1. Introduction
Soybean [
Glycine max (L.) Merrill] is a prominent legume crop for vegetable protein and oil [
1]. It provides about 59% of vegetable oil and 70% plant protein for human and poultry consumption worldwide [
2]. To meet the demands of an increasing human population and the quest for a better living standard, it is estimated that global soybean production should at least double by 2050. However, soybean breeding and functional genome studies are significantly hampered by its paleopolyploid genetic background and low transformation efficiency. Virus-induced gene silencing (VIGS) is a widely used RNA-mediated gene knock-down technology [
3]. This technology has great significance for crops that are difficult to genetically manipulate, such as soybeans and barley. At present, several soybean-infecting viruses have been engineered into VIGS vectors, e.g., bean pod mottle virus (BPMV), soybean yellow common mosaic virus (SYCMV), cucumber mosaic virus (CMV), tobacco rattle virus (TRV), and apple latent spherical virus (ALSV) [4-8]. However, the application of these VIGS vectors to soybeans is always limited by severe viral symptoms, a narrow infectivity spectrum (they can only infect part of the soybean cultivar), low silencing efficiency, difficult manipulation, etc. For instance, BPMV causes severe viral symptoms in most soybean cultivars, which may significantly affect the observation of the phenotype caused by the silenced gene [5, 9]. Understanding the molecular mechanism behind these phenomena can obviously benefit the application of ALSV to soybeans and the pathogenesis of these viruses as well.
ALSV is the typical member of the genus
Cheravirus in the family
Secoviridae [
10]. Its genome is composed of two positive-sense single-stranded RNA (+ssRNA) molecules that are encapsulated in isometric viral particles of approximately 25 nm in diameter [
11]. Both RNA molecules are polyadenylated at the 3′-end, but their 5′-end is not capped with m7G; instead, it is covalently linked to a viral protein called viral-protein genome-linked protein (VPg). The large +ssRNA (RNA1) encodes only one open reading frame (ORF), the product of which is proteolyzed by the viral cysteine proteinase (Pro) into four mature proteins, namely, helicase (Hel), VPg, Pro, and RNA-dependent RNA polymerase (Pol) [
11]. The small +ssRNA (RNA2) is also moncistronic, and the encoded polyprotein is proteolyzed by Pro into movement protein (MP) and three capsid proteins (Vp25, Vp20, and Vp24) [
11]. Although ALSV was isolated from apple [
12], it has a wide host range and can infect many plant species in laboratory conditions, such as tobacco, tomato, potato, cucumber, soybean, pea, broad bean, grapevine, cowpea, and even medicinal plants (
Lithospermum erythrorhizon) [13-17]. As a latent virus, ALSV usually causes no to very mild symptoms in most hosts, can stably survive in host plants for a long period of time, and can be transmitted by seed and/or pollen in some hosts [18, 19]. These advantages make ALSV an ideal VIGS vector for plant genetic research. Indeed, ALSV has been modified into a gene silencing vector for gene function analysis on many crops [13-17]. On soybeans, ALSV causes no or negligible symptoms, has high silencing efficiency, and can invade the seeds [8, 20, 21]. However, the infectivity and silencing efficiency of ALSV vary with soybean varieties, and even many soybean varieties exhibit full resistance to ALSV [8, 20, 21]. Understanding the resistance mechanism of soybean to ALSV will benefit the application of ALSV for functional genomic studies in soybean. Currently, there is no study on the genetic basis of the infectivity of ALSV on soybeans, and no resistance gene or locus against ALSV has been reported yet. In this study, we report the mapping of the resistance loci and prediction of resistance candidates in Heinong 84, a soybean cultivar from Northeast China, using a hybrid soybean population and high-throughput sequencing-assisted bulk-segregation analyses (HTS
‒BSA).
2. Materials and Methods
2.1. Soybean verities and Growth Conditions
Soybean varieties Heihe 43, Dongsheng 7, Hefeng 55, ZYD00006, Dongnongdou 252, Suinong 14, Dongnongdou 254, Heinong 84, Zhonghuang 13, and Dongnong 50 were grown in pot in a growth chamber with 50% humidity and 16:8 (light: dark) photoperiod at 26℃. The hybrid population from the crossing between Heinong 84 and Zhonghuang 13 has been reported previously [
22].
2.2. ALSV inoculation
The ALSV infectious clone on the pCB301 backbone has been reported previously [
20]. Soybeans were sap-inoculated as described earlier with a few modifications [
20]. In brief, the two plasmids pALSV-R1 and pALSVR2-PDSi harboring RNA1 and RNA2 of ALSV, respectively, were transformed into the
Agrobacterium tumefaciens strain GV3101 (plus pSoup-p19) by electroporation. An equal amount of the bacteria harboring pALSV-R1 and pALSVR2-PDSi were mixed and infiltrated into
Nicotiana benthamiana leaves. At 20 days post-inoculation, the systemic leaves were harvest for viral particle enrichment as described [
20]. The virion solution was used for the subsequent sap inoculation of the first true leaf of soybean seedlings that had been pre-dusted with 600 mesh carborundum powder. After inoculation, the leaf was rinsed with distilled water and covered with a prewetted paper towel to prevent dehydration. The seedlings were then put back into the growth chamber with normal care measures.
2.3. RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT‒qPCR)
Total RNA was extracted from soybean leaves using the Eastep® Super Total RNA Extraction Kit [Cat# LS1040; Promega (Beijing) Biotech Co., Ltd, Beijing, China], following the supplied instructions. First-strand complementary DNA (cDNA) was synthesized using the HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) with random hexamers and Oligo-dT
20. qPCR was conducted in a 20 μL reaction volume, comprising 1 μL of 200 ng/μL cDNA, 0.4 μL each of 10 mmol/μL forward and reverse primers, 10 μL of 2×ChamQ Universal SYBR qPCR Master Mix, and 8.2 μL sterilized ultrapure water. All primers used in the present study are listed in
Table 1.
2.4. DNA extraction, DNA pool preparation, and high throughput sequencing
Total genome DNA was isolated using the FastPure Plant DNA Isolation Mini Kit (Cat#: DC104; Vazyme Biotech Co., Ltd, Nanjing, China). DNA pools were prepared as described earlier with few modifications [
22]. The susceptible and resistant pools included 30 susceptible and resistant soybean DNA, with 6 μg DNA per sample, respectively. High throughput sequencing was performed by the Illumina HiSeqTM 2500 platform in Hangzhou Lianchuan Biotechnology Co., Ltd. (Hangzhou, Zhejiang, China).
2.5. Bulk Segregation Analysis (BSA)
The adapter sequence on reads from the HiSeqTM 2500 platform was trimmed and low-quality reads were discarded using a Trimmomatic v0.39 [
23]. The resulting high-quality reads were mapped to the reference genome of Zhonghuang 13 (CNCB accession GWHAAEV00000000.1) using HiSat2 v2.2.1 with parameter end-to-end [
24]. Single nucleotide polymorphism (SNP) was called using samtools v1.15 after removing duplication [
25]. QTLseqr (v0.7.5.2) was used to localize susceptible loci (SNPs with a sample depth below 10 or total depth below 30 were removed)[
26].
2.6. Design and validation of primers for cleaved amplified polymorphic sequences (CAPS)
The mapping results from HiSat2 were utilized to identify single nucleotide polymorphisms (SNPs) inside the resistance locus using bedtools v2.30 [
27]. The SNP2CAPS v0.6 software was employed to transform the single nucleotide polymorphisms (SNPs) into CAPS markers [
28]. CAPS primers were designed using SnapGene 4.1.9 with the default parameters. The polymerase chain reaction (PCR) was carried out in a 20-μl volume system in a T30D tri-block super-gradient PCR system (LongGene, Hangzhou, Zhejiang, China). The thermal cycle contains a pre-denaturation step at 95°C for 3 minutes, 30 cycles of denaturation at 95°C for 30 seconds, annealing at the primer melting temperature (Tm) for 30 seconds, extension at 72°C for 15 seconds, and a final extension step at 72°C for 5 minutes. Restriction enzyme digestion was performed in a 20 µL system, comprising 2.0 µL of 10 × CutSmart buffer, 0.5 µL of restriction enzyme, e.g.,
Hind Ⅲ,
EcoR V, and
Nhe I (New England Biolabs, Beijing, China), 10 µL of PCR product, and 7.5 µL of ddH
2O. The restriction enzyme digestion mixture was incubated at 37°C for 30 minutes and then treated at 85°C for 5 seconds to deactivate the enzyme. Subsequently, the mixtures of PCR and restriction enzyme digestion were analyzed by electrophoresis on a 1% agarose gel. ImageJ was used to analyze the amplified bands, and further statistical analyses were performed using Fisher's Exact test.
2.7. SA and H2O2 quantification
The Plant Salicylic Acid (SA) ELISA Kit (Cat#: MBS9314138; Spbio, Wuhan, Hubei, China) was used to determine the concentration of SA according to the manufactural instructions. The BioTech Epoch Full Wavelength Enzyme Labeler was used to read the optical density at a wavelength of 450 nm (OD450). Every sample was technically triple replicated, and the OD450 read was corrected by the value of the blank control and then compared to the OD450 value of the healthy control (leaf tissue inoculated with buffer).
H2O2 content was measured using the Hydrogen Peroxide Assay Kit [Cat#: AKAO009; Boxbio Biotech (Beijing) Co., Ltd, Beijing, China] following the supplied manual. In brief, about 0.1 g of leaf tissue was homogenized in 1 mL of solution I, the resulting homogenate was clarified by centrifugation at 8000 g for 30 minutes at 4°C. The supernatant was transferred to a new test tube, and added 100 µL of solution II, 200 µL of solution III, and 1 mL of solution IV were added sequentially and then mixed well through a vortex. After incubation at room temperature for 5 minutes, 1 ml of the reaction solution was transferred to a measuring cuvette, and the absorbance at 415 nm was read by a Thermo Fisher UV spectrophotometer. Finally, the value at OD415 was plotted against the standard curve to calculate the concentration of H2O2.
3. Results
3.1. Screening of susceptible soybean varieties in Northeast China
The Northeast region is the most significant soybean-producing area in China. To explore the soybean cultivars suitable for VIGS application in this area, we mechanically inoculated the first true leaf of twelve-day-old seedlings of ten major soybean varieties from the Northeast region of China with ALSV-PDSi, an ALSV infectious clone harboring a fragment of the soybean
PDS gene between MP and VP25 for tracking virus infection [
20]. Zhonghuang 13, which has been confirmed to be susceptible to ALSV, was included as a positive control [
20]. As expected, a photobleaching symptom was observed on the upper uninoculated leaves of the Zhonghuang 13 at 20 days post-inoculation (dpi) (
Figure 1A). Moreover, the seed coat and seeds of Zhonghuang 13 also showed a photobleaching symptom (
Figure 1B), suggesting that ALSV may be seed-transmittible in Zhonghuang 13. However, no visible photobleaching symptom was observed on the upper uninoculated leaves of the rest of the soybean varieties throughout their growth period (
Figure 1A). The presence of ALSV-PDSi on the upper uninoculated leaves was confirmed by reverse transcription-polymerase chain reaction (RT-PCR). Results showed that none of these soybean varieties are susceptible to ALSV-PDSi (
Figure 1C), which further confirms that only a portion of soybean varieties are susceptible to ALSV.
3.2. Genetic analysis of the resistance of Heinong 84 to ALSV
Previously, we constructed a soybean hybrid population by crossing Heinong 84 and Zhonghuang 13 to locate the resistance gene against soybean mosaic virus strain N3 in Heinong 84 [
22]. We thus decided to take advantage of this hybrid population to analyze the genetic basis of the trait of ALSV resistance. A total of 100 F2-generation seedlings were mechanically inoculated by ALSV-PDSi. The infection of ALSV-PDSi on each plant was confirmed by both the photobleaching phenotype and RT-PCR. Results showed that 40 out of the 100 F2-generation seedlings displayed a photobleaching phenotype and viral genomic RNA on the upper systemic leaves. The remaining 60 F2-generation seedlings showed no photobleaching phenotype throughout the growth period and were absent of viral genomic RNA on the upper systemic leaves (
Table 2). The segregation ratio did not match 3:1 (χ2 = 4.4672;
p = 0.03445), but matched 9:7 (χ
2 = 0.185;
p = 0.6673;
Table 2), indicating that this resistance trait of Heinong 84 may be controlled by two genes.
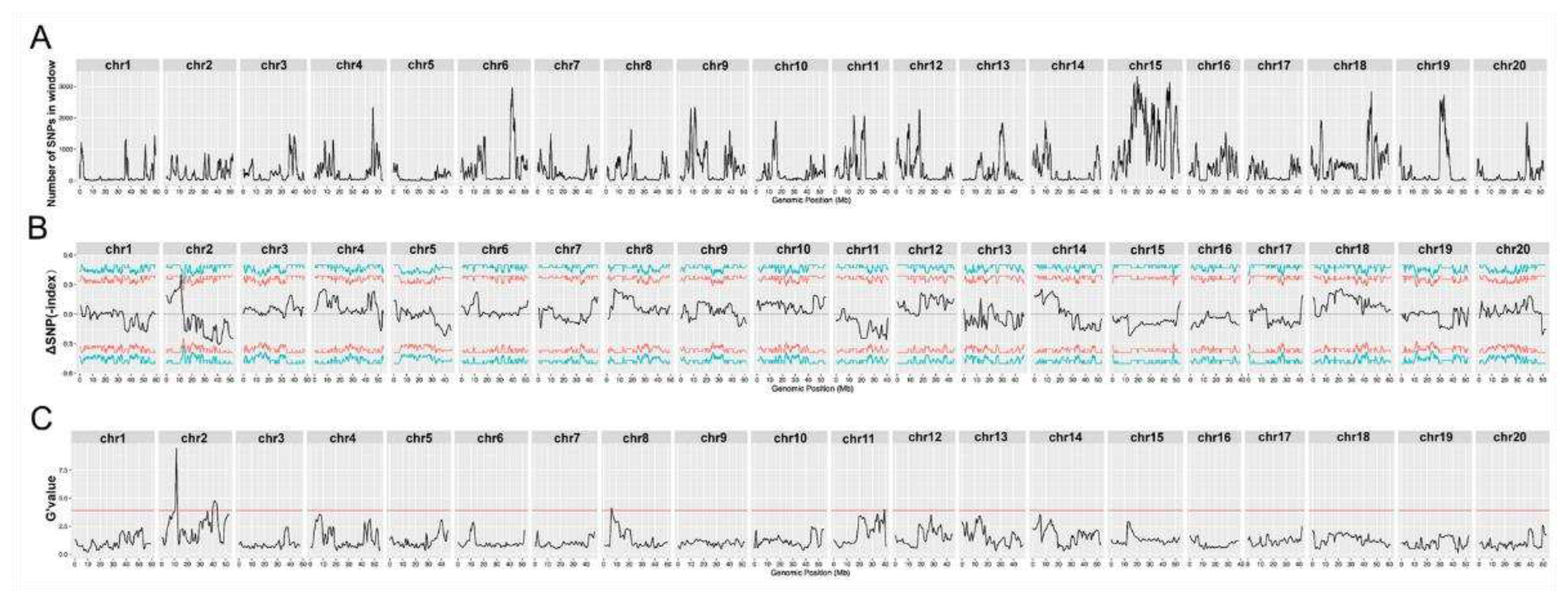
3.3. Location of the resistance loci in Heinong 84 by HGS-BSA
We inoculated the F3-generation of the hybrid population and used the same standard to select resistant and susceptible offspring. A total of 30 susceptible individuals (showing a photobleaching phenotype) and 30 resistant individuals were selected for further analyses. Noticeably, the resistant pool may include a very small number of susceptible individuals since it is impossible to achieve 100% inoculation efficiency. Equal amounts of genomic DNA from individuals who were resistant or susceptible were pooled as the resistant or susceptible pool, respectively. The two pools and the genomic DNA of Heinong 84 were then sequenced. Following the trimming of adaptors and quality control measures, we obtained 29.8, 30.7, and 27.3 billion high-quality reads (quality score ≥ 30) of the resistant pool, susceptible pool, and Heinong 84, respectively. These data were mapped to the reference genome of Zhonghuang 13, and single-nucleotide polymorphism (SNP) sites were thereby retrieved. A total of 366,633 SNPs distributed on the 20 chromosomes of Zhonghuang 13 were found. Two genomic intervals, one 4.62 Mb genomic interval on chromosome 2 (39,213,950‒43,829,476 bp;
p = 0.00004) with 99% confidence and another 6.41 Mb genomic interval on chromosome 11 (19,104,714‒25,514,536 bp;
p = 0.00040), were identified as the potential quantitative trait loci (QTLs) (
Figure 2;
Table 3). These data thus confirm that the resistance of Heinong 84 to ALSV may be controlled by two genes, hereinafter referred to as
resistance to ALSV locus 1 (
RALSV-L1) and
RALSV-L2, respectively.
3.4. Dissection the genetic basis of RALSV-L1 and RALSV-L2 using CAPS markers
The CAPS marker-based test is a rapid, reliable, and co-dominant method that has been successfully applied for QTL mapping, molecular marker-assisted breeding, and inheritance analysis [28, 29]. Three pairs of CAPS primers per locus were designed based on the SNPs in each locus (
Table 1). PCR using the total DNA extracted from the parents followed by restriction enzyme digestion showed that the primers based on SNP3999 and SNP2115 resulted partial digestion (Supplementary Fig. 1), and thus were excluded in the subsequent experiments. All individuals in the susceptible and resistance pools were then analyzed by these CAPS primers. Results showed that the primers based on SNP4209 on the locus of chromosome 2 (C2-SNP4209-F and C2-SNP4209-R) had the highest cosegregation confidence (
p = 0.001115; Supplementary
Figure 2-3), while the two pairs of primers on chromosome 11 had similar performance (Supplementary
Figure 4-5). We then determined the genotype of each plant in the susceptible and resistance pools based on the results of the CAPS assay. For statistical purposes, the ALSV-associated loci on chromosome 2 in Heinong 84 and Zhonghuang 13 were defined as
A1 and
a1, respectively. Partially digested samples were recognized as heterozygous. Results showed that most individuals that are homologous to
A1 or
a1 were resistant and susceptible to ALSV, respectively (
Table 4), indicating that
A1 is a dominant resistance gene. Interestingly, half of heterozygous individuals are resistant and half are susceptible to ALSV (
Table 4), suggesting that
A1 may be a dose-dependent dominant resistance gene and/or that the genotype of the other locus together determines the phenotype when this locus is heterozygous.
Similar genotype analyses were performed for the locus on chromosome 11, which was defined as
B2 and
b2 in Heinong 84 and Zhonghuang 13, respectively. Results showed that the majority of individuals that are homologous to
B2 or
b2 were susceptible and resistant to ALSV, respectively (
Table 4), indicating that the ALSV-associated locus on chromosome 11 may encode a recessive resistance gene, and this gene, even if homozygous, can only provide partial resistance. Interestingly, only one heterozygous plant was identified in this study (
Table 4), thus it could not be further analyzed.
We further performed a correlation analysis of the two loci (
Table 4). The Fisher's exact test confirmed that the two ALSV-associated loci were significantly associated with the resistance phenotype (
p = 0.003263). As expected, almost all individuals with the genotypes
A1A1b2b2 and
A1a1b2b2 are resistant (
Table 4), further confirming that A1 is a dominant resistance gene, while b2 is a recessive resistance gene. The majority of individuals with the genotypes
a1a1B2b2,
a1a1b2b2, and
a1a1B2B2 are also susceptible (
Table 4), suggesting that
b2 cannot provide full resistance without the
A1 gene. Interestingly, about half of the individuals with the genotypes of
A1A1B2B2 or
A1a1B2B2 are resistant and half are susceptible to ALSV (
Table 4), confirming that A1 is a dominant gene that may function by directly inhibiting viral proliferation, while
B2 may encode a host factor that is required for viral proliferation.
3.5. Prediction of candidate genes in the resistance loci RALSV-L1 and RALSV-L1
Using the reference genome of Zhonghuang 13, a total of 266 protein-coding, miRNA, and tRNA genes were identified in the resistance interval on chromosome 2, among which 235 genes exhibited differences between Heinong 84 and Zhonghuang 13. Considering the effects of the SNP on the gene where it is located, these genes were divided into three groups: low-potential candidates, moderate-potential candidates, and high-potential candidates. The low-potential candidates had SNPs that caused synonymous mutations; the moderate-potential candidates had SNPs that caused missense mutations and variation in the untranslated region (UTR) and/or intron; while the high-potential candidates contained SNPs that caused splice acceptor variants and intron variants, stop codon loss, frameshift variants, and stop codon gain. Based on this criterion, a total of 69 and 10 moderate and high potential candidates were identified in the resistance locus, respectively (Supplementary
Table 1). The same analyses were performed on the resistance locus on chromosome 11. A total of 238 protein-coding, miRNA, and tRNA genes were identified in the resistance interval on chromosome 11, among which 191 and 29 were moderate and high potential candidates (Supplementary
Table 2). In general, the candidates of
RALSV-L1 and
RALSV-L2 belongs to various pathways and have varied biological functions. Nevertheless, we noticed that several candidates for the
RALSV-L1 and
RALSV-L2 belong to the same protein complex or biological pathways, including the cleavage and polyadenylation specificity factor (CPSF) complex, leucine-rich repeat-containing protein, protein ubiquitination, and microtubule motors (Supplementary
Table 1 and 2).
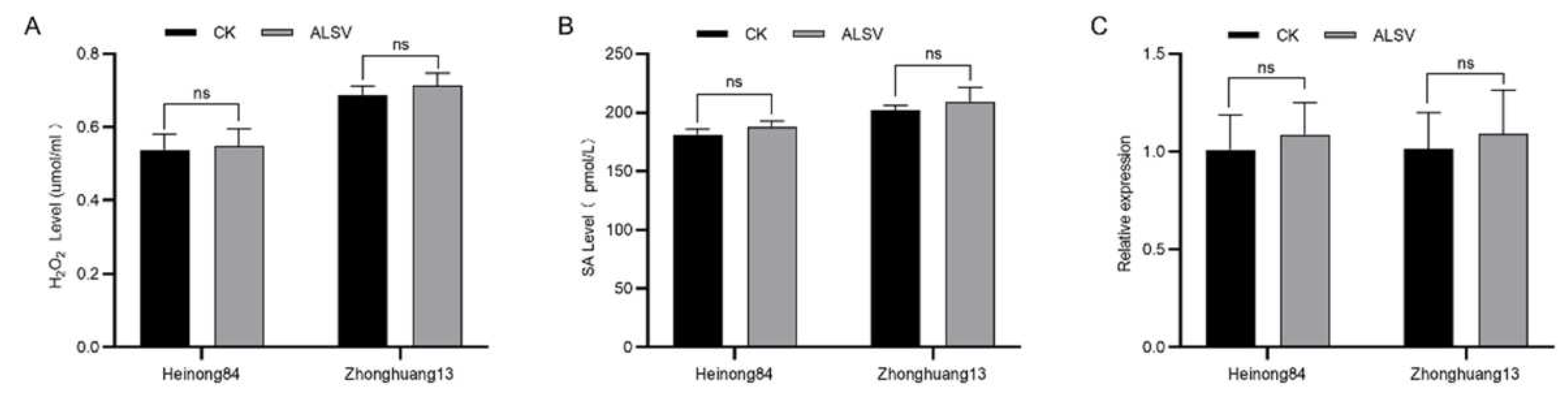
Figure 3.
The resistance of Heinong 84 to ALSV is not associated with innate immunity. (A) The accumulation of H2O2 in soybean plants inoculated by ALSV or buffer at 48 hpi. (B) SA levels in soybean plants inoculated by ALSV or buffer at 48 hpi. (C) The transcript levels of PR1 in soybean plants inoculated by ALSV or buffer at 48 hpi. The GmCons6 gene (Glyma.12G051100) was used as the internal control. Ns indicates p > 0.05 in the Student’s t-test. Experiments were replicated three times with consistent results.
Figure 3.
The resistance of Heinong 84 to ALSV is not associated with innate immunity. (A) The accumulation of H2O2 in soybean plants inoculated by ALSV or buffer at 48 hpi. (B) SA levels in soybean plants inoculated by ALSV or buffer at 48 hpi. (C) The transcript levels of PR1 in soybean plants inoculated by ALSV or buffer at 48 hpi. The GmCons6 gene (Glyma.12G051100) was used as the internal control. Ns indicates p > 0.05 in the Student’s t-test. Experiments were replicated three times with consistent results.
3.6. The resistance is not associated with innate immunity
To test whether the dominant locus
RALSV-L1 in Heinong 84 is associated with innate immunity, we compared the content of hydrogen peroxide, a marker signal of biotic stresses in plants. Thus, seedlings of Heinong 84 and Zhonghuang 13 were inoculated with ALSV or buffer, and the content of hydrogen peroxide in the inoculated leaf tissue was determined at 48 hours post-inoculation (hpi). Results showed that there was no significant difference in the contents of hydrogen peroxide in the leaves of Heinong 84 and Zhonghuang 13 inoculated by ALSV or buffer at 48 hpi (Fig. 3A), indicating that the inoculation of ALSV does not induce the accumulation of hydrogen peroxide in Heinong 84 and Zhonghuang 13. We also compared the content of salicylic acid (SA), the key phytohormone of biotic stresses in plants, with the content of ALSV infection. Results showed that the SA content in Heinong 84 and Zhonghuang 13 was not significantly upregulated after the inoculation of ALSV at 48 hpi (Fig. 3B). Finally, we directly compared the expression of
pathogenesis-related genes 1 (
PR1), the marker gene of innate immunity, in Heinong 84 and Zhonghuang 13 inoculated by ALSV or buffer at 48 hpi. RT‒qPCR results showed that there was no significant difference in the expression of
PR1 in the leaves of Heinong 84 and Zhonghuang 13 inoculated by ALSV or buffer at 48 hpi (Fig. 3C). Together, we concluded that the resistance in Heinong 84 to ALSV is not controlled by innate immunity instead may be controlled by non-immune-related mechanisms, e.g., RNA silencing, translation repression, essential host factors for virus proliferation, or atypical dominant viral resistance protein [
30].
3.7. There are resistance genes other than RALSV-L1 or RALSV-L2 in soybeans
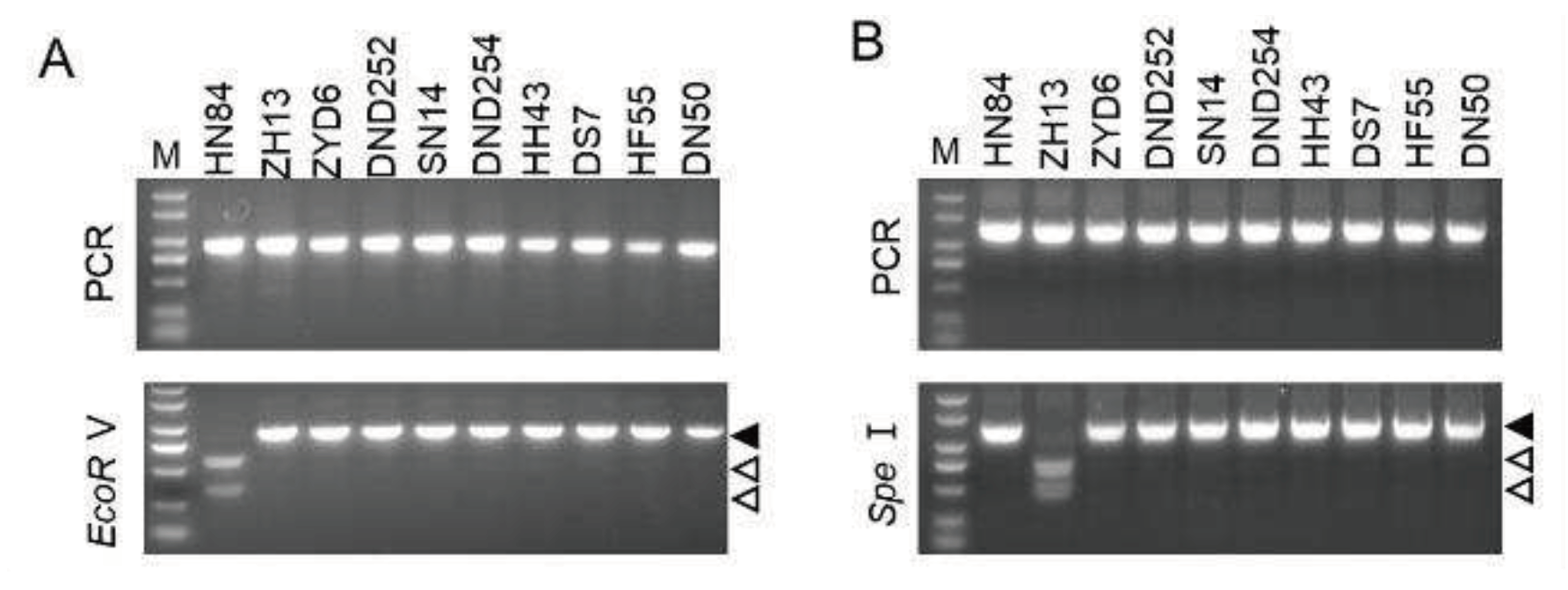
We further analyzed the resistance of the other eight soybean varieties, namely, Heihe 43, Dongsheng 7, Hefeng 55, ZYD00006, Dongnongdou 252, Suinong 14, Dongnongdou 254, and Dongnong 50, using the two pairs of CAPS primers. Results showed that the ALSV resistance-associated locus on chromosome 2 of all eight soybean cultivars had the same genotype as Zhonghuang 13, while the ALSV resistance-associated locus on chromosome 11 of these soybean cultivars was the same as Heinong 84 (Fig. 4). These data suggest that the resistance of these cultivars may be controlled by additional resistance genes, and these primers are not suitable for dissecting the resistance of these soybean cultivars to ALSV.
Figure 4.
CAPS results of the eight soybean cultivars. (A‒B) Gel electrophoresis results of the PCR products of primer pairs based on C2-SNP4209 (A) and C11-SNP2130 (B) before and after restriction enzyme digestion. Lanes 1 to 10 represent Heinong 84 (HN84), Zhonghuang 13 (ZH13), ZYD00006 (ZYD6), Dongnongdou 252 (DND252), Suinong 14 (SN14), Dongnongdou 254 (DND254), Heihe 43 (HH43), Dongsheng 7 (DS7), Hefeng 55 (HF55), and Dongnong 50 (DN50), respectively. The PCR amplicons are indicated by solid arrowheads and digested fragments are indicated by hollow arrowheads.
Figure 4.
CAPS results of the eight soybean cultivars. (A‒B) Gel electrophoresis results of the PCR products of primer pairs based on C2-SNP4209 (A) and C11-SNP2130 (B) before and after restriction enzyme digestion. Lanes 1 to 10 represent Heinong 84 (HN84), Zhonghuang 13 (ZH13), ZYD00006 (ZYD6), Dongnongdou 252 (DND252), Suinong 14 (SN14), Dongnongdou 254 (DND254), Heihe 43 (HH43), Dongsheng 7 (DS7), Hefeng 55 (HF55), and Dongnong 50 (DN50), respectively. The PCR amplicons are indicated by solid arrowheads and digested fragments are indicated by hollow arrowheads.
4. Discussion
Despite the advantages of ALSV as a VIGS vector for soybean genome study and some soybean cultivars, such as Wyandot, Magellan, Jack, Qihuang 34, Andou 203, Nannong 1138-2, Nannong 47, Zhonghuang 13, Shanning 29, and Xiangdou 4, having been identified as susceptible cultivars [8, 20, 21], many soybean cultivars display complete resistance to ALSV. We also found that all ten major soybean cultivars from the Northeast region of China were completely resistant to ALSV. Understanding the genetic mechanisms underlying ALSV resistance is of great significance for the application of ALSV to soybeans. However, no resistance-associated genetic locus or DNA marker has been characterized. Based on simple genetic background comparisons, it was speculated that the resistance was determined by one or several genes [
21]. In this study, the resistance of Heinong 84 to ALSV was detailedly analyzed by a hybrid population crossed from the resistant cultivar Heinong 84 and the susceptible cultivar Zhonghuang 13. Our BSA and CAPS assay data clearly showed that the resistance of Heinong 84 is associated with two genetic loci located on chromosomes 2 and 11, respectively. These data allow us to understand, for the first time, the genetic basis of soybean resistance to ALSV. Interestingly, the results of the CAPS assay of the other eight soybean varieties using the same primers suggest that there may be other resistance loci in soybean. Thus, the resistance of soybean to ALSV is more complex than previously thought.
CAPS primers were also designed for rapid identification of resistance loci and for dissecting the genetic basis of the resistance. The data from the CAPS assays further confirmed that the resistance of ALSV is controlled by two loci: one dominant locus on chromosome 2 and another recessive locus on chromosome 11. Interestingly, detailed dissection of the relationship between the genotype and resistance phenotype suggests that the two loci have very complex genetic relationships: that the locus on chromosome 2 plays a dominant but dose-dependent role in resisting ALSV, while the locus on chromosome 11 only has an auxiliary role in resisting ALSV. Based on these observations, it is possible that
RALSV-L1 may encode atypical dominant viral resistance protein (ADVRP), and
RALSV-L2 belongs to a key host factor required for ALSV proliferation. Indeed, the inoculation of ALSV did not induce the accumulation of H
2O
2 or SA and did not stimulate the expression of pathogenesis-related genes, e.g.,
PR1. At present, several ADVRPs have been characterized, such as the restricted TEV movement 1 (RTM1) that confers resistance to several potyviruses [
31], the jacalin-type lectin required for potexvirus resistance 1 (JAX1) that confers broad-spectrum resistance to potexviruses [
32], the tomato Tm-1, which confers resistance to tomato mosaic virus (ToMV, a tobamovirus) [33, 34], and h-type thioredoxin (ZmTrxh), which provides maize with sugarcane mosaic virus (SCMV, a potyvirus) resistance [
35]. However, no candidate was found to be homologous to these ADVRPs. Thus, the
RALSV-L1 may encode a novel ADVRP. Viruses are obligate intracellular parasites and require many host factors to accomplish their infection cycle, e.g., protein expression, genome replication, and intercellular movement. The incompatible or unfavorable interaction between viral proteins or genomes will cause a delay or even complete failure of the infection [
36]. The eukaryotic initiation factor 4E (eIF4E) and eIFiso4e are the two most documented recessive resistance genes [
37]. However, we did not find either eIF4E or eIFiso4E in the candidates. No candidate was found to be homologous to other antiviral recessive resistance genes, such as essential for potexvirus accumulation 1 (EXA1) and the translationally controlled tumor protein TCTP [38, 39]. Thus, further investigations are needed to fully illustrate the function of the recessive gene in the
RALSV-L2 locus.
5. Conclusions
In conclusion, our BSA and CAPS assay data suggest that the resistance of Heinong 84 to ALSV is associated with genetic loci on chromosomes 2 and 11, respectively. Our data also imply that the resistance is possibly controlled by the complex interaction between an ADVRP and the host factor required for ALSV proliferation.
Supplementary Materials
The following supporting information can be downloaded at: Preprints.org, Figure: Gel electrophoresis results of the PCR and restriction digestion products of CAPS primers; Figure S2: Gel electrophoresis results of the PCR products of primer pairs based on SNP4209 on chromosome 2; Figure S3: Gel electrophoresis results of the PCR products of primer pairs based on SNP4232 on chromosome 2; Figure S4: Gel electrophoresis results of the PCR products of primer pairs based on SNP2130 on chromosome 11; Figure S5: Gel electrophoresis results of the PCR products of primer pairs based on SNP2298 on chromosome 11; Table S1: Candidates of the RALSV-L1 locus; Table S2: Candidates of the RALSV-L2 locus.
Author Contributions
Conceptualization, Xiaofei Cheng and Xiaoyun Wu; Data curation, Jiaxing Sun; Funding acquisition, Xiaofei Cheng; Investigation, Tingshuai Ma, Ying Zhang, Yu Zhao, Kekely Attiogbe and Xinyue Fan; Methodology, Yong Li; Project administration, Weiqin Ji and Xiaofei Cheng; Software, Yong Li; Supervision, Xiaofei Cheng and Xiaoyun Wu; Validation, Kekely Attiogbe, Wenqian Fan, Yalou Luo and Xinwei Yu; Visualization, Yong Li; Writing – original draft, Tingshuai Ma and Ying Zhang; Writing – review & editing, Weiqin Ji, Xiaofei Cheng and Xiaoyun Wu.
Funding
This research was funded by the National Natural Foundation of China, grant number 32022071.
Data Availability Statement
All data are available within the Article and Supplementary Files. All constructs are available upon request.
Acknowledgments
We would like to express our special thanks to Dr. Xiaorong Tao and Dr. Yi Xu from the Nanjing Agricultural University for sharing the ALSV infectious clone. We also grateful to Dr. Xiaoyan Luan, Xinlei Liu, and Yongguo Xue from the Soybean Research Institute of Heilongjiang Academy of Agricultural Science for the assistance in the production of the hybrid soybean population.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hartman, G. L.; West, E. D.; Herman, T. K. , Crops that feed the World 2. Soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Security.
- SoyStat http://www.soystats.com/.
- Senthil-Kumar, M.; Anand, A.; Uppalapati, S. R.; Mysore, K. S. , Virus-induced gene silencing and its applications. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources.
- Zhang, C.; Yang, C.; Whitham, S. A.; Hill, J. H. , Development and Use of an Efficient DNA-Based Viral Gene Silencing Vector for Soybean. Molecular Plant-Microbe Interactions.
- Zhang, C.; Ghabrial, S. A. , Development of Bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 2006, (2), 401–411. [Google Scholar] [CrossRef]
- Lim, S.; Nam, M.; Kim, K. H.; Lee, S.-H.; Moon, J.-K.; Lim, H.-S.; Choung, M.-G.; Kim, S.-M.; Moon, J. S. , Development of a new vector using Soybean yellow common mosaic virus for gene function study or heterologous protein expression in soybeans. Journal of Virological Methods 2016, 228, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, K. H.; Lim, S.; Kang, Y. J.; Yoon, M. Y.; Nam, M.; Jun, T. H.; Seo, M.-J.; Baek, S.-B.; Lee, J.-H.; Moon, J.-K.; Lee, S.-H.; Lee, S.-H.; Lim, H.-S.; Moon, J. S.; Park, C.-H. , Optimization of a virus-induced gene silencing system with Soybean yellow common mosaic virus for gene function studies in soybeans. Plant Pathol J 2016, (2), 112–122. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, N.; Yoshikawa, N. , Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Molecular Biology 2009, (1), 15–24. [Google Scholar] [CrossRef]
- Zhang, C.; Bradshaw, J. D.; Whitham, S. A.; Hill, J. H. , The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant physiology 2010, (1), 52–65. [Google Scholar] [CrossRef]
- Le Gall, O.; Sanfaçon, H.; Ikegami, M.; Iwanami, T.; Jones, T.; Karasev, A.; Lehto, K.; Wellink, J.; Wetzel, T.; Yoshikawa, N. , Cheravirus and Sadwavirus: two unassigned genera of plant positive-sense single-stranded RNA viruses formerly considered atypical members of the genus Nepovirus (family Comoviridae). Archives of virology 2007, 152, 1767–1774. [Google Scholar] [CrossRef]
- Li, C.; Yoshikawa, N.; Takahashi, T.; Ito, T.; Yoshida, K.; Koganezawa, H. , Nucleotide sequence and genome organization of Apple latent spherical virus: a new virus classified into the family Comoviridae. Journal of General Virology 2000, (2), 541–547. [Google Scholar] [CrossRef]
- Koganezawa, H. , An isometric viruslike particle isolated from russet ring-diseased apple. Ann Phytopath Soc Jpn 1985, 51, 363. [Google Scholar]
- Li, C.; Yamagishi, N.; Kasajima, I.; Yoshikawa, N. , Virus-induced gene silencing and virus-induced flowering in strawberry (Fragaria× ananassa) using apple latent spherical virus vectors. Horticulture research 2019, 6. [Google Scholar] [CrossRef]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A.; Yaegashi, H.; Yamagishi, N.; Takahashi, T.; Isogai, M. , Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 2009, (2), 407–416. [Google Scholar] [CrossRef]
- Satoh, N.; Kon, T.; Yamagishi, N.; Takahashi, T.; Natsuaki, T.; Yoshikawa, N. , Apple latent spherical virus vector as vaccine for the prevention and treatment of mosaic diseases in pea, broad bean, and eustoma plants by bean yellow mosaic virus. Viruses 2014, (11), 4242–4257. [Google Scholar] [CrossRef] [PubMed]
- Izuishi, Y.; Isaka, N.; Li, H.; Nakanishi, K.; Kageyama, J.; Ishikawa, K.; Shimada, T.; Masuta, C.; Yoshikawa, N.; Kusano, H.; Yazaki, K. , Apple latent spherical virus (ALSV)-induced gene silencing in a medicinal plant, Lithospermum erythrorhizon. Scientific Reports 2020, (1), 13555. [Google Scholar] [CrossRef]
- Maeda, K.; Kikuchi, T.; Kasajima, I.; Li, C.; Yamagishi, N.; Yamashita, H.; Yoshikawa, N. , Virus-induced flowering by Apple latent spherical virus vector: effective use to accelerate breeding of grapevine. Viruses 2020, (1), 70. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, N.; Sasaki, S.; Yamagata, K.; Komori, S.; Nagase, M.; Wada, M.; Yamamoto, T.; Yoshikawa, N. , Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the Apple latent spherical virus vector. Plant Molecular Biology 2011, 75, 193–204. [Google Scholar] [CrossRef]
- Nakamura, K.; Yamagishi, N.; Isogai, M.; Komori, S.; Ito, T.; Yoshikawa, N. , Seed and pollen transmission of Apple latent spherical virus in apple. Journal of General Plant Pathology 2011, (1), 48–53. [Google Scholar] [CrossRef]
- Don, Y.; Wei, Q.; Hong, H.; Huang, Y.; Zhao, Y.; Feng, M.; Dou, D.; Xu, Y.; Tao, X. , Establishment of ALSV-induced gene silencing in Chinese soybean cultivars. Scientia Agricultura Sinica 2022, (9), 1710–1722. [Google Scholar]
- Gedling, C. R.; Ali, E. M.; Gunadi, A.; Finer, J. J.; Xie, K.; Liu, Y.; Yoshikawa, N.; Qu, F.; Dorrance, A. E. , Improved apple latent spherical virus-induced gene silencing in multiple soybean genotypes through direct inoculation of agro-infiltrated Nicotiana benthamiana extract. Plant Methods 2018, (1), 19. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Deng, W.; Liu, J.; Fang, Y.; Liu, Y.; Ma, T.; Zhang, Y.; Xue, Y.; Tang, X.; Cao, D.; Zhu, Z.; Luan, X.; Cheng, X. , Fine mapping the soybean mosaic virus resistance gene in soybean cultivar Heinong 84 and development of CAPS markers for rapid identification. Viruses 2022, (11), 2533. [Google Scholar] [CrossRef]
- Bolger, A. M.; Lohse, M.; Usadel, B. , Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England), 2114. [Google Scholar]
- Kim, D.; Paggi, J. M.; Park, C.; Bennett, C.; Salzberg, S. L. , Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 2019, (8), 907–915. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J. K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M. O.; Whitwham, A.; Keane, T.; McCarthy, S. A.; Davies, R. M.; Li, H. , Twelve years of SAMtools and BCFtools. GigaScience.
- Mansfeld, B. N.; Grumet, R. , QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. Plant Genome.
- Quinlan, A. R. , BEDTools: the Swiss-army tool for genome feature analysis. Current protocols in bioinformatics.
- Thiel, T.; Kota, R.; Grosse, I.; Stein, N.; Graner, A. , SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res 2004, (1), e5. [Google Scholar] [CrossRef]
- Yeam, I.; Kang, B. C.; Lindeman, W.; Frantz, J. D.; Faber, N.; Jahn, M. M. , Allele-specific CAPS markers based on point mutations in resistance alleles at the pvr1 locus encoding eIF4E in Capsicum. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 2005, (1), 178–86. [Google Scholar] [CrossRef]
- Wu, X.; Valli, A.; García, J. A.; Zhou, X.; Cheng, X. , The tug-of-war between plants and viruses: great progress and many remaining questions. Viruses 2019, (3), 203. [Google Scholar] [CrossRef]
- Chisholm, S. T.; Mahajan, S. K.; Whitham, S. A.; Yamamoto, M. L.; Carrington, J. C. , Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proceedings of the National Academy of Sciences of the United States of America 2000, (1), 489–94. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Maejima, K.; Komatsu, K.; Shiraishi, T.; Okano, Y.; Himeno, M.; Sugawara, K.; Neriya, Y.; Minato, N.; Miura, C.; Hashimoto, M.; Namba, S. , Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 2012, (2), 778–793. [Google Scholar] [CrossRef]
- Ishibashi, K.; Kezuka, Y.; Kobayashi, C.; Kato, M.; Inoue, T.; Nonaka, T.; Ishikawa, M.; Matsumura, H.; Katoh, E. , Structural basis for the recognition–evasion arms race between Tomato mosaic virus and the resistance gene Tm-1. Proceedings of the National Academy of Sciences 2014, (33), E3486–E3495. [Google Scholar] [CrossRef]
- Ishibashi, K.; Masuda, K.; Naito, S.; Meshi, T.; Ishikawa, M. , An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proceedings of the National Academy of Sciences 2007, (34), 13833–13838. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, H.; Gong, Y.; Tao, Y.; Jiang, L.; Zuo, W.; Yang, Q.; Ye, J.; Lai, J.; Wu, J.; Lübberstedt, T.; Xu, M. , An atypical thioredoxin imparts early resistance to Sugarcane mosaic virus in Maize. Molecular plant 2017, (3), 483–497. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. , Recessive Resistance to Plant Viruses: Potential Resistance Genes Beyond Translation Initiation Factors. Front Microbiol 2016, 7, 1695. [Google Scholar] [CrossRef]
- Robaglia, C.; Caranta, C. , Translation initiation factors: a weak link in plant RNA virus infection. Trends in Plant Science 2006, (1), 40–45. [Google Scholar] [CrossRef]
- Bruckner, F. P.; da Silva Xavier, A.; de Souza Cascardo, R.; Otoni, W. C.; Zerbini, F. M.; Alfenas-Zerbini, P. , Translationally controlled tumor protein (TCTP) from tomato and Nicotiana benthamiana is necessary for successful infection by A Potyvirus. Molecular Plant Pathology 2017, (5), 672–683. [Google Scholar] [CrossRef]
- Hashimoto, M.; Neriya, Y.; Keima, T.; Iwabuchi, N.; Koinuma, H.; Hagiwara-Komoda, Y.; Ishikawa, K.; Himeno, M.; Maejima, K.; Yamaji, Y.; Namba, S. , EXA1, a GYF domain protein, is responsible for loss-of-susceptibility to plantago asiatica mosaic virus in Arabidopsis thaliana. The Plant Journal 2016, (1), 120–131. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).