1. Introduction
Currently, the use of IHC and/or ISH is the standard of care for routine assessment of biomarkers in breast cancers. This assessment allows subdivision of BC into several groupings; hormone receptor (ER/PR) positive/negative, HER2 positive equivocal and negative and triple negative breast cancers (TNBC).
Studies by Perou et. al. [
1] reported that BC can also be classified into several molecular subgroups based on gene expression profiles; luminal, HER2, basal-like and normal breast-like cancers. Subsequently these groups have been shown to have distinct clinical behaviours and responses to chemotherapy [
2]. As most histology laboratories have not had direct access to molecular testing, IHC tests have been used as surrogate of gene expression analysis to reproduce these gene expression profiles [
3].
However, it is accepted that BC gene expression profiles are more complex than this and this has led to the development of RNA expression profile tests such as OncotypeDX, Endopredict and Prosigna (based on PAM50). These tests are all now included in the NHSE Cancer Test Directory (NHS England Website) on surgically resected BC tissue in a particular cohort of patients. They utilise the expression data from between 12-50 genes, depending on which profiling test is being run. Unsurprisingly, there are significant discordances between sub-typing with these molecular tests and IHC-based testing [
5].
In an attempt to further refine IHC sub-typing, the proliferation marker Ki-67 was introduced. However, there are well-known limitations in the use of this marker; these include inter-laboratory variability even among expert breast pathologists and a lack of consensus regarding the optimal cut-off point between positivity and negativity [
6].
There is still some controversy over the most appropriate method to define ER and PR status and the reproducibility for the new diagnostic category of ‘HER-2 low’ BC is poor even among expert breast pathologists [
7]. These obstacles have generated a search for novel avenues for BC sub-typing. To overcome the IHC/ISH and cut-off issues, the proposal to use mRNA-based assay has been suggested as a possible alternative in routine practice. The use of RT-qPCR, particularly in the form of the Xpert BC STRAT4 assay, has been successfully validated. This assay has been used in the Europe-wide EQA study and has shown correlation and reproducibility with IHC/ISH in several European laboratories, however, the number of cases assessed so far has been limited [
8].
The recent development of the APIS Breast Cancer Subtyping Kit presents a realistic molecular alternative to IHC/ISH. It is an RNA-based diagnostic assay that assesses mRNA expression of the standard IHC biomarkers HER2, ER & PR. In addition, Ki67 plus another three genes provide further data used to generate a proliferation signature for every sample. This assessment can be performed on pre-operative core needle biopsy (CNB) or resected formalin-fixed paraffin-embedded (FFPE) breast tumour tissue. Laboratory personnel can perform the assessment of cancer samples using this kit; there is no requirement for the involvement of specialist Histopathologist.
We present the first clinical study of 100 consecutive, prospective, pre-operative BC core biopsies analysed using the APIS mRNA assay. Samples were assessed in parallel with the routine IHC/ISH workflows thus enabling patient discussion of all the data during the routine breast MDT meetings.
2. Material and Methods
2.1. Samples
98 samples of FFPE pre-operative breast core biopsies and 2 core biopsies of lymph nodes were assessed histologically and the presence and grade of cancer confirmed. Current routine processes assessed all the samples by IHC/ISH and they were simultaneously assessed using the APIS mRNA-based assay.
The mRNA data generated by the APIS assay was included in the patient pathological report and clinical discussion of the data occurred. It is important to note that national guidelines were followed and only the IHC/ISH results were employed in relation to patient management.
2.2. Immunohistochemistry and In situ Hybridisation
Immunohistochemistry for HER2, ER, PR and Mib-1 (Ki67) was performed using an automated staining module as per standard protocols for routine assessment of these samples. Nuclear staining for ER and PR was scored using the Allred scoring system for BC based on assessment of intensity of nuclear staining and proportion of immuno-stained cells [
8].
ER IHC was divided into negative (0/8) and strongly positive (7-8/8) groups; no cases of weak or intermediate nuclear staining were recorded. PR IHC was divided into three groups based on the staining seen; negative (0/8), weak-moderate (2-6/8) and strong (7-8/8).
Ki67 IHC was divided in two groups low (<20%) and high (>20%). The threshold for positivity of >1% was adopted as described previously ASCO/CAP ER/PR [
9].
For HER-2, the standard ASCO⁄CAP scoring system was applied, 0, 1+, 2+, 3+.
ISH was used to further assess BCs exhibiting Her2 2+ IHC.
All IHC slides from each BC case underwent histological evaluation by an experienced BC pathologist
2.3. RNA Extraction
Total RNA was isolated from FFPE tissue sections measuring 10 µm in thickness, ensuring an area of the specimen with tumour content of ≥20% was analysed. The automated extraction of RNA was performed using Maxwell48 RSC RNA FFPE Kit following the manufacturer's recommended protocol. Subsequently, the RNA content within each eluate was quantified and normalized to a concentration of 2.5 ng/µL. Samples were stored at -80°C until required.
2.4. Gene Expression by RT-qPCR
The quantification of mRNA expression levels was performed using the APIS Breast Cancer Subtyping Kit. This analysis uses RT-PCR for the measurement of ESR1, PGR, ERBB2, MKI67, as well as three additional targets found within the proliferation signature (CCNA2, KIF23, and PCNA), along with two reference genes (IPO8 and PUM1).
The assay was set-up as per the manufacturer’s protocol and the RT-qPCR reaction was analysed using the QuantStudio™ 5 Dx real-time PCR system (QS5™Dx; Thermo Fisher Scientific).
The protocol requires each specimen to undergo duplicate analysis for each gene assay. A maximum of 10 patient samples and a positive and negative control could be analysed at any one time.
Proprietary software was used to determine levels of gene expression for each sample utilising the reference genes to generate a ΔCt. Briefly, this is achieved through the subtraction of the average cycle threshold (Ct) value of the duplicate measurements of the target of interest from the mean Ct value obtained from duplicate measurements of the reference genes. Binary target calls (positive/negative) were reported based on previously established ΔCt cut-off values specific to each target gene.
A logistic model using ΔCt values for MKI67, CCNA2, PCNA, and KIF23 was employed to calculate a proliferation score within the range of 0 to 1.
A score below 0.5 indicates low proliferation, while a score above 0.5 indicates high proliferation. The molecular subtype classification was determined by considering the combined statuses of ESR1, PGR, ERBB2 and MKI67.
3. Results
3.1. APIS Molecular Characteristics
The run validity of RNA results were available for 99 cases tested. One sample failed to produce sufficient RNA for analysis.
Using the BC Subtyping Kit Analysis software developed by APIS Assay Technologies the results calling produced: 76 samples ER positive (above -1.98 cutoff value), 65 were PR positive (above -0.63), 14 samples were Her2 positive (above 2.00). The proliferation signature (MKI67, PCNA, CCNA2, KIF23) produced a
‘Low’ category in 21 cases versus a High category in 78 cases. See
Table 1.
Molecular classification according to this software produced the following subtypes: 39 samples Luminal A, 45 Luminal B (11 Her-2 +ve and 34 Her-2–ve), 11 triple negative (TNBC) and 4 HER2 enriched.
3.2. Pathological and IHC Characteristics
In this consecutive series of 100 cases 3 BCs were Grade 1, 57 were Grade 2 and 38 were Grade 3. Two were nodal metastases. The large majority (89) were invasive carcinoma of no special type (formerly known as Invasive ductal carcinomas (IDC)), 9 were invasive lobular carcinoma (ILC) and 2 were mucinous carcinoma.
The large majority of BCs were ER positive (Allred score 7/8 in 8 cases and 8/8 in 78 cases), 14 cases were ER-negative. PR values were as follows: 0/8 in 14 cases, low (from 2-6/8) in 21 and high (from 7-8/8) in 75 cases.
HER-2 IHC was negative (score 0) in 59 cases, 1+ in 20 cases, 2+/ISH negative in 5 cases and 15 cases were scored positive (3+).
IHC Ki67-proliferation was high (>20%) in 48 cases and low (<20%) in 52 cases.
3.3. Comparison of APIS mRNA Scores for ER, PR, HER-2, Ki-67 and IHC Based Results
Each biomarker was recorded as either concordant or discordant when IHC and mRNA results were compared (See table 2).
Briefly, there was 96% overall concordance between IHC and APIS across ER/PR & HER2 assays; 97% ER, 89% PR and 100% HER2.
Discordant Cases: 3/100 cases had results that were discordant between ER-IHC and ER-APIS. One sample was scored by IHC as luminal (ER score 7/8) but was ER-APIS negative in both of two subsequent biopsies (case 6 and 71). This sample was also discordant for both biopsies in the PR assays. A further sample was also scored ER-IHC positive (score 8/8) and ER-APIS negative; case 60, a recurrent IDC in which most of the specimen was scar tissue with a very small amount of cancer cells. This sample was also discordant for PR (IHC-PR positive, PR-APIS negative). One sample (case 93, ILC with mixed features; both classical-ER+/HER-2 negative and pleomorphic ER-/HER-2 +) was ER-IHC positive but ER-APIS negative.
There were an additional 9 cases that were discordant PR-IHC versus PR-APIS. 3% cases were discordant between Ki67 and APIS results when compared with MKI67 alone, but this rose to 24% discordance when the four-gene RNA proliferation signature was compared to Ki67-IHC
4. Discussion
The pandemic resulted in a huge backlog of patients awaiting surgery and subsequently a large number of specimens requiring histological evaluation; this has put an enormous strain on histopathology laboratories across the UK and unfortunately, a consequence being that some cancers were taking longer to be diagnosed.
The APIS Breast Cancer Subtyping Kit provides data for all the biomarkers routinely used in breast cancer staging allowing patient management based on robust and reproducible results. So far those results have been provided through IHC and microscopic examination by highly trained Histopathologist. The use of an RNA-based assay reduces the need for the involvement of this cohort of specialists, thereby releasing their time for other essential work.
Moreover eliminating the subjective interpretation of IHC test produces molecular results that can be used by healthcare professionals to tailor each breast cancer patient according to their molecular signature.
The data reported here indicates that there is an extremely high level of concordance in the results produced by two different methodologies but it also identified a small number of clinically relevant discordances.
In the evaluation of our discordant cases, we categorised them into clinically relevant discordances or discordances requiring improvements/refinement.
4.1. Clinically Relevant Discordances
Case 1 was assessed by histology as grade 3 IDC with basal-like features being noted, it was recorded by IHC as luminal subtype (ER score 7/8) with high proliferation. However this sample was reported as a triple negative (basal-like) molecular sub-type by the APIS RNA assessment on two biopsies from the same patient (cases 6 and 71). Figure 1
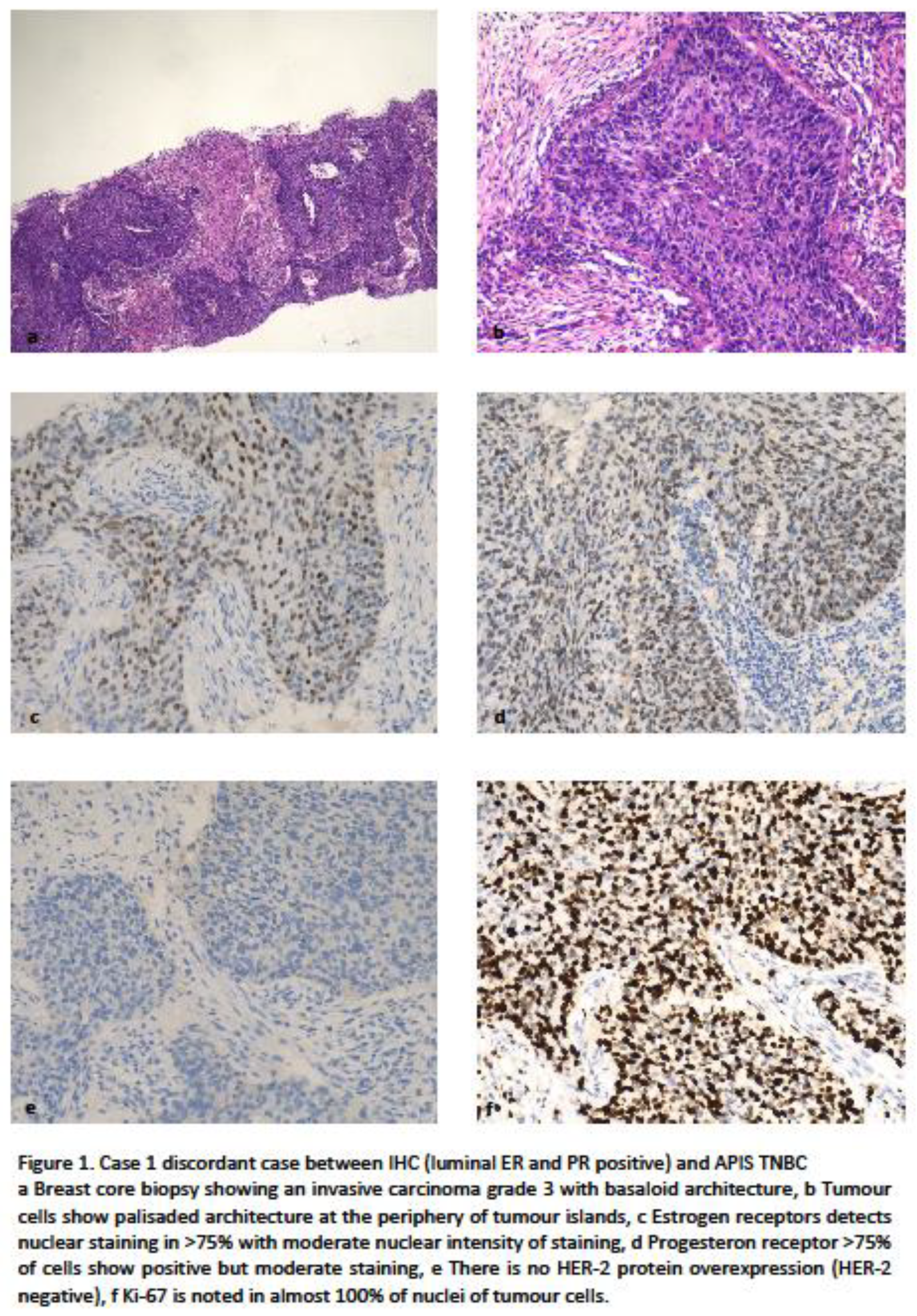
It is important to note that histologically Luminal BCs classified as basal-like by expression analysis have been demonstrated to be those more responsive to chemotherapy, as shown by the GIADA neoadjuvant study [
12]. This case is similar to one previously reported by Kim et al [
5] in which clinical follow-up showed worse overall survival than would be expected for a luminal sub-type BC.
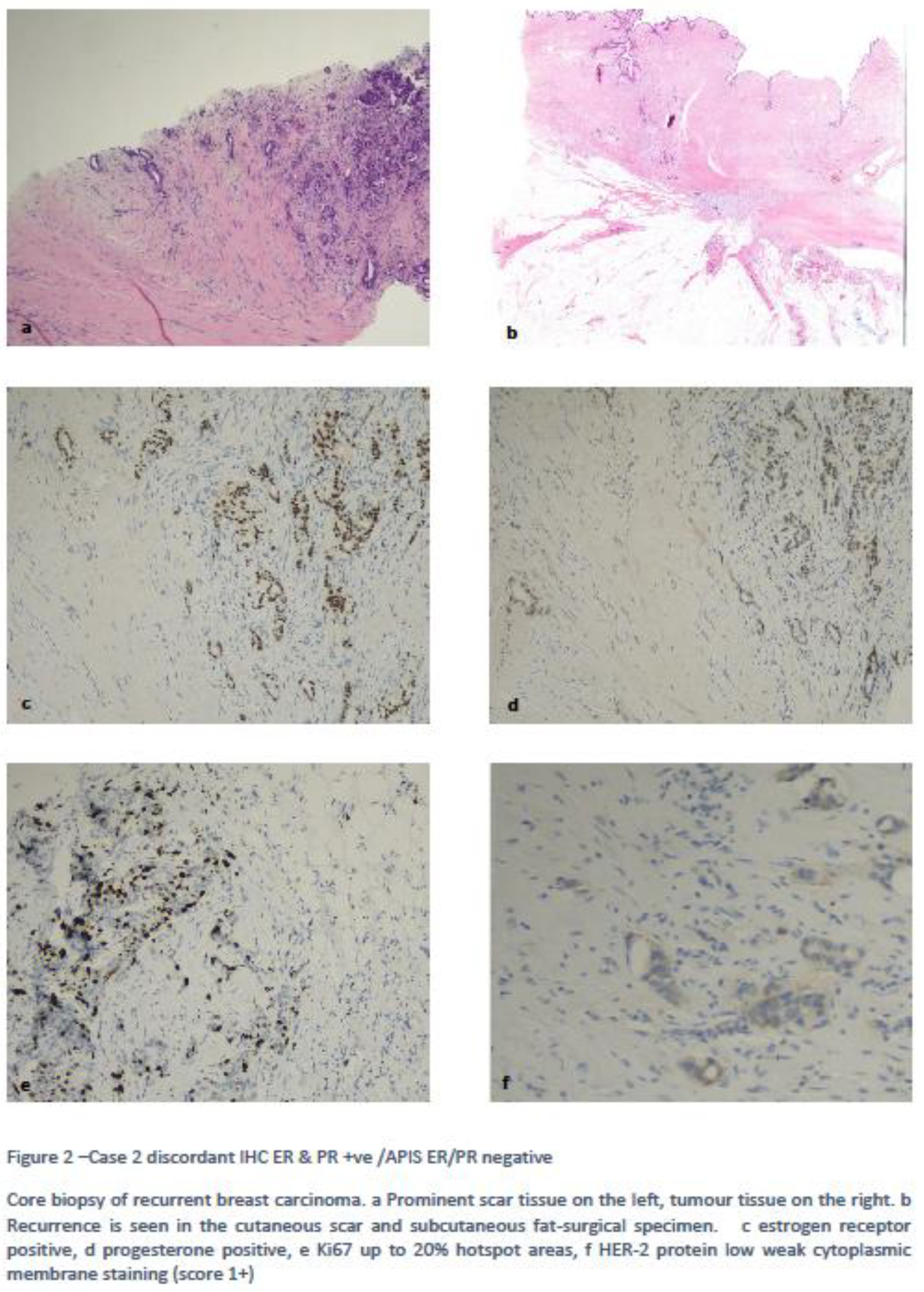
Case 2 was ER-IHC positive but ER-APIS negative. The patient had a previous hormone receptor positive HER-2 negative IDC with a recurrence occurring in the scar tissue. Review of the slide showed most of the specimen was scar tissue with limited amount of IDC, hence the possible cause of discordant results was due to the low tumour cellularity of the sample. Figure 2
Case 3 was an ILC with mixed features: a small proportion of classical-ER+/HER-2 negative ILC cells and larger proportion of pleomorphic, ER negative/HER-2 positive cells. The APIS assay scored this ER and PR negative/Her-2 positive. Given that the latter component was more prevalent in the sample, the APIS negative result it is not surprising i.e. heterogeneity of the sample has to be taken into consideration when using assessing samples with the APIS assay. Figure 3.
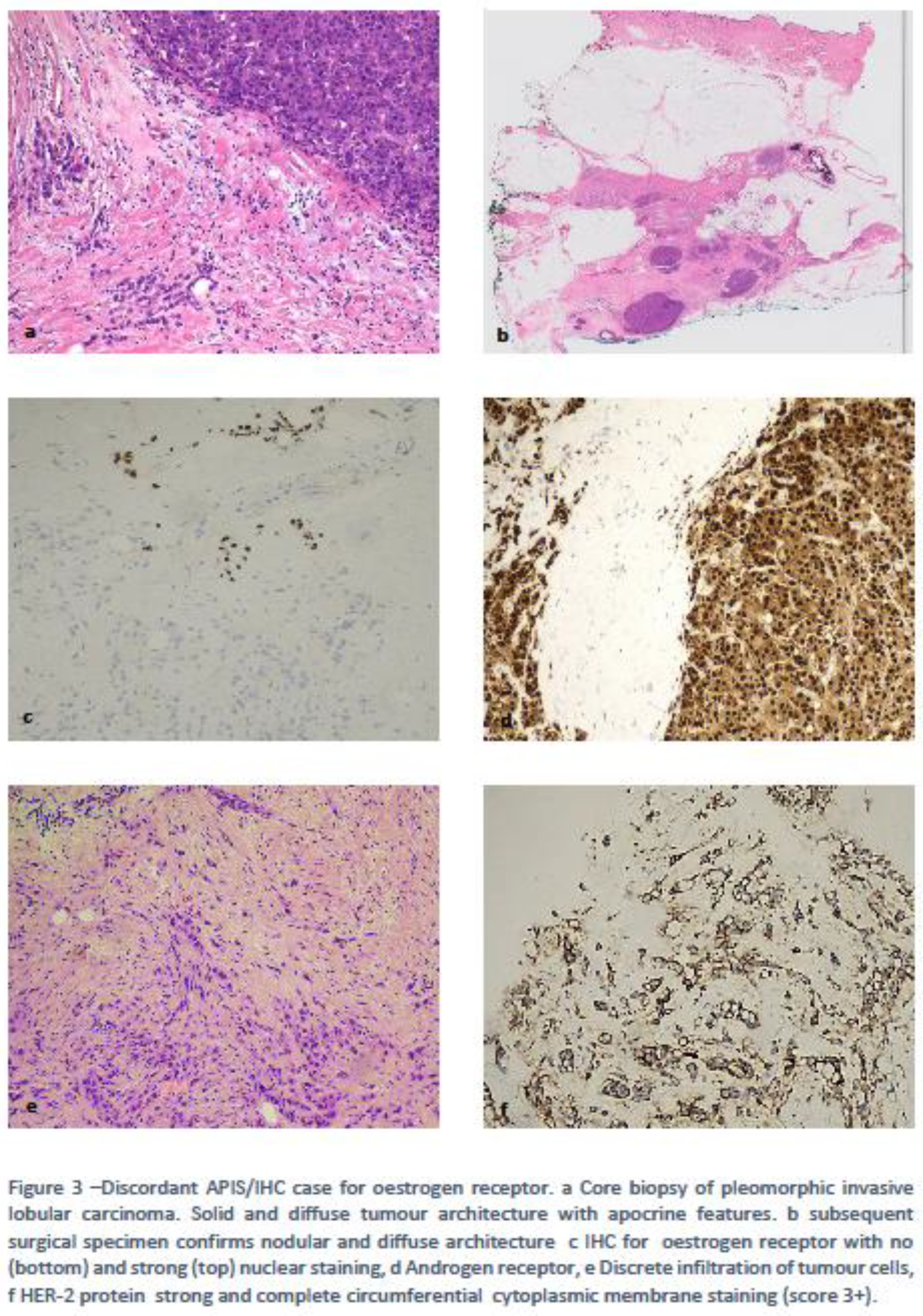
4.2. Discordances Requiring Improvements/Refinement.
When discussing those areas of discordance that we defined “discordances requiring improvements/refinement” this mainly involved the APIS PR results compared to IHC and Ki-67 IHC analysis compared to the results of the proliferation signature; these represent the majority of discordant cases in this study (10/100, 24/100 respectively).
Of the 9 PR discordant cases, 7 had weak and moderate nuclear staining producing a score between 2 to 6 on IHC, but were all scored negative by the APIS assay. It has been widely noted that the behavior of BCs with low levels of hormone receptors is more consistent with features of triple negative breast carcinomas rather than with Luminal carcinomas. Even the ASCO/CAP recommendations suggest to include a specific comments when encountering low levels of hormone receptors [
10].
Currently the discrepancy between the final 2 cases is unexplained although one had very few tumour cells in the samples and this may contribute to the negative PR result in the APIS assay.
The limitations of measuring proliferation markers Ki-67 are well known. They include inter-laboratory variability even among expert breast pathologists and there is a lack of consensus (?) around Ki67 values which represent optimal cut off points for positivity and negativity [
6].
The assessment of the proliferation signature (MK67, PCNA, CCNA2 & KIF23) is most likely the reason for the larger number of discrepant results as these markers take into account genes involved in aspects of the cell cycle other than that of Ki-67 i.e. PCNA [
14], Kinesin 23 (KIF23) [
15] and Cyclin A2 (CCNA2) [
16].
It is accepted that a gene signature better reflects the proliferative nature of a sample than just a single marker (KI67) [
17].
Our study considered the challenges of the implementation of the APIS assay into clinical practice, taking into account, particularly, the time-effectiveness of the assay; the assay could produce data in a similar time-frame and similar costs to that produced by IHC. The introduction of the APIS BC Subtyping Kit into routine analysis in place of IHC, could potentially save pathologists time and alleviate pressure on IHC pathology laboratory which has been significantly impacted by post-COVID backlog cases and led to delays in patient management. The application on core biopsies has also potential implications on the use of this assay in the context of the best selection of neoadjuvant treatment in Luminal and TNB carcinomas.
Nevertheless, the assay does have limitations particularly around morphological heterogeneity and tumour cellularity of samples. There are clinical implications such as a patient might be treated as having non-luminal BC and therefore deprived of the benefit of hormone therapy if a heterogeneous tumour was assessed incorrectly as in Case 2 in our study. In situ analysis are still superior to in vitro nucleic acid based analysis in this respect, i.e. when the specimen is scarce in cellularity or morphologically heterogeneous.
It is possible that IHC would still have to be used in cases where the histology indicates a heterogeneous sample or one with low tumour burden.
Moreover, the cohort of BCs analysed in this study is mostly grade 2 and 3 BC, hence the impact of this mRNA assessment in wider cohort of BC remains to be determined.
The assay currently only provides a binary (positive-negative) mRNA result. It has been recommended to the manufacturers that they design a report where the level of ER, PR and HER-2 is graded in a way that mirrors the IHC based reports. In other words, where ER & PR are reported with a score from 2 to 8, HER-2 scores of low (1+, 2+/ISH) and positive 2+ISH+ & 3+, thereby truly offering a like-for-like replacement to IHC/ISH assessment.
Funding
All APIS BC Subtyping Kit were kindly provided by APIS BC Subtyping group who have also provided funding for overtime work done by the molecular biology staff. The group was not involved in the case selection or final data analysis.
Ethic approval
Not requested as laboratory improvement project.
Patient consent
Not requested as part of diagnostic reporting.
Acknowledgments
We are grateful to the histology lab staff for using their time to perform the tests. Our Thanks go to Alex Breeze for tables drawings. .
Conflicts of Interest
None.
References
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747-52.
- Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135:301-6.
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222-32.
- Di Palma S, Simpson RH, Marchiò C, et al. Salivary duct carcinomas can be classified into luminal androgen receptor-positive, HER2 and basal-like phenotypes. Histopathology. 2012;61(4):629-643. [CrossRef]
- Kim HK, Park KH, Kim Y, et al. Discordance of the PAM50 Intrinsic Subtypes Compared with Immunohistochemistry-Based Surrogate in Breast Cancer Patients: Potential Implication of Genomic Alterations of Discordance. Cancer Res Treat. 2019;51(2):737-747. [CrossRef]
- Polley MY, Leung SC, McShane LM, et al. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013;105(24):1897-1906. [CrossRef]
- Zaakouk, M., Quinn, C., Provenzano, E., Boyd, C., Callagy, G., Elsheikh, S., Flint, J., Millican-Slater, R., Gunavardhan, A., Mir, Y., Makhija, P., Di Palma, S., Pritchard, S., Tanchel, B., Rakha, E., Atallah, N. M., Lee, A. H. S., Pinder, S., & Shaaban, A. M. (2023). Concordance of HER2-low scoring in breast carcinoma among expert pathologists in the United Kingdom and the republic of Ireland -on behalf of the UK national coordinating committee for breast pathology. Breast (Edinburgh, Scotland), 70, 82–91. [CrossRef]
- Erber R, Hartmann A, Fasching PA, et al. Reproducibility of mRNA-Based Testing of ESR1, PGR, ERBB2, and MKI67 Expression in Invasive Breast Cancer-A Europe-Wide External Quality Assessment. Cancers (Basel). 2021;13(18):4718. Published 2021 Sep 21. [CrossRef]
- Ahmad Fauzi MF, Wan Ahmad WSHM, Jamaluddin MF, et al. Allred Scoring of ER-IHC Stained Whole-Slide Images for Hormone Receptor Status in Breast Carcinoma. Diagnostics (Basel). 2022;12(12):3093. Published 2022 Dec 8. [CrossRef]
- Allison KH, Hammond MEH, Dowsett M, et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol. 2020;38(12):1346-1366. [CrossRef]
- Wolff AC, Somerfield MR, Dowsett M, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: ASCO-College of American Pathologists Guideline Update. J Clin Oncol. 2023;41(22):3867-3872. [CrossRef]
- Dieci MV, Guarneri V, Tosi A, et al. Neoadjuvant chemotherapy and immunotherapy in luminal B-like breast cancer: results of the phase II GIADA trial. Clin Cancer Res 2022; 28: 308–317.
- Ibrahim A, Toss MS, Makhlouf S, Miligy IM, Minhas F, Rakha EA. Improving mitotic cell counting accuracy and efficiency using phosphohistone-H3 (PHH3) antibody counterstained with haematoxylin and eosin as part of breast cancer grading. Histopathology. 2023;82(3):393-406. [CrossRef]
- Liu F, Wu Y, Mi Y, Gu L, Sang M, Geng C. Identification of core genes and potential molecular mechanisms in breast cancer using bioinformatics analysis. Pathol Res Pract. 2019;215(7):152436. [CrossRef]
- Li T-F, Zeng H-J, Shan Z, Ye R-Y, Cheang T-Y, Zhang Y-J, et al. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer cell international 2020;20: 123.
- Lu Y, Yang G, Xiao Y, et al. Upregulated cyclins may be novel genes for triple-negative breast cancer based on bioinformatic analysis. Breast Cancer. 2020;27(5):903-911. [CrossRef]
- Sotiriou, C., & Piccart, M. J. (2007). Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care?. Nature reviews. Cancer, 7(7), 545–553. [CrossRef]
Table 1.
ΔCt cut off values for the APIS BC Subtyping Kit targets.
Table 1.
ΔCt cut off values for the APIS BC Subtyping Kit targets.
| Target |
Positive/negative Cut off values |
| ESR1 |
-1.98 |
| PGR |
-0.63 |
| ERBB2 |
2.00 |
| MKI67 |
-0.64 |
Table 2.
| Immunohistochemistry |
APIS Breast Cancer Subtyping Kit Analysis |
| Serial No |
Type/Met |
ER |
PR |
HER-2 |
MIB1 IHC |
ESR1 |
PGR |
ERBB2 |
MK167 |
Proliferation |
Molecula Subtype |
| 1 |
IDC G2 |
8/8 |
8/8 |
Low 2+ ISH -neg |
Low |
+ve |
+ve |
-ve |
Low |
Low |
Luminal A |
| 2 |
IDC G2 |
8/8 |
8/8 |
Low 2+ ISH -ve |
Low |
+ve |
+ve |
-ve |
high |
high |
Luminal B Her-2 -ve |
| 3 |
IDC G3 |
8/8 |
8/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
high |
high |
Luminal B Her2 -ve |
| 4 |
IDC G3 |
0/8 |
6/8 |
-ve |
high |
-ve |
-ve |
-ve |
high |
high |
TNBC |
| 5 |
IDC G3 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
high |
high |
Luminal B her-2 -ve |
| 6 |
IDC G3 |
7/8 |
7/8 |
-ve |
high |
-ve |
-ve |
-ve |
high |
high |
TNBC |
| 7 |
IDC G2 |
8/8 |
8/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
high |
high |
Luminal B her-2 -ve |
| 8 |
IDC G3 |
8/8 |
8/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B her-2 -ve |
| 9 |
IDC G2 |
8/8 |
0/8 |
-ve |
low |
+ve |
-ve |
-ve |
low |
high |
Luminal A |
| 10 |
IDC G2 |
8/8 |
0/8 |
Low 1+ |
low |
+ve |
-ve |
-ve |
low |
Low |
Luminal A |
| 11 |
IDC G3 |
0/8 |
0/8 |
+ve 3+ |
low |
-ve |
-ve |
+ve |
Low |
High |
HER-2 Enriched |
| 12 |
IDC G2 |
8/8 |
0/8 |
-ve |
low |
+ve |
-ve |
-ve |
Low |
High |
Luminal A |
| 13 |
IDC G2 |
8/8 |
0/8 |
-ve |
low |
NA |
NA |
NA |
NA |
NA |
NA |
| 14 |
IDC G2 |
8/8 |
4/8 |
-ve |
high |
+ve |
-ve |
-ve |
high |
High |
Luminal B her-2 -ve |
| 15 |
IDC G1 |
7/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
Low |
Luminal A |
| 16 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 17 |
IDC G3 |
0/8 |
0/8 |
-ve |
high |
-ve |
-ve |
-ve |
high |
high |
Triple negative |
| 18 |
IDC G3 |
8/8 |
7/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 19 |
IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
high |
Luminal B |
| 20 |
IDC G3 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her 2 -ve |
| 21 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 22 |
IDC G2 |
8/8 |
0/8 |
-ve |
low |
+ve |
-ve |
-ve |
low |
High |
Luminal A |
| 23 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 24 |
IDC G3 |
8/8 |
4/8 |
-ve |
high |
+ve |
-ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 25 |
IDC G2 |
8/8 |
6/8 |
Low 1+ |
low |
-ve |
+ve |
-ve |
low |
High |
Luminal A |
| 26 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
Low |
High |
Luminal A |
| 27 |
IDC G3 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 28 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 29 |
IDC G3 |
0/8 |
0/8 |
+ve 3+ |
low |
-ve |
-ve |
-ve |
low |
High |
HER-2 Enriched |
| 30 |
IDC G2 |
8/8 |
7/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 31 |
IDC G3 |
8/8 |
8/8 |
+ve 3+ |
high |
+ve |
+ve |
+ve |
High |
high |
Luminal B her-2 +ve |
| 32 |
IDC G3 |
0/8 |
2/8 |
-ve |
high |
-ve |
-ve |
-ve |
low |
high |
TNBC |
| 33 |
IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 34 |
IDC G2 |
8/8 |
8/8 |
Low 2+ ISH -ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her 2 -ve |
| 35 |
IDC G2 |
8/8 |
0/8 |
-ve |
low |
+ve |
-ve |
-ve |
low |
High |
Luminal A |
| 36 |
IDC G3 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 37 |
IDC G2 |
0/8 |
0/8 |
-ve |
low |
-ve |
-ve |
-ve |
low |
low |
Triple negative |
| 38 |
IDC G2 |
7/8 |
8/8 |
Low 1+ |
low |
+ve |
+ve |
neg |
low |
low |
Luminal A |
| 39 |
IDC G3 |
7/8 |
8/8 |
Low 1+ |
high |
positive |
positive |
neg |
high |
high |
luminal B her-2 -ve |
| 40 |
IDC G3 |
0/8 |
0/8 |
+ve 3+ |
high |
-ve |
|
+ve |
High |
High |
luminal B Her2 +ve |
| 41 |
IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her2 -ve |
| 42 |
IDC G3 |
8/8 |
8/8 |
+ve 3+ |
high |
+ve |
+ve |
+ve |
High |
High |
luminal B Her-2 +ve |
| 43 |
IDC G3 |
0/8 |
3/8 |
Low 1+ |
high |
-ve |
-ve |
-ve |
High |
High |
TNBC |
| 44 |
IDC G2 |
8/8 |
6/8 |
2+ISH -ve |
low |
+ve |
-ve |
-ve |
low |
low |
Luminal A |
| 45 |
ILC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
negative |
low |
high |
luminal A |
| 46 |
ILC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 47 |
IDC G2 |
8/8 |
8/8 |
Low +1 |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 48 |
IDC G3 |
0/8 |
3/8 |
-ve |
high |
-ve |
-ve |
-ve |
High |
High |
TNBC |
| 49 |
ILC G2 |
8/8 |
8/8 |
Low 1+ |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 50 |
IDC G3 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her 2 -ve |
| 51 |
IDC G3 |
8/8 |
6/8 |
+ve 3+ |
high |
+ve |
-ve |
+ve |
High |
High |
Luminal B Her-2 +ve |
| 52 |
IDC G3 |
8/8 |
8/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her 2 -ve |
| 53 |
IDC G3 |
0/8 |
2/8 |
+ve 3+ |
high |
-ve |
+ve |
+ve |
High |
High |
Luminal B Her-2 +ve |
| 54 |
IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 55 |
IDC G3 |
8/8 |
8/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 56 |
IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 57 |
IDC G2 |
0/8 |
3/8 |
+ve 3+ |
high |
-ve |
+ve |
+ve |
High |
High |
Luminal B Her 2 +ve |
| 58 |
IDC G3 |
8/8 |
8/8 |
+ve 3+ |
high |
+ve |
+ve |
+ve |
High |
High |
Lum B Her-2 +ve |
| 59 |
mucinous ca G2 |
8/8 |
8/8 |
Low 1+ |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 60 |
IDC G2 |
8/8 |
8/8 |
low 2+, ISH -ve |
low |
-ve |
-ve |
-ve |
low |
High |
Triple negative |
| 61 |
IDC G2 |
8/8 |
8/8 |
+ve 3+ |
low |
+ve |
+ve |
+ve |
low |
High |
Lum B Her-2 +ve |
| 62 |
IDC G2 |
8/8 |
8/8 |
Low 1+ |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 63 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 64 |
IDC G3 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
high |
high |
luminal B |
| 65 |
IDC G2 |
8/8 |
2/8 |
-ve |
low |
+ve |
-ve |
-ve |
low |
High |
Luminal A |
| 66 |
IDC G1 |
8/8 |
5/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 67 |
IDC G3 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B her-2 -ve |
| 68 |
IDC G2 |
8/8 |
8/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her-2 -ve |
| 69 |
IDC G3 |
0/8 |
0/8 |
-ve |
High |
-ve |
-ve |
-ve |
High |
High |
TNBC |
| 70 |
ILC G2 |
8/8 |
8/8 |
Low 1+ |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 71 |
IDC G3 |
7/8 |
7/8 |
-ve |
high |
-ve |
-ve |
-ve |
High |
High |
TNBC |
| 72 |
ILC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 73 |
ILC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
high |
luminal B her-2 -ve |
| 74 |
Nodal met IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B her-2 -ve |
| 75 |
IDC G3 |
8/8 |
5/8 |
Low 1+ |
high |
+ve |
+ve |
-ve |
High |
High |
Luminal B Her2 -ve |
| 76 |
IDC G3 |
8/8 |
3/8 |
-ve |
high |
+ve |
-ve |
-ve |
High |
High |
Luminal B Her2 -ve |
| 77 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 78 |
ILC G2 |
8/8 |
7/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
high |
Luminal A |
| 79 |
IDC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 80 |
IDC G2 |
8/8 |
3/8 |
-ve |
low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 81 |
ILC G2 |
8/8 |
3/8 |
-ve |
low |
+ve |
-ve |
-ve |
low |
high |
Luminal A |
| 82 |
IDC G3 |
7/8 |
7/8 |
-ve |
high |
+ve |
+ve |
+ve |
High |
High |
Luminal B Her-2 -ve |
| 83 |
IDC G3 |
8/8 |
8/8 |
+ve 3+ |
High |
+ve |
+ve |
+ve |
High |
High |
Luminal B Her-2 +ve |
| 84 |
IDC G2 |
8/8 |
0/8 |
-ve |
low |
+ve |
-ve |
-ve |
low |
low |
Luminal A |
| 85 |
IDC G2 |
8/8 |
8/8 |
Low 1+ |
low |
-ve |
+ve |
-ve |
low |
high |
Luminal A |
| 86 |
IDC G3 |
8/8 |
8/8 |
-ve |
Low |
+ve |
+ve |
-ve |
low |
High |
Luminal A |
| 87 |
IDC N met |
0/8 |
0/8 |
2+ISH -ve |
High |
-ve |
-ve |
-ve |
High |
High |
TNBC |
| 88 |
IDC G1 |
8/8 |
4/8 |
-ve |
Low |
+ve |
+ve |
-ve |
low |
low |
Luminal A |
| 89 |
IDC G3 |
8/8 |
5/8 |
+ve 3+ |
high |
+ve |
-ve |
+ve |
High |
high |
Luminal B Her-2 +ve |
| 90 |
IDC G2 |
8/8 |
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
High |
High |
luminal B her-2 -ve |
| 91 |
Muc G2 |
8/8 |
8/8 |
-ve |
Low |
+ve |
+ve |
-ve |
low |
low |
luminal A |
| 92 |
IDC G2 |
8/8 |
8/8 |
-ve |
High |
+ve |
+ve |
-ve |
High |
High |
Luminal B her-2 -ve |
| 93 |
Pleom ILC |
8/8 |
3/8 |
+ve 3+ |
low |
-ve |
-ve |
+ve |
low |
High |
HER-2 Enriched |
| 94 |
IDC G3 |
7/8 |
6/8 |
low 1+ |
high |
+ve |
+ve |
-ve |
high |
high |
luminal B her-2 -ve |
| 95 |
IDC G3 |
7/8 |
8/8 |
low 1+ |
high |
+ve |
+ve |
-ve |
high |
High |
Luminal B her-2 -ve |
| 96 |
IDC G2 |
8/8 |
4/8 |
-ve |
low |
+ve |
-ve |
-ve |
high |
High |
luminal B her-2 -ve |
| 97 |
IDC G2 8/8 |
|
8/8 |
-ve |
high |
+ve |
+ve |
-ve |
igh hig |
High |
Lum B Her-2 -ve |
| 98 |
IDC G2 |
8/8 |
8/8 |
Low 1+ |
low |
+ve |
+ve |
-ve |
low |
high |
luminal A |
| 99 |
IDC G3 |
0/8 |
3/8 |
-ve |
high |
-ve |
-ve |
+ve |
high |
high |
HER-2 enrcihed |
| 100 |
ILC G2 |
8/8 |
8/8 |
-ve |
low |
+ve |
+ve |
neg |
low |
low |
luminal A |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).







