. (2021) Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 9;373(6551):eabi5273. [CrossRef]
- Chung, E. , Chow, E.J., Wilcox, N.C., Burstein, R., Brandstetter, E., Han, P.D., et al. (2021) Comparison of Symptoms and RNA Levels in Children and Adults With SARS-CoV-2 Infection in the Community Setting. JAMA Pediatr. 175(10):e212025. [CrossRef]
- Madera, S. , Crawford, E., Langelier, C., Tran, N.K., Thornborrow, E., Miller, S., et al. (2021) Nasopharyngeal SARS-CoV-2 viral loads in young children do not differ significantly from those in older children and adults. Sci. Rep. 4;11(1):3044. [CrossRef]
- Jia, R. , Wang, X., Liu, P., Liang, X., Ge, Y., Tian, H., et al. (2020) Mild Cytokine Elevation, Moderate CD4+ T Cell Response and Abundant Antibody Production in Children with COVID-19. Virol Sin. 35(6):734-743. [CrossRef]
- Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, et al.. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 Mar;603(7902):679-686.
- Tegally, H. , Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Moyo S., Amoako D.G., Baxter C., et al. Continued Emergence and Evolution of Omicron in South Africa: New BA.4 and BA.5 lineages. Nat. Med. 2022;28:1785–1790. [CrossRef]
- Setiabudi D, Sribudiani Y, Hermawan K, Andriyoko B, Nataprawira HM. The Omicron variant of concern: The genomics, diagnostics, and clinical characteristics in children. Front Pediatr. 2022 Aug 2;10:898463. [CrossRef]
- Oliveira EA, Oliveira MCL, Silva ACSE, Colosimo EA, Mak RH, Vasconcelos MA, Silva LR, Martelli DB, Pinhati CC, Martelli-Júnior H. Clinical Outcomes of Omicron Variant (B.1.1.529) Infection in Children and Adolescents Hospitalized With COVID-19 in Brazil With Observational Data on the Efficacy of the Vaccines in Adolescents. Pediatr Infect Dis J. 2023 Mar 1;42(3):218-225. [CrossRef]
- Araujo da Silva AR, de Carvalho BRR, Esteves MDM, Teixeira CH, Souza CV. The Role of COVID-19 Vaccinal Status in Admitted Children during OMICRON Variant Circulation in Rio de Janeiro, City-Preliminary Report. Vaccines (Basel). 2022 Apr 15;10(4):619. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).