Submitted:
03 January 2024
Posted:
04 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Identification of ZIP genes in C. sativus
2.2. Gene structure and motif analysis
2.3. Phylogenetic analysis and multiple sequence alignment
2.4. Gene duplication analysis and genome distribution
2.5. Analysis of promoter regions of CsZIP genes
2.6. Analysis of transcriptome data
2.7. Plant materials and growth conditions
2.8. RNA isolation and real-time PCR analysis
3. Results
3.1. Identification of CsZIP genes from C. sativus pan-genome
3.2. Characterization of CsZIP genes from Chinese long 9930
| Accession name | Accession group | Accession name | Accession group |
|---|---|---|---|
| 9930 | East Asian cultivated accession | Hx117 | Indian cultivated accession |
| XTMC | East Asian cultivated accession | Hx14 | Indian cultivated accession |
| Cu2 | East Asian cultivated accession | W4 | Indian wild accession |
| Cuc37 | Eurasian cultivated accession | W8 | Indian wild accession |
| Gy14 | Eurasian cultivated accession | Cuc64 | Indian wild accession |
| 9110gt | Eurasian cultivated accession | PI183967 | Indian wild accession |
| Cuc80 | Xishuangbanna cultivated accession |
| Protein name | 9930 | XTMC | Cu2 | Cuc80 | Cuc37 | Gy14 | 9110gt | PI183967 | Cuc64 | W4 | W8 | Hx14 | Hx117 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CsZIP1 | 249 | 350 | 350 | - | 350 | 350 | 350 | 350 | 350 | 350 | 350 | 350 | 350 | |
| CsZIP2 | 417 | 417 | 417 | 417 | 417 | 441 | 417 | 417 | 417 | 417 | 417 | 408 | 417 | |
| CsZIP3 | 228 | 275 | 275 | 275 | 275 | 306 | 275 | 275 | 306 | 275 | 275 | 275 | 275 | |
| CsZIP4 | 594 | 594 | 639 | 594 | 594 | 594 | 594 | 594 | 594 | 594 | 594 | 594 | 594 | |
| CsZIP5 | 367 | 367 | 367 | 365 | 367 | 367 | 367 | 367 | 369 | 367 | 365 | 369 | 815 | |
| CsZIP6 | 463 | 463 | 463 | 463 | 463 | 407 | 463 | 463 | 463 | 463 | 463 | 463 | 463 | |
| CsZIP7 | 335 | 335 | 335 | 335 | 335 | 335 | 335 | 335 | 335 | 302 | 335 | 335 | 346 | |
| CsZIP8 | 349 | 349 | 349 | 349 | 349 | 349 | 349 | 349 | 349 | 349 | 349 | 349 | 1096 | |
| CsZIP9 | 334 | 334 | 334 | 334 | 334 | 337 | 334 | - | 338 | 334 | 334 | 334 | 337 | |
| CsZIP10 | 354 | 354 | 354 | 354 | 354 | 354 | 354 | 354 | 354 | 354 | 354 | 354 | 354 |
| Gene | Introns | Chromosomal site | Gene length | protein length | Protein MW(kDa) | Isoelectric point | Subcellular location predicted |
|---|---|---|---|---|---|---|---|
| CsZIP1 | 2 | 1 | 2955 | 249 | 27.2 | 6.88 | Cell membrane |
| CsZIP2 | 3 | 2 | 3491 | 417 | 44.5 | 6.09 | Chloroplast |
| CsZIP3 | 9 | 4 | 9406 | 228 | 24.6 | 7.02 | Cell membrane |
| CsZIP4 | 4 | 4 | 5063 | 594 | 62.0 | 6.27 | Cell membrane |
| CsZIP5 | 2 | 4 | 2777 | 367 | 38.8 | 5.98 | Cell membrane |
| CsZIP6 | 10 | 5 | 21173 | 463 | 49.9 | 5.97 | Cell membrane |
| CsZIP7 | 1 | 6 | 2038 | 334 | 35.7 | 5.73 | Chloroplast |
| CsZIP8 | 0 | 6 | 1490 | 348 | 38.0 | 5.58 | Cell membrane |
| CsZIP9 | 2 | 7 | 7007 | 334 | 35.9 | 6.02 | Cell membrane |
| CsZIP10 | 2 | 7 | 4076 | 354 | 37.2 | 8.53 | Cell membrane |
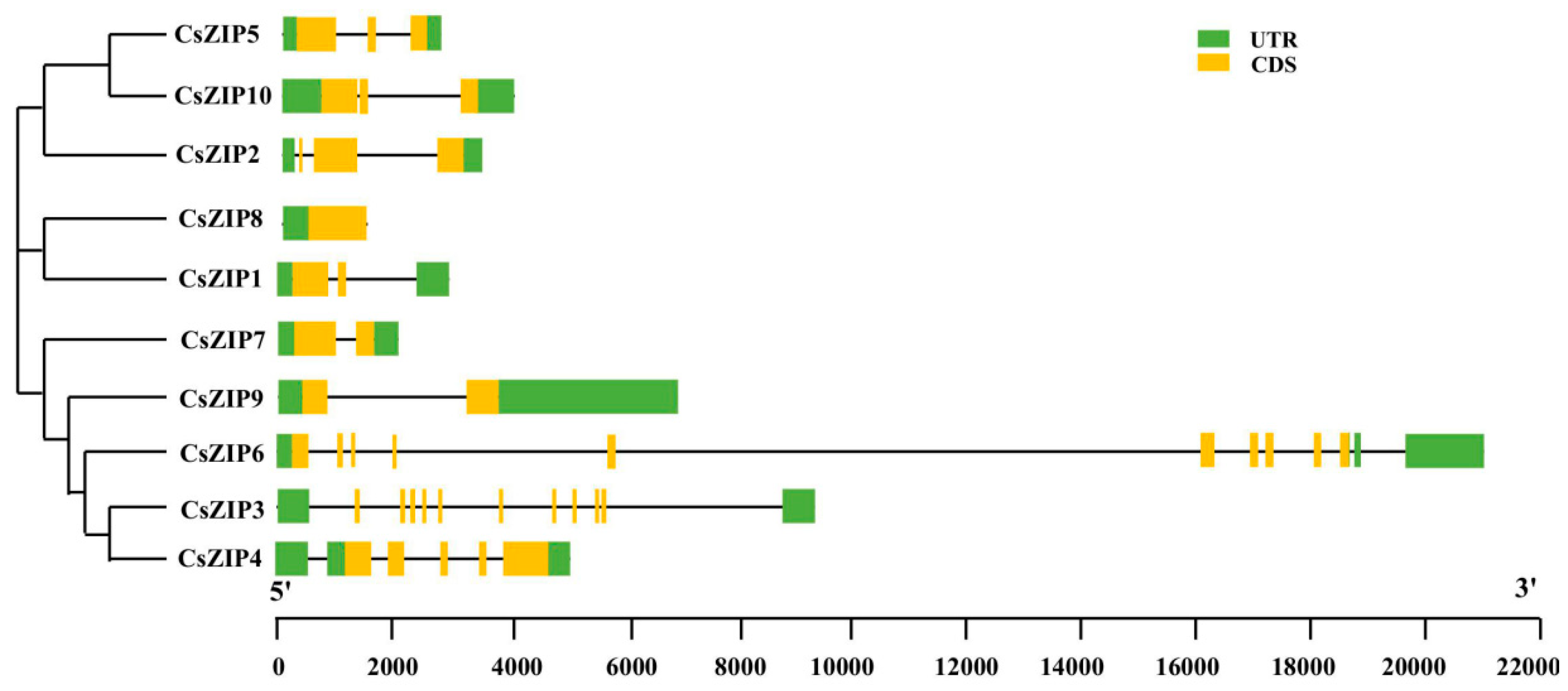
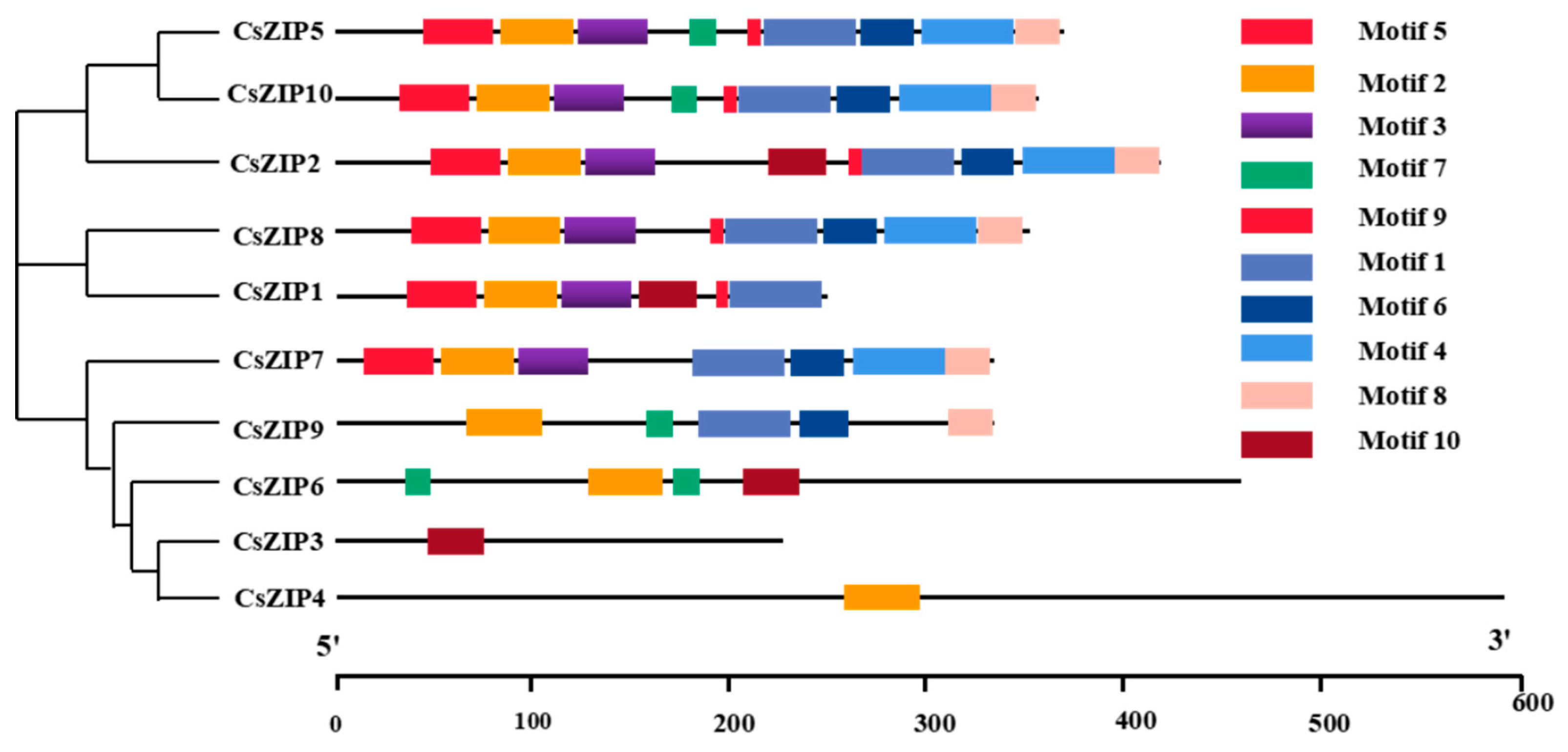
3.3. Analysis of phylogenetic relationship and gene structure and protein motif
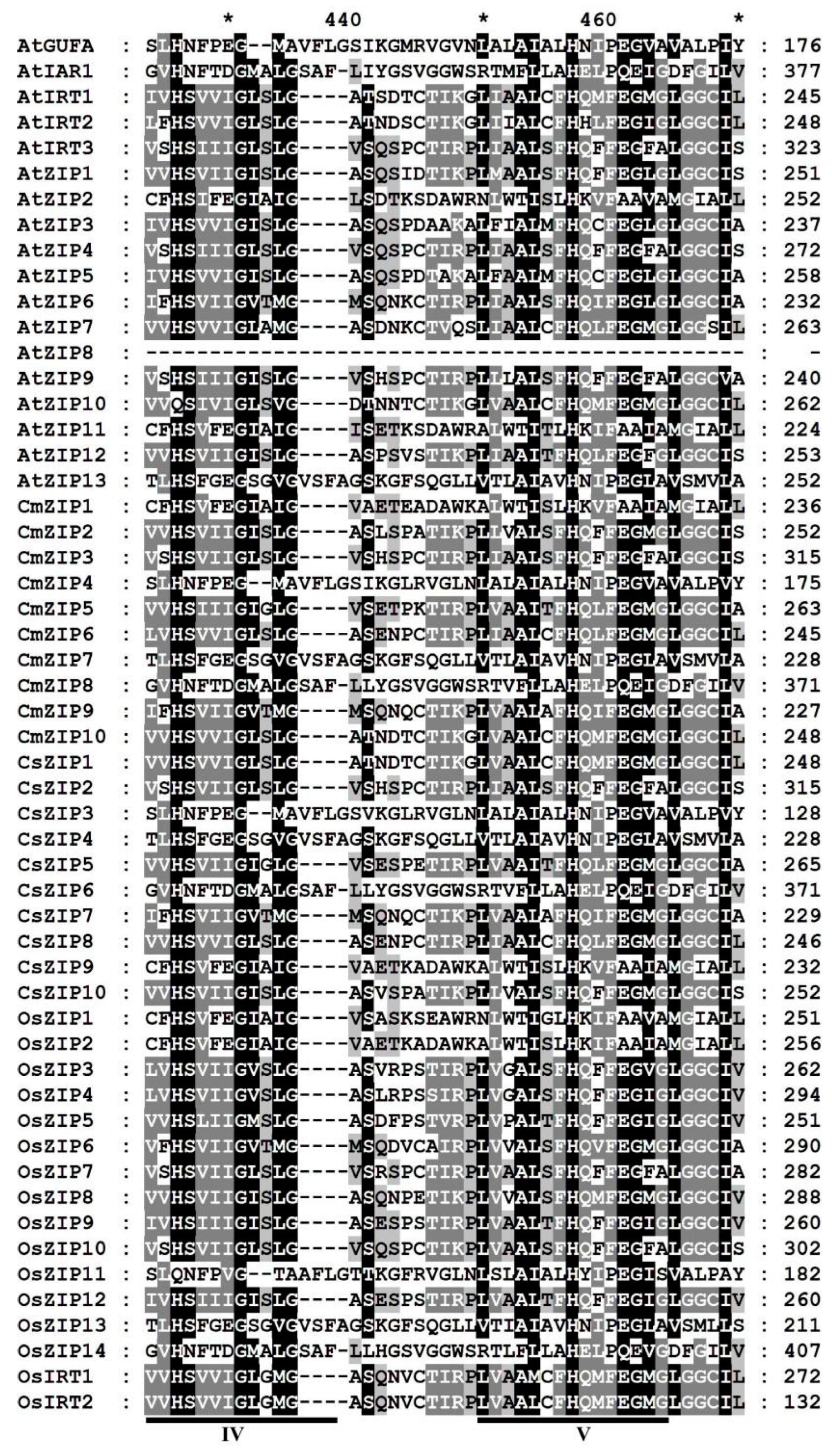
3.4. Multiple sequence alignment
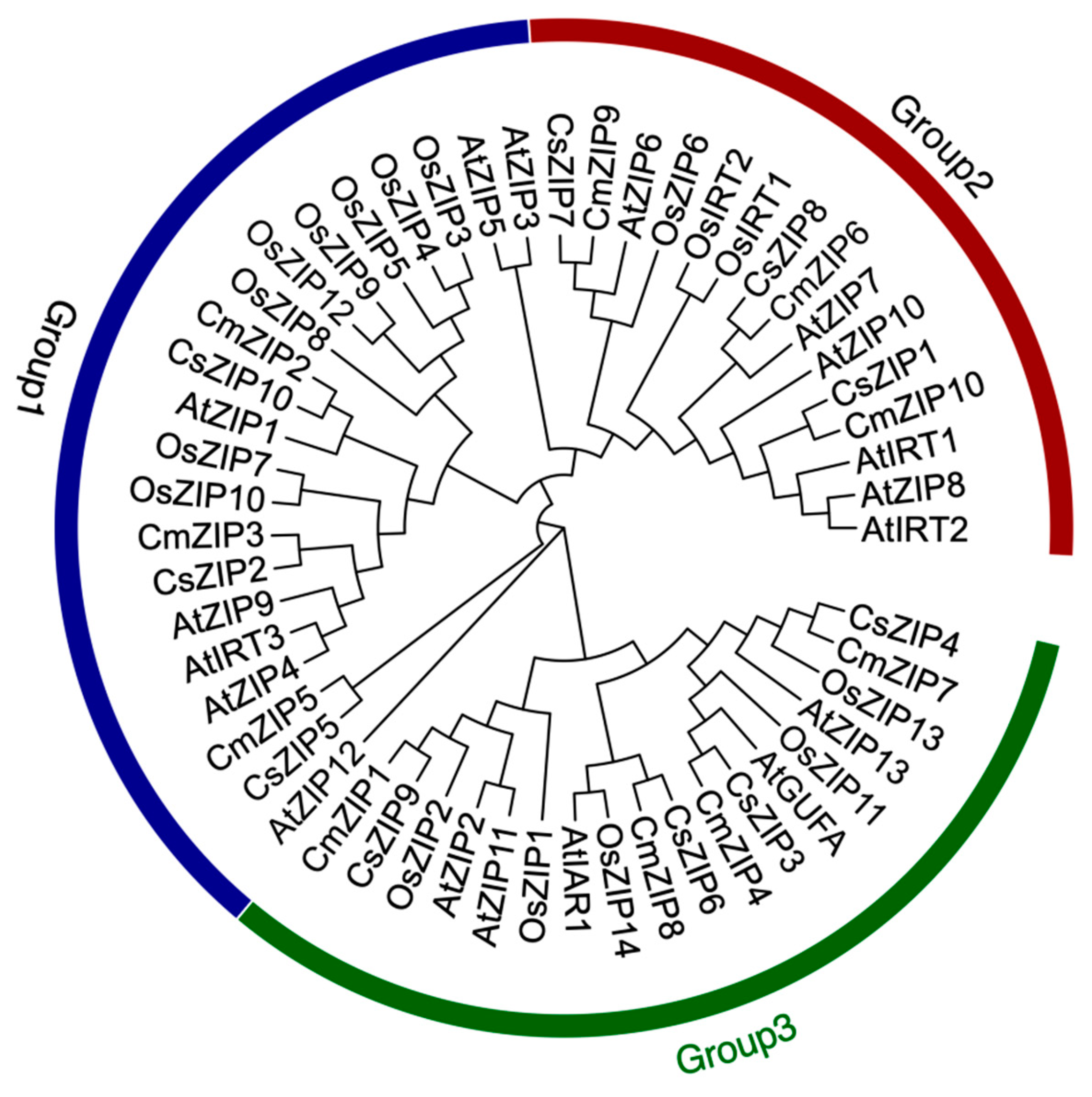
3.5. Phylogenetic analysis among different species
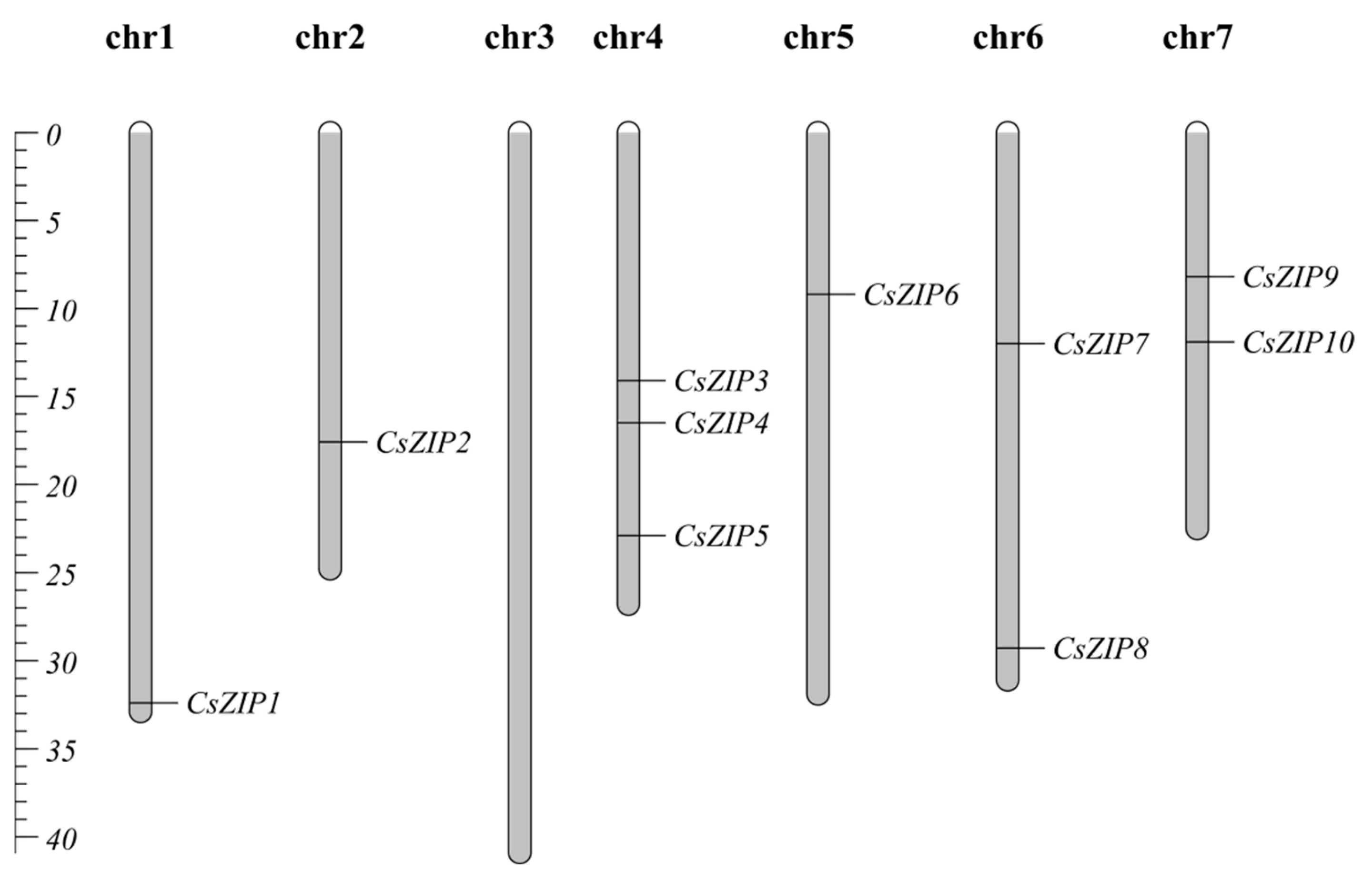
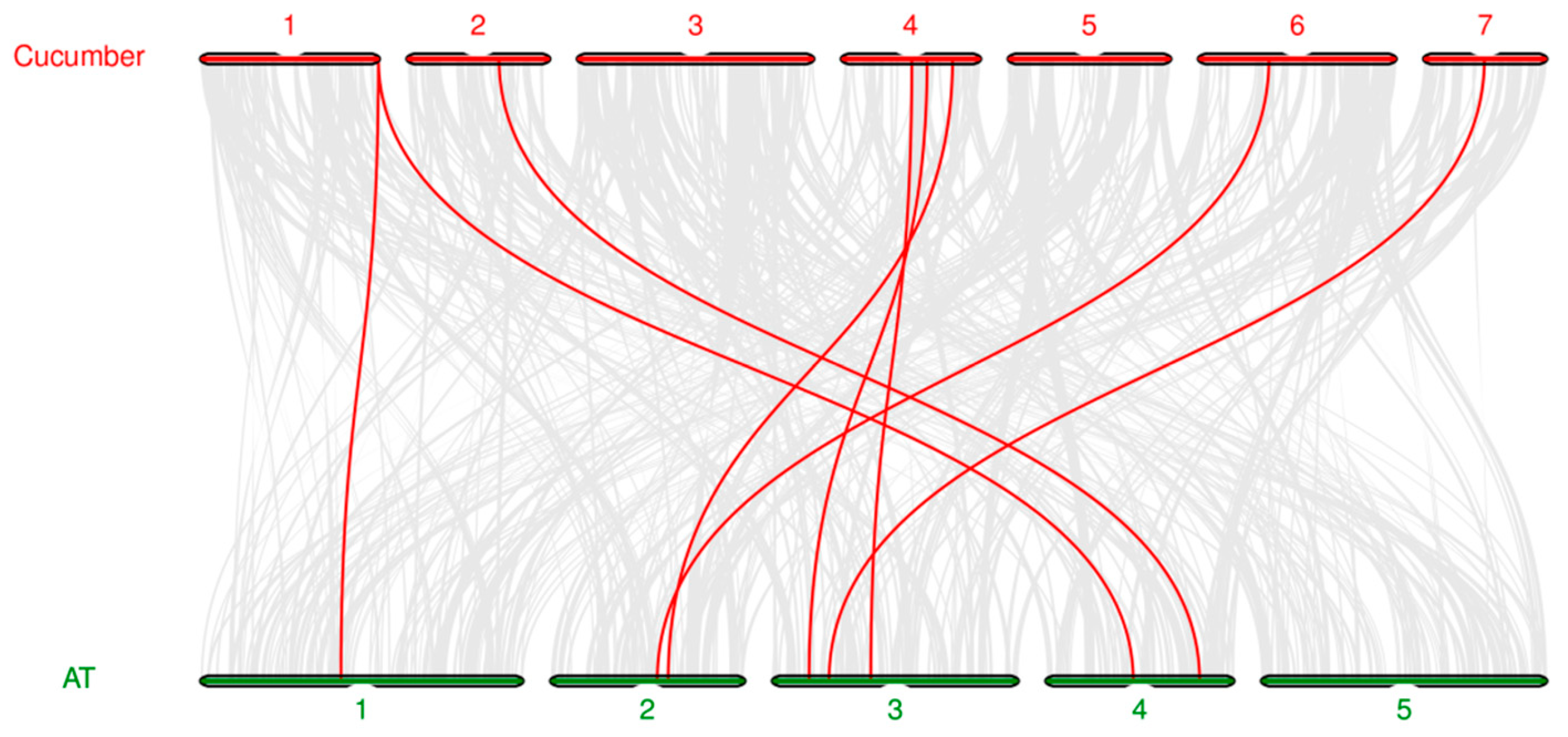
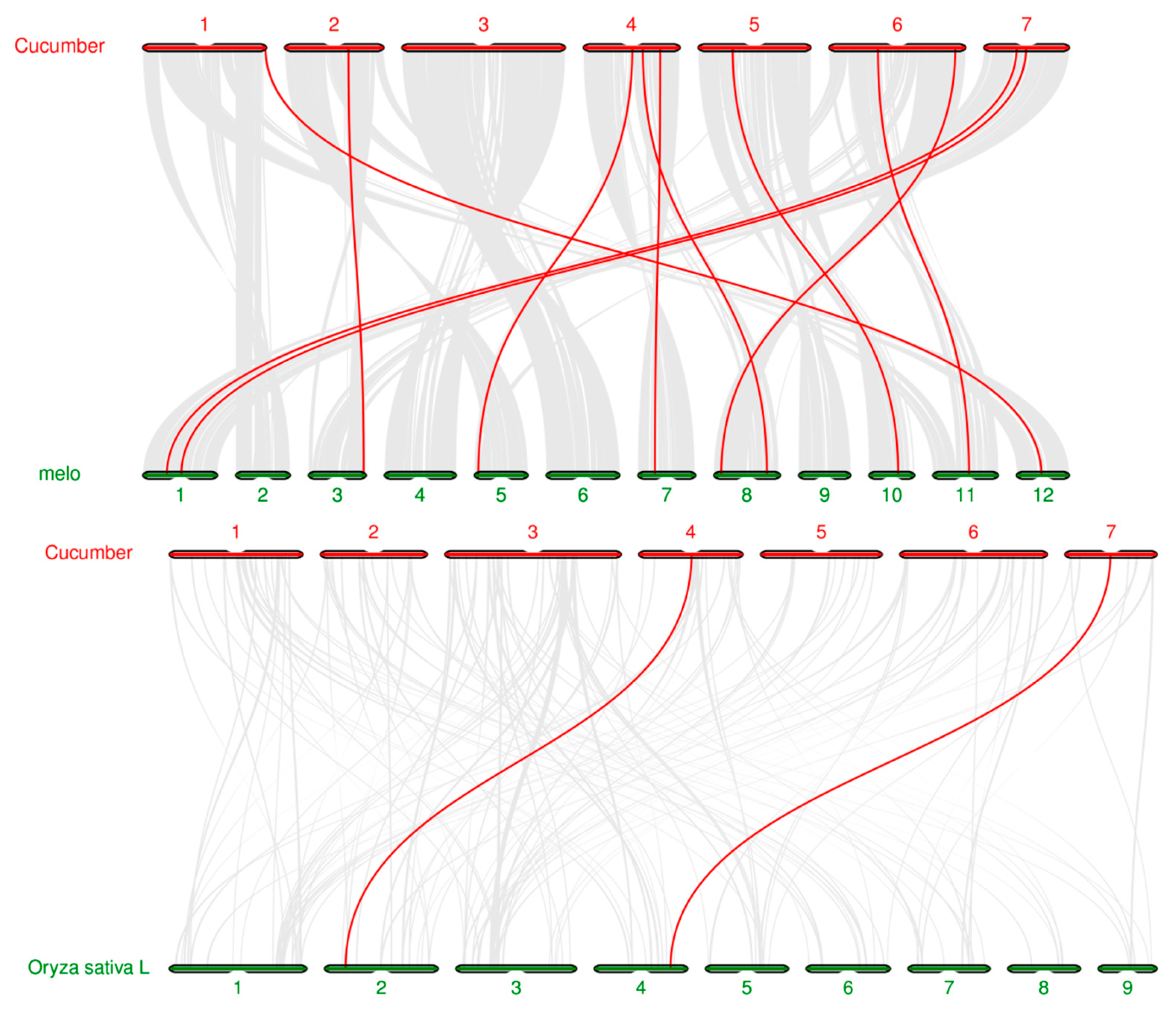
3.6. Chromosome localization and collinearity analysis
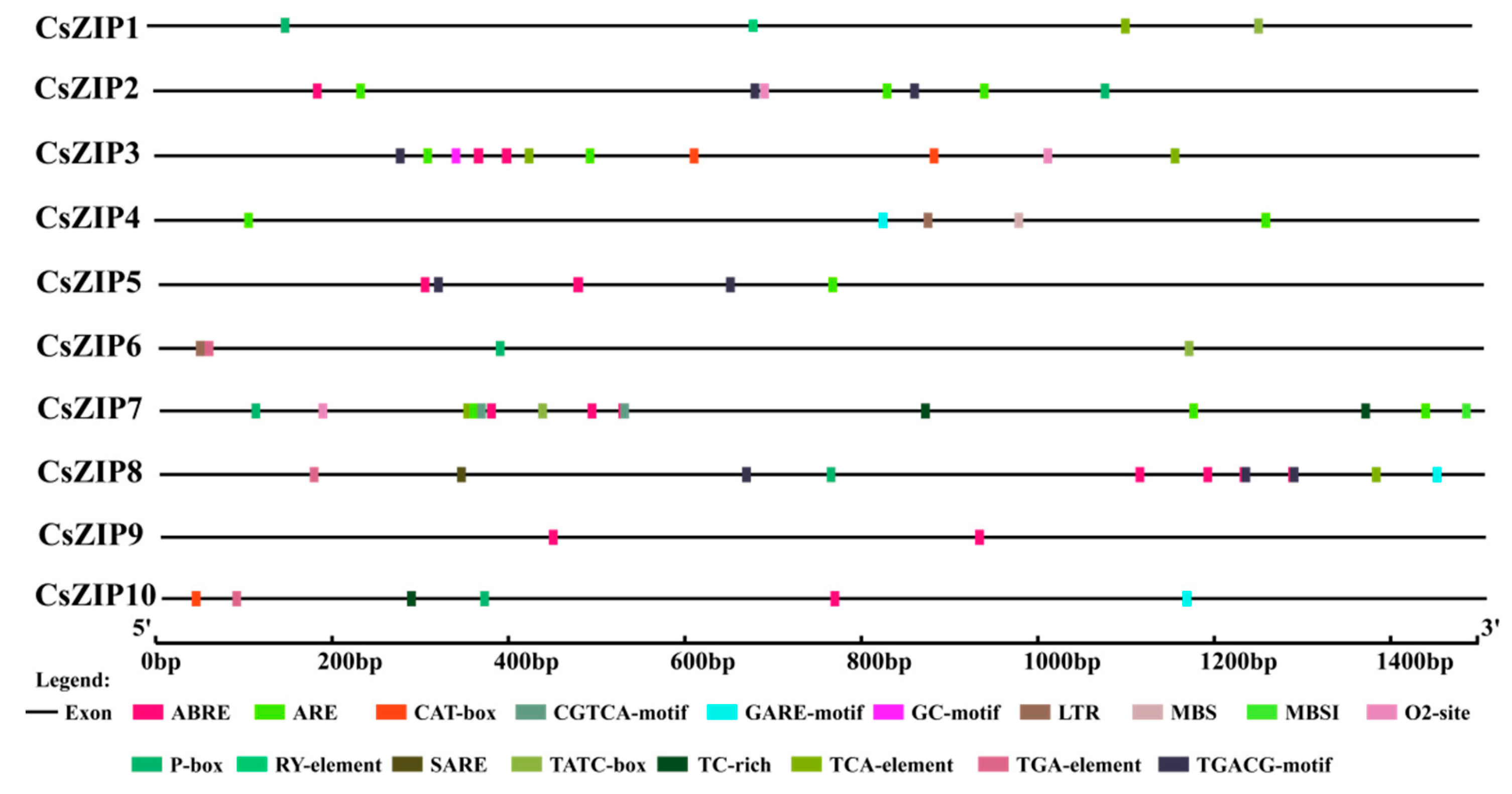
3.7. Analysis of cis-acting elements in promoter region of CsZIP genes
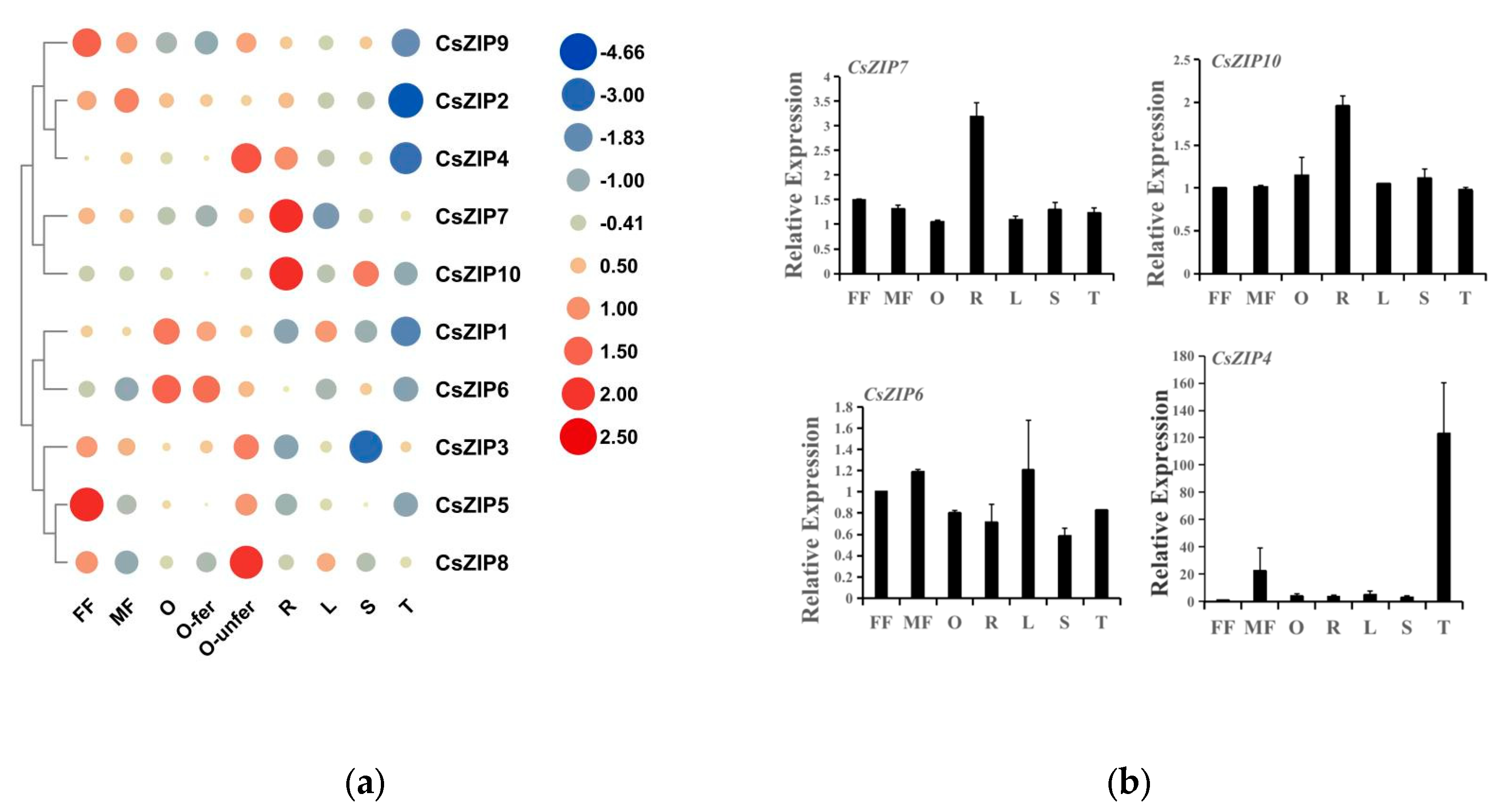
3.8. Expression of CsZIP genes in different tissues
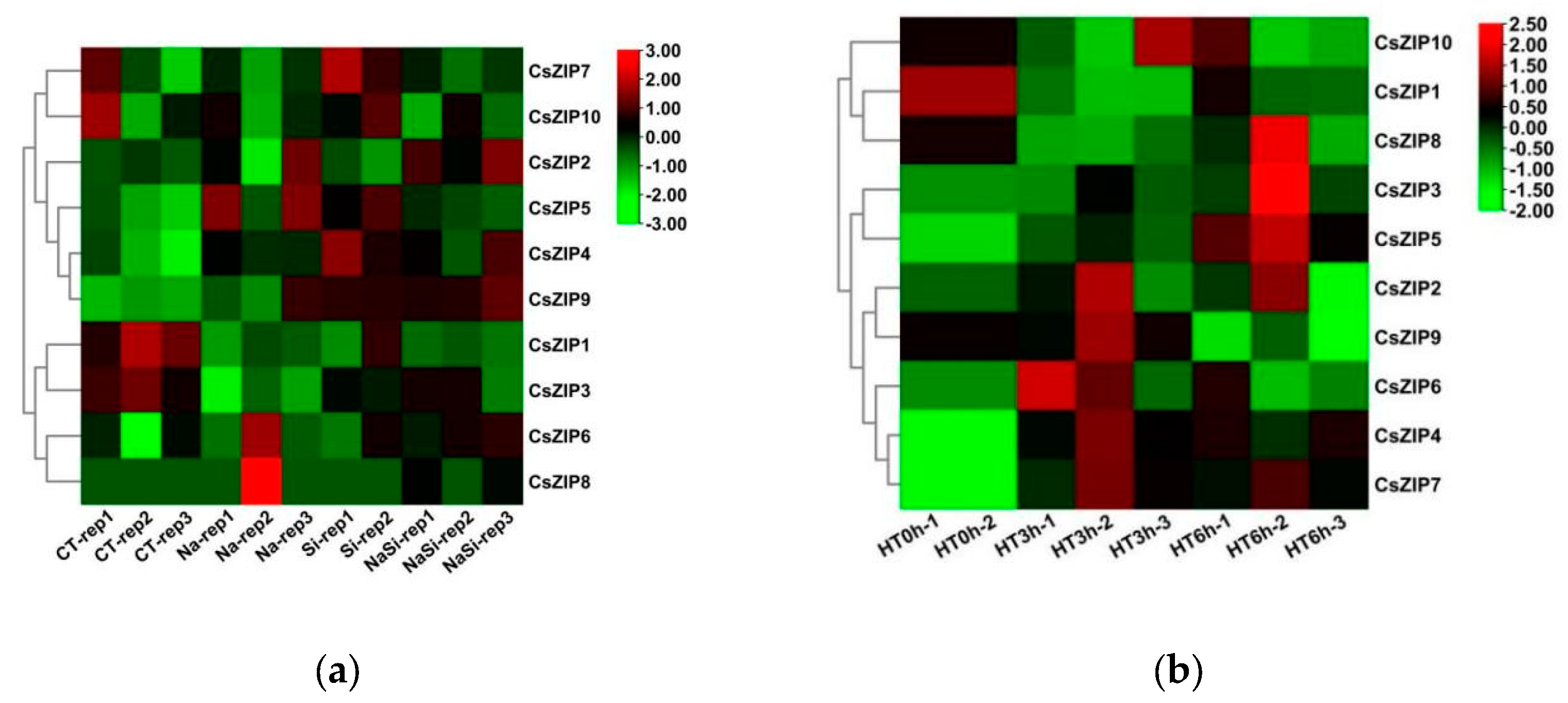
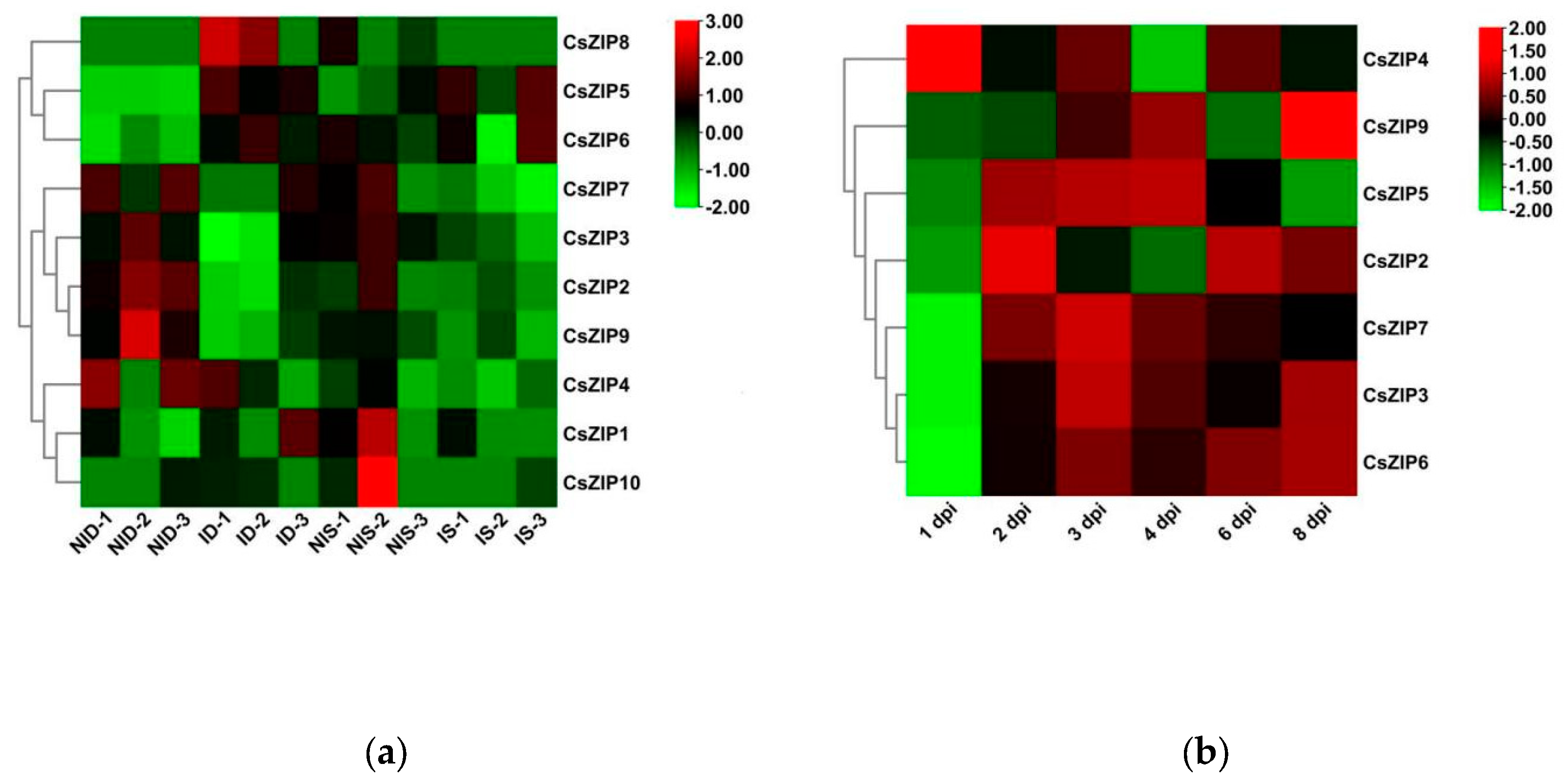
3.9. Expression profiles of CsZIP genes under abiotic and biotic stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grotz N, Guerinot ML. Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim Biophys Acta 2006 Jul;1763(7):595-608. [CrossRef]
- Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals 2001 Sep-Dec;14(3-4):251-70. [CrossRef]
- Guerinot, ML. The ZIP family of metal transporters. Biochim Biophys Acta 2000 May 1;1465(1-2):190-8. [CrossRef]
- Nishida S, Tsuzuki C, Kato A, Aisu A, Yoshida J, Mizuno T. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol 2011 Aug;52(8):1433-42. [CrossRef]
- Krausko M, Labajová M, Peterková D, Jásik J. Specific expression of AtIRT1 in phloem companion cells suggests its role in iron translocation in aboveground plant organs. Plant Signal Behav 2021 Sep 2;16(9):1925020. [CrossRef]
- Vert G, Barberon M, Zelazny E, Séguéla M, Briat JF, Curie C. Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta 2009 May;229(6):1171-9. [CrossRef]
- Milner MJ, Seamon J, Craft E, Kochian LV. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J Exp Bot 2013 Jan;64(1):369-81. [CrossRef]
- Gaitán-Solís E, Taylor NJ, Siritunga D, Stevens W, Schachtman DP. Overexpression of the transporters AtZIP1 and AtMTP1 in cassava changes zinc accumulation and partitioning. Front Plant Sci 2015 Jul 9;6:492. [CrossRef]
- Ramegowda, Y. , Venkategowda, R., Jagadish, P. et al. Expression of a rice Zn transporter, OsZIP1, increases Zn concentration in tobacco and finger millet transgenic plants. Plant Biotechnol Rep 2013, 7, 309–319. [Google Scholar] [CrossRef]
- van de Mortel JE, Almar Villanueva L, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Ver Loren van Themaat E, Koornneef M, Aarts MG. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2006 Nov;142(3):1127-47. [CrossRef]
- Itai RN, Ogo Y, Kobayashi T, Nakanishi H, Nishizawa NK. rice genes involved in phytosiderophore biosynthesis are synchronously regulated during the early stages of iron deficiency in roots. rice (N Y) 2013 Jun 25;6(1):16. [CrossRef]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 2006 Feb;45(3):335-46. [CrossRef]
- Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ 2009 Apr;32(4):408-16. [CrossRef]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006, 52, 464–469. [Google Scholar] [CrossRef]
- Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc Natl Acad Sci U S A 2007 May 1;104(18):7373-8. [CrossRef]
- Liu XS, Feng SJ, Zhang BQ, Wang MQ, Cao HW, Rono JK, Chen X, Yang ZM. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol 2019 Jun 27;19(1):283. [CrossRef]
- shimaru Y, Masuda H, Suzuki M, Bashir K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J Exp Bot 2007;58(11):2909-15, 2909. [CrossRef]
- Lee S, Jeong HJ, Kim SA, Lee J, Guerinot ML, An G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol Biol 2010 Jul;73(4-5):507-17. [CrossRef]
- Lee S, Kim SA, Lee J, Guerinot ML, An G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol Cells 2010 Jun;29(6):551-8. [CrossRef]
- Wu X, Zhu ZB, Chen JH, Huang YF, Liu ZL, Zou JW, Chen YH, Su NN, Cui J. Transcriptome analysis revealed pivotal transporters involved in the reduction of cadmium accumulation in pak choi (Brassica chinensis L.) by exogenous hydrogen-rich water. Chemosphere 2019 Feb;216:684-697. [CrossRef]
- Guo J, Li K, Zhang X, Huang H, Huang F, Zhang L, Wang Y, Li T, Yu H. Genetic properties of cadmium translocation from straw to brown rice in low-grain cadmium rice line. Ecotoxicol Environ Saf 2019 Oct 30;182:109422. [CrossRef]
- Yu R, Li D, Du X, Xia S, Liu C, Shi G. Comparative transcriptome analysis reveals key cadmium transport-related genes in roots of two pak choi (Brassica rapa L. ssp. chinensis) cultivars. BMC Genomics 2017 Aug 8;18(1):587. [CrossRef]
- Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res 2017 Jan 4;45(D1):D313-D319. [CrossRef]
- Fan W, Liu C, Cao B, Qin M, Long D, Xiang Z, Zhao A. Genome-Wide Identification and Characterization of Four Gene Families Putatively Involved in Cadmium Uptake, Translocation and Sequestration in Mulberry. Front Plant Sci 2018 Jun 29;9:879. [CrossRef]
- Palusińska M, Barabasz A, Kozak K, Papierniak A, Maślińska K, Antosiewicz DM. Zn/Cd status-dependent accumulation of Zn and Cd in root parts in tobacco is accompanied by specific expression of ZIP genes. BMC Plant Biol 2020 Jan 22;20(1):37. [CrossRef]
- Liu M, Zhang C, Duan L, Luan Q, Li J, Yang A, Qi X, Ren Z. CsMYB60 is a key regulator of flavonols and proanthocyanidans that determine the colour of fruit spines in cucumber. J Exp Bot 2019 Jan 1;70(1):69-84. [CrossRef]
- Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc 2008;3(2):153-62. [CrossRef]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2015 Apr 15;31(8):1296-7. [CrossRef]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W369-73. [CrossRef]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 2004 Jun;14(6):1188-90, 1188. [CrossRef]
- Voorrips, RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 2002 Jan-Feb;93(1):77-8. [CrossRef]
- Li L, Stoeckert CJ Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003 Sep;13(9):2178-89, 2178. [CrossRef]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res 2009 Sep;19(9):1639-45. [CrossRef]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 2000 Oct;17(10):1483-98. [CrossRef]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002 Jan 1;30(1):325-7. [CrossRef]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020 Aug 3;13(8):1194-1202. [CrossRef]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001 Dec;25(4):402-8. [CrossRef]
- Li H, Wang S, Chai S, Yang Z, Zhang Q, Xin H, Xu Y, Lin S, Chen X, Yao Z, Yang Q, Fei Z, Huang S, Zhang Z. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nat Commun 2022 Feb 3;13(1):682. [CrossRef]
- Eng BH, Guerinot ML, Eide D, Saier MH Jr. Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol 1998 Nov 1;166(1):1-7. [CrossRef]
- Fu XZ, Zhou X, Xing F, Ling LL, Chun CP, Cao L, Aarts MGM, Peng LZ. Genome-Wide Identification, Cloning and Functional Analysis of the Zinc/Iron-Regulated Transporter-Like Protein (ZIP) Gene Family in Trifoliate Orange (Poncirus trifoliata L. Raf.). Front Plant Sci 2017 Apr 19;8:588. [CrossRef]
- Zhang, H. , Zhao, S., Li, D. et al. Genome-Wide Analysis of the ZRT, IRT-Like Protein (ZIP) Family and Their Responses to Metal Stress in Populus trichocarpa. Plant Mol Biol Rep 35, 534–549 (2017). [CrossRef]
- Zhang Z, Chen N, Zhang Z, Shi G. Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency. Tolerance. Plants (Basel) 2022 Mar 16;11(6):786. [CrossRef]
- Gindri RG, Navarro BB, da Cruz Dias PV, Tarouco CP, Nicoloso FT, Brunetto G, Berghetti ÁLP, da Silva LOS, Fett JP, Menguer PK, Ricachenevsky FK. Physiological responses of rice oszip7 loss-of-function plants exposed to varying Zn concentrations. Physiol Mol Biol Plants 2020 Jul;26(7):1349-1359. [CrossRef]
- Tan L, Zhu Y, Fan T, Peng C, Wang J, Sun L, Chen C. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem Biophys Res Commun 2019 Apr 23;512(1):112-118. [CrossRef]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002 Jun;14(6):1223-33. [CrossRef]
- Akther MS, Das U, Tahura S, Prity SA, Islam M, Kabir AH. Regulation of Zn uptake and redox status confers Zn deficiency tolerance in tomato. Scientia Horticulturae 2020; 273. [CrossRef]
- Kabir AH, Tahura S, Elseehy MM, El-Shehawi AM. Molecular characterization of Fe-acquisition genes causing decreased Fe uptake and photosynthetic inefficiency in Fe-deficient sunflower. Sci Rep 2021 Mar 10;11(1):5537. [CrossRef]
- Cheong JJ, Choi YD. Methyl jasmonate as a vital substance in plants. Trends Genet 2003 Jul;19(7):409-13. [CrossRef]
- Castillo-Gonzalez J, Ojeda-Barrios D, Hernandez-Rodriguez A, Gonzalez-Franco AC, Robles-Hernandez L, Lopez-Ochoa GR. ZINC METALLOENZYMES IN PLANTS. Interciencia 2018, 43, 242–248.
- Eriksson S, Böhlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 2006 Sep;18(9):2172-81. [CrossRef]
- Preciado-Rangel P, Reyes-Pérez JJ, Ramírez-Rodríguez SC, Salas-Pérez L, Fortis-Hernández M, Murillo-Amador B, Troyo-Diéguez E. Foliar Aspersion of Salicylic Acid Improves Phenolic and Flavonoid Compounds, and Also the Fruit Yield in Cucumber (Cucumis sativus L.). Plants (Basel) 2019 Feb 16;8(2):44. [CrossRef]
- Li S, Liu X, Zhou X, Li Y, Yang W, Chen R. Improving Zinc and Iron Accumulation in Maize Grains Using the Zinc and Iron Transporter ZmZIP5. Plant Cell Physiol 2019 Sep 1;60(9):2077-2085. [CrossRef]
- Lee S, Kim SA, Lee J, Guerinot ML, An G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol Cells. 2010 Jun;29(6):551-8. [CrossRef]
- Pedas P, Schjoerring JK, Husted S. Identification and characterization of zinc-starvation-induced ZIP transporters from barley roots. Plant Physiol Biochem. 2009 May;47(5):377-83. [CrossRef]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem. 2003 Nov 28;278(48):47644-53. [CrossRef]
- GONG Yuanyong, ZHAO Lihua, YAN Fei. Genome-Wide Identification and Expression Pattern Analysis of the Tomato ZIP Gene Family[J].Northeast Agricultural Science, 48(02), 42-48+109.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





