Submitted:
07 January 2024
Posted:
10 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
Study design and site
Isolate Culture, Colistin susceptibility and DNA extraction
Genome sequencing, Sequence analysis and Virulence gene detection
Study Approvals and consent to participate
Data availability
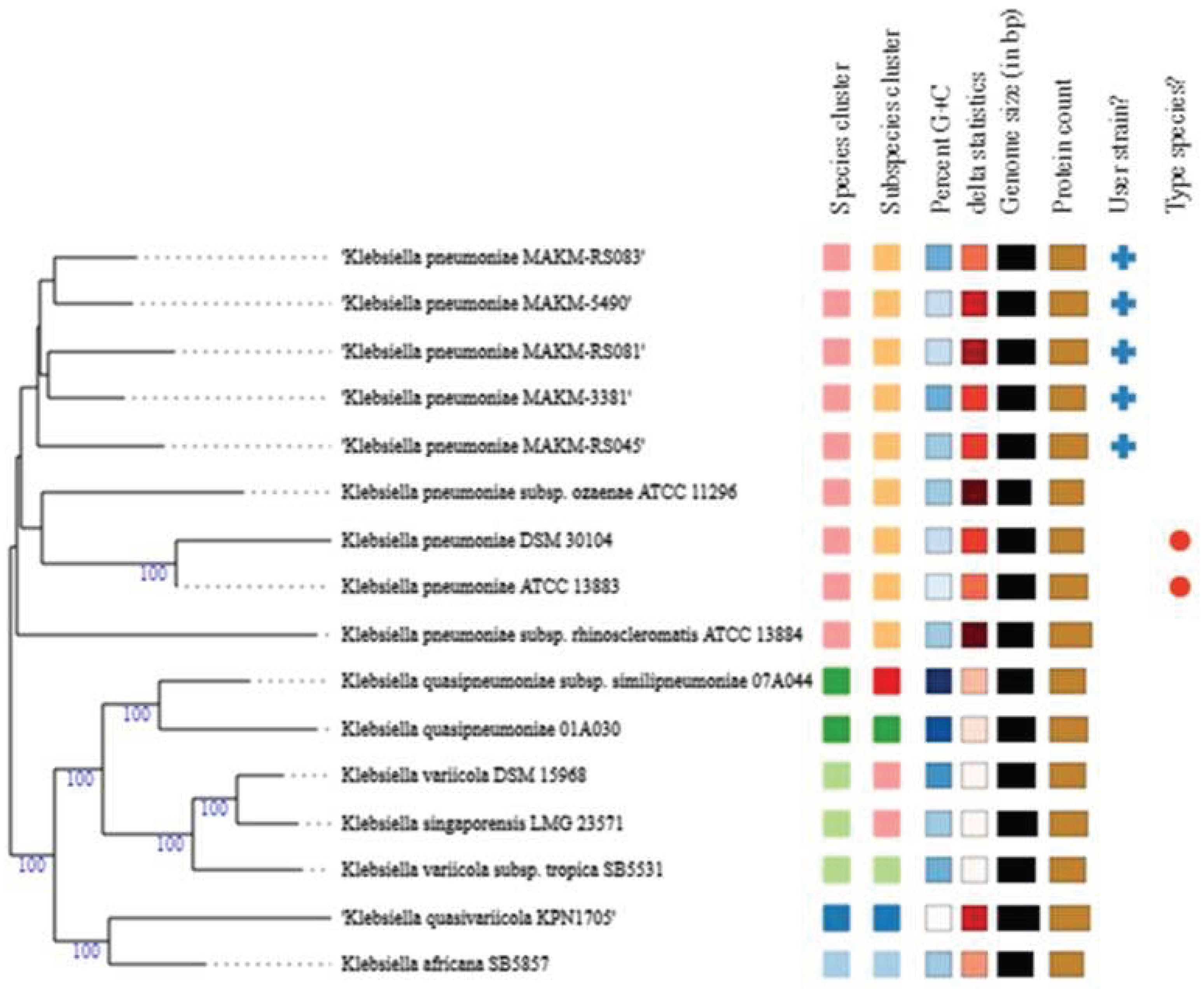
3. Results and discussion
4. Conclusion
Author Contributions
Funding
Ethical Approval
Competing Interests
References
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiology Reviews 2017, 41, 252–275. [CrossRef]
- Panigrahi K, Pathi BK, Poddar N, Sabat S, Pradhan S, Pattnaik D, et al. Colistin Resistance Among Multi-Drug Resistant Gram-Negative Bacterial Isolates From Different Clinical Samples of ICU Patients: Prevalence and Clinical Outcomes. Cureus 2020. [CrossRef]
- Gorrie CL, Mirčeta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clinical Infectious Diseases 2017, 65, 208–215. [CrossRef] [PubMed]
- Maroncle N, Balestrino D, Rich C, Forestier C. Identification of Klebsiella pneumoniae Genes Involved in Intestinal Colonization and Adhesion Using Signature-Tagged Mutagenesis. Infection and Immunity 2002, 70, 4729–4734. [CrossRef] [PubMed]
- Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018, 8, 4. [CrossRef]
- Willner D, Daly J, Whiley D, Grimwood K, Wainwright CE, Hugenholtz P. Comparison of DNA Extraction Methods for Microbial Community Profiling with an Application to Pediatric Bronchoalveolar Lavage Samples. PLOS ONE 2012, 7, e34605. [CrossRef]
- Guerra MES, Destro G, Vieira B, Lima AS, Ferraz LFC, Hakansson AP, et al. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Frontiers in Cellular and Infection Microbiology 2022, 12. [CrossRef]
- March C, Moranta D, Regueiro V, Llobet E, Tomás A, Garmendia J, et al. Klebsiella pneumoniae Outer Membrane Protein A Is Required to Prevent the Activation of Airway Epithelial Cells *. Journal of Biological Chemistry 2011, 286, 9956–9967. [CrossRef] [PubMed]
- Merciecca T, Bornes S, Nakusi L, Theil S, Rendueles O, Forestier C, et al. Role of Klebsiella pneumoniae Type VI secretion system (T6SS) in long-term gastrointestinal colonization. Sci Rep 2022, 12, 16968. [CrossRef]
- Wang H, Guo Y, Liu Z, Chang Z. The Type VI Secretion System Contributes to the Invasiveness of Liver Abscess Caused by Klebsiella pneumoniae. The Journal of Infectious Diseases 2023, 228, 1127–1136. [CrossRef]
| Strain | Sequence type (ST) | Contigcount | N50 | Largest Contig |
Total Size |
Genome size (Mb) | No. of genes | K_type | O type | Virulence genes |
|---|---|---|---|---|---|---|---|---|---|---|
| MAKM-3381 | ST1119 | 106 | 340514 | 657442 | 5443383 | 5.3 | 6304 | Unknown (KL110) | O3b |
yagX/ecpC, yagW/ecpD, yagZ/ecpA, yagY/ecpB, yagV/ecpE, ompA, ykgK/ecpR, fepA, entB, entA, fepC, entE, fepG, entS, entF, fimD, xcpR, fepB, mgtC, fepD, iroN, iutA, entC, fimA, fimB, tssH-5/clpV, waaC, galU, gspE, fimC, gspG, fes, algW, fimE, pchD, fimH, fleQ,IlpA, waaA, luxS, sfaF, focD, fimI, gspF, clpV1, waaF, algA, pilB, htpB |
| MAKM-5490 | ST39 | 90 | 250679 | 431665 | 5512705 | 5.4 | 5875 | K62 | O1 | fimD, fimH, fimC, fimF, fimI, fimG, fimA, fimE, entD, sfaF, focD, icl, tssH-5/clpV, fimB, clpV1, wzi62 |
| MAKM-RS045 | ST540-1LV | 101 | 221857 | 757815 | 5576186 | 5.4 | 5908 | K13 | O1 |
yagX/ecpC, yagW/ecpD, yagZ/ecpA, yagY/ecpB, yagV/ecpE, ompA, ykgK/ecpR, fepA, entB, entA, fepC, entE, entS, fepG, fimD, entF, xcpR, fepB, mgtC,fepD, iroN, iutA, entC, fimA, fimB, tssH-5/clpV, waaC, galU, fimC, gspG, fes, fimE, gspE, pchD, sfaF, focD, gspF, fleQ, IlpA, algW, waaA, mbtA, luxS, algA, fimI, clpV1, waaF, lpxC, pilB, wzi13 |
| MAKM-RS081 | ST231 | 100 | 374001 | 1481664 | 5617318 | 5.3 | 6031 | K51 | O1 |
yagX/ecpC, yagW/ecpD, yagZ/ecpA, yagY/ecpB, yagV/ecpE, ompA, ykgK/ecpR, fepA, entB, entA, entE, fepC, entS, fepG, entF, fimD, xcpR, fepB, mgtC, fepD, iutA, iroN, entC, fimA, fimB, fimC, waaC, galU, tssH-5/clpV, gspG, fes, fimE, gspE, pchD, gspF, clpV1, fleQ, IlpA, algW, waaA, mbtA, luxS, wbaP/rfbP, sfaF, focD, waaF, lpxC, fimH, pilB, wzi104 |
| MAKM-RS083 | ST17 | 78 | 329440 | 596846 | 5399823 | 5.2 | 5694 | K25 | O5 |
yagX/ecpC, yagW/ecpD, yagZ/ecpA, yagY/ecpB, yagV/ecpE, ompA, ykgK/ecpR, fepA, entB, entA, fepC, entE, fepG, entF, pla, fimD, entS, xcpR, fepB, mgtC, fepD, iroN, iutA, entC, fimA, fimB, waaC, galU, tssH-5/clpV, fimC, gspE, gspG, fes, algW, fimE, pchD, gspF, clpV1, algA, fleQ, IlpA, waaA, mbtA, luxS, sfaF, focD, fimI,waaF, pilB, wzi141 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





