Submitted:
11 January 2024
Posted:
12 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
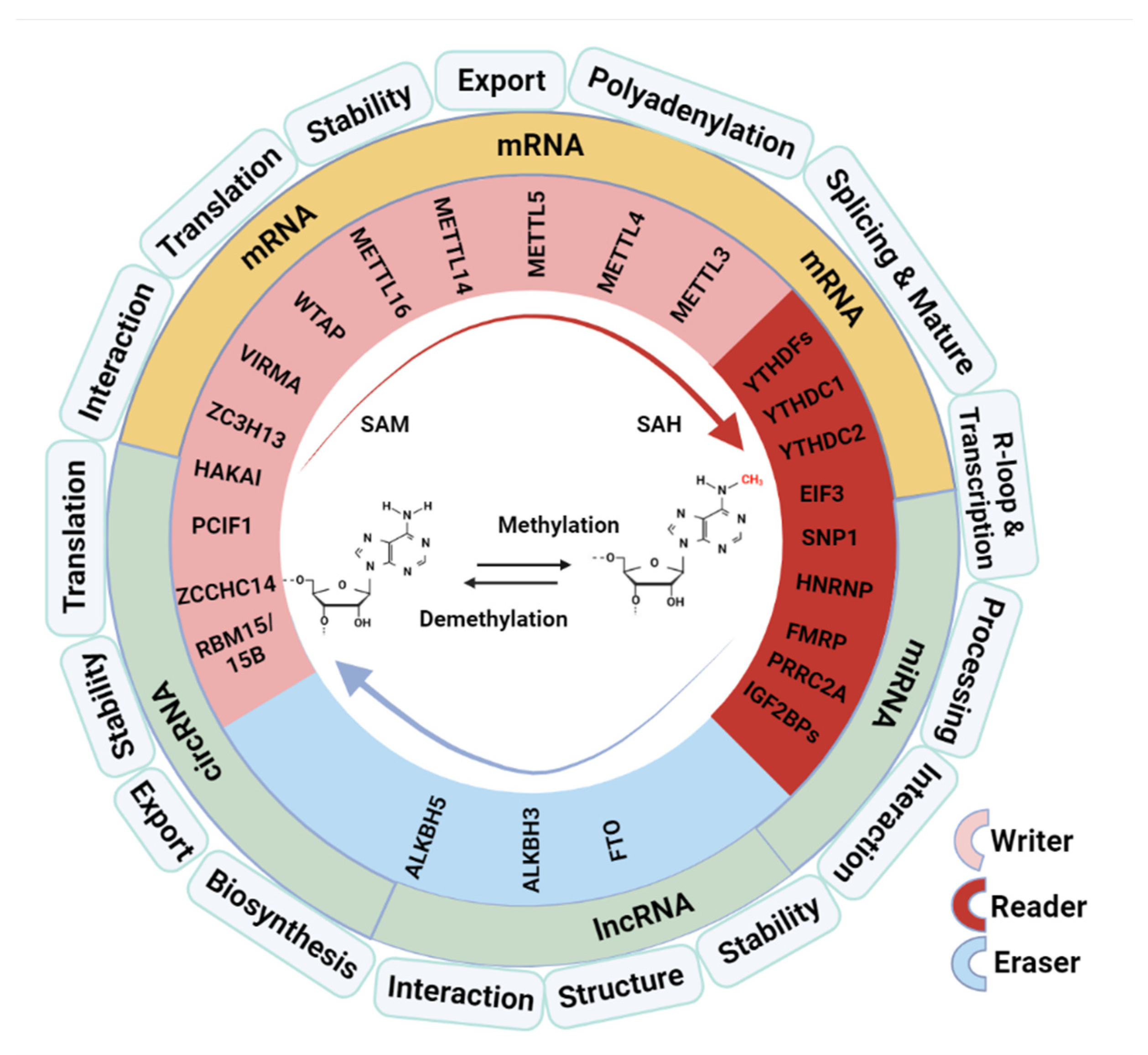
2. The Role of M6A Modification in RNA Metabolism and Functions
2.1. Regulate Pre-mRNA Splicing and RNA Nuclear Export
2.2. Affect mRNA Stability and Translation Efficiency
2.3. Involve in Processing of microRNAs
2.4. Influence the Structure and Function of LncRNA
3. RNA M6A Methylation in Radiation-Induced Cellular Response and Tissue Reactions
3.1. Impacts of Radiation on RNA M6A Methylation
3.2. M6A Methylation in Radiation-Induced DNA Damage Response to Determin Cellular Radiosensitivity
3.3. M6A Methylation in Radiation-Induced Injury of IR Sensitive Hematopoietic Tissue
3.4. M6A Methylation in Radiation-Induced Toxics of Organs
4. RNA M6A Methylation in Cellular Responses in Cancer Radiotherapy
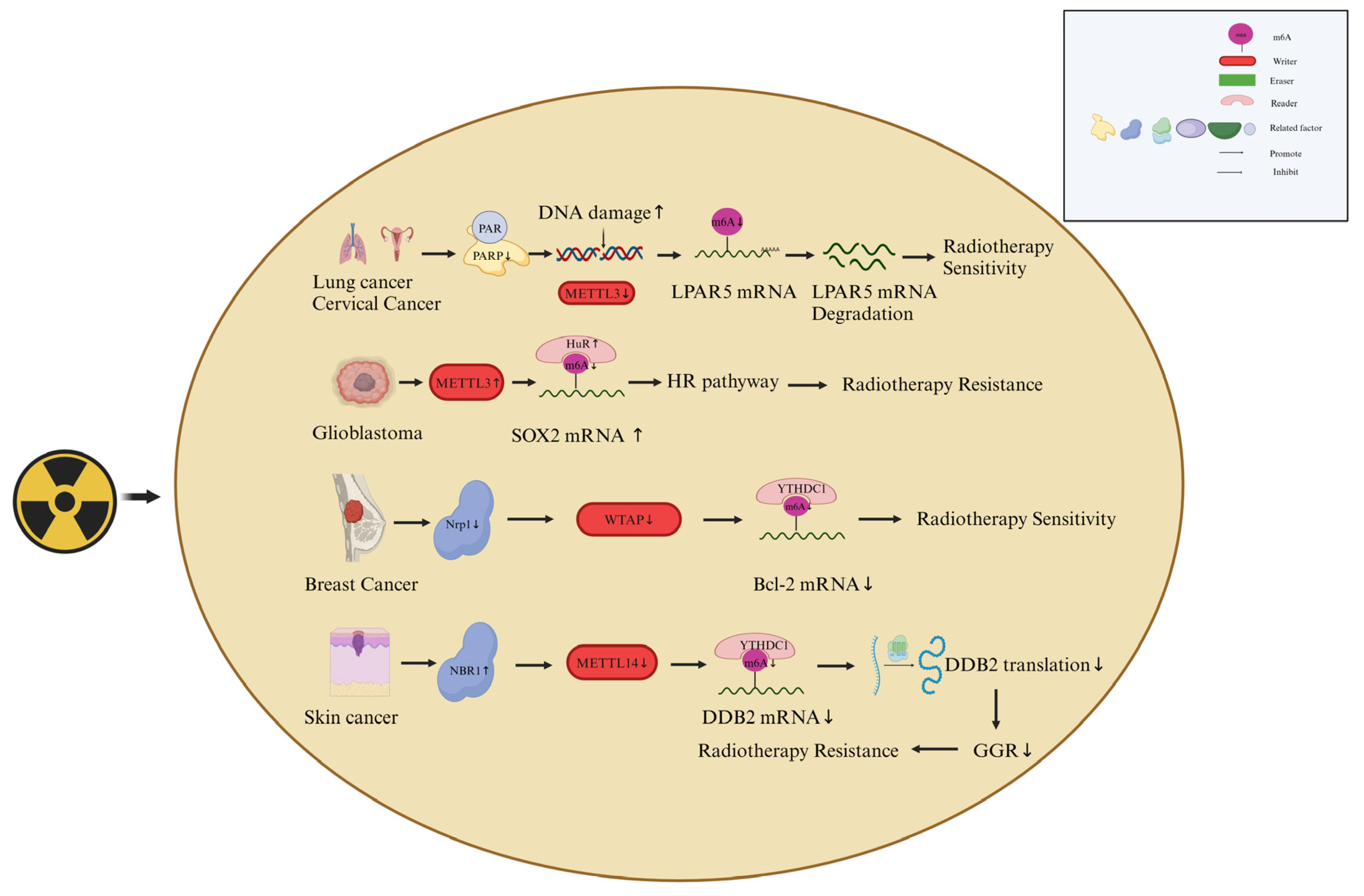
4.1. M6A Writers Are Involved in the Responses of Cancer Cells to Radiotherapy
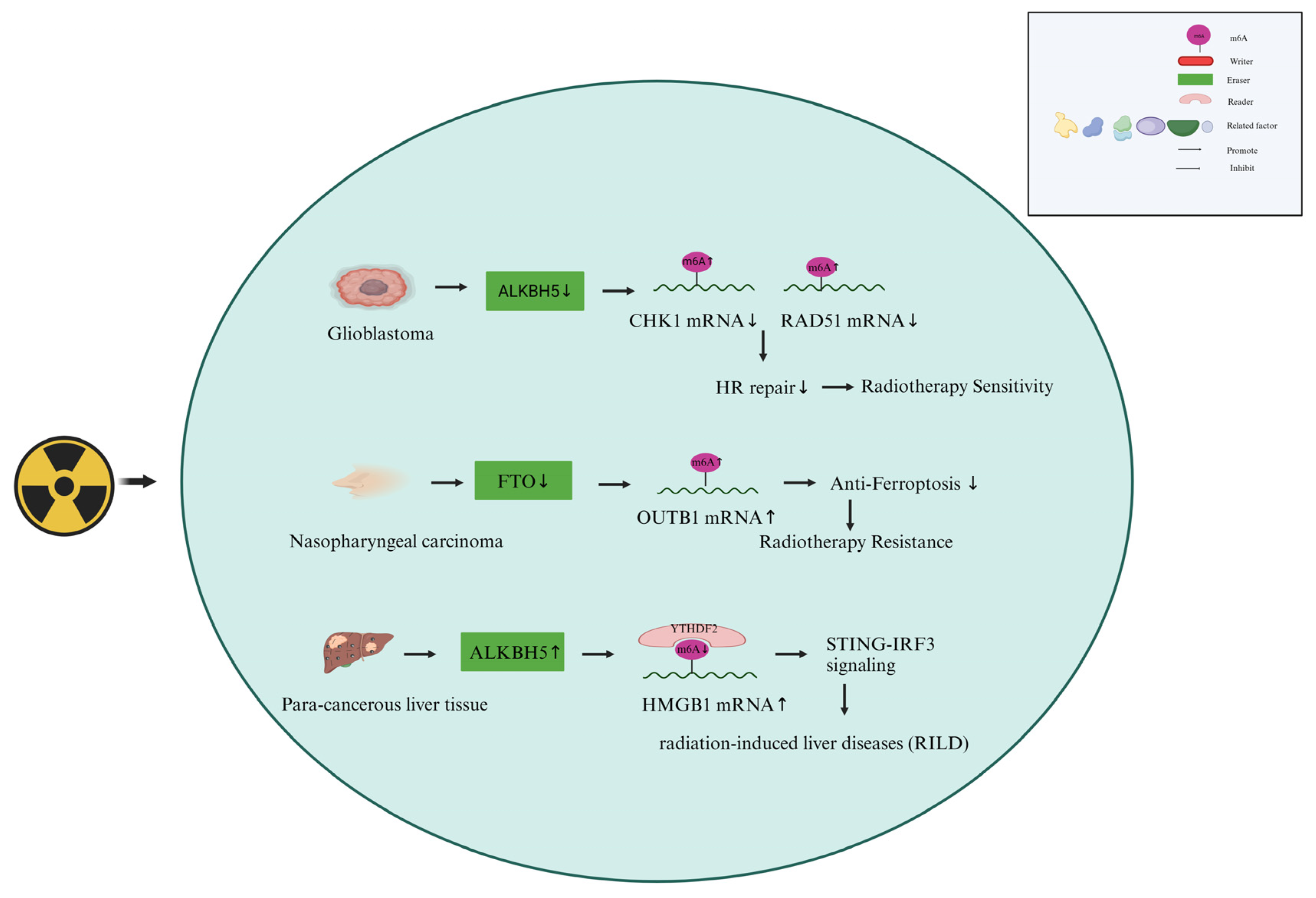
4.2. M6A Erasers Are Involved in the Responses of Cancer Cells to Radiotherapy
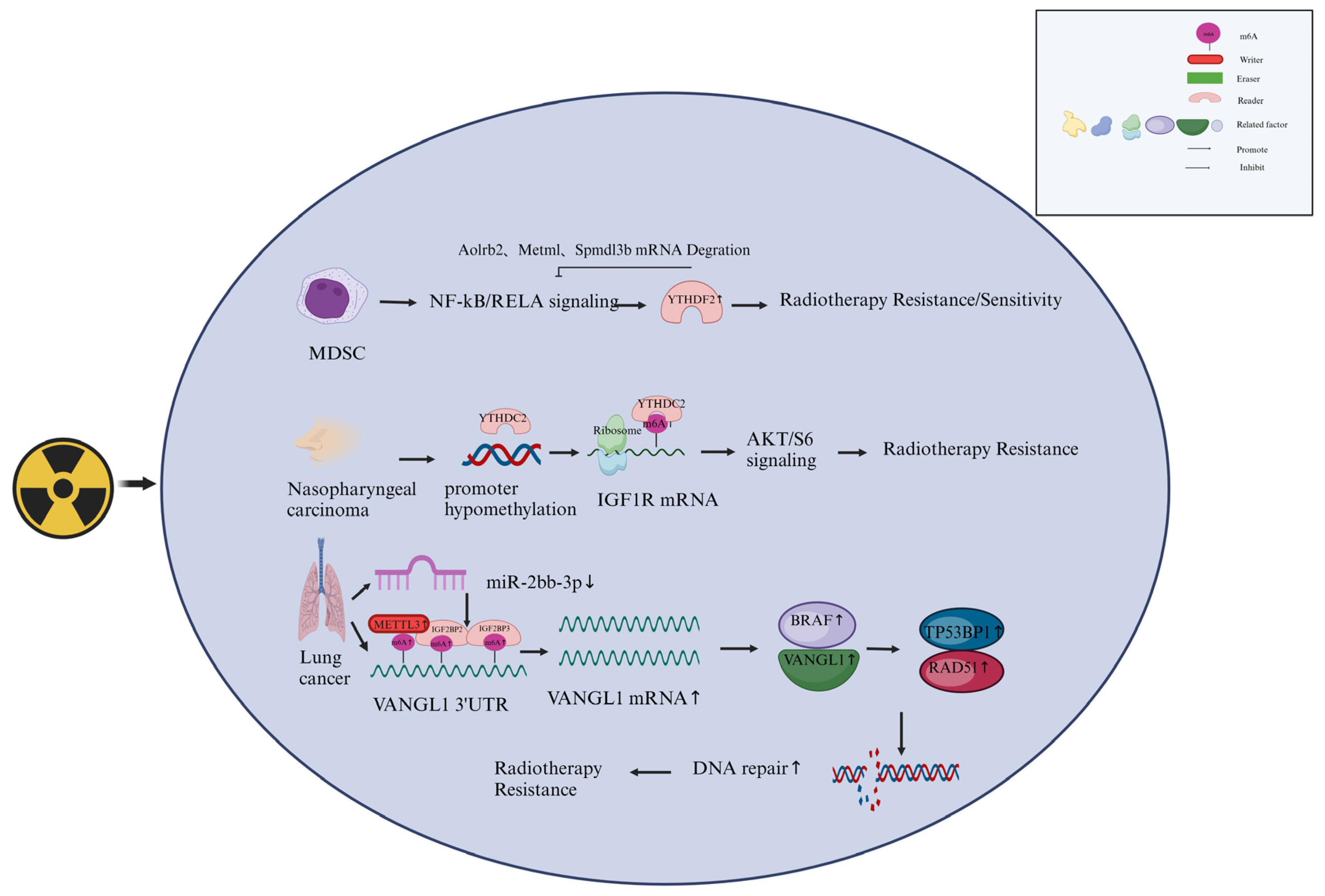
4.3. M6A Readers Involved in Cancer radiotherapy
5. Conclusion and Perspective
Author Contributions
Funding
References
- Ma, J.; Zhang, L.; Chen, S.; Liu, H. A Brief Review of Rna Modification Related Database Resources. Methods 2021, 203, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.; Liu, Y.; Zhao, Y.; Yao, Y.; Wu, R.; Liu, Q.; Wang, Y.; Wang, X. A Dynamic Reversible Rna N(6) -Methyladenosine Modification: Current Status and Perspectives. J Cell Physiol 2019, 234, 7948–7956. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Lin, W.-J.; Ren, M.; Qiu, J.; Yang, C.; Wang, X.; Li, N.; Zeng, T.; Sun, K.; You, L.; et al. M(6)a Reader Ythdc1 Modulates Autophagy by Targeting Sqstm1 in Diabetic Skin. Autophagy 2021, 18, 1318–1337. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine Modification of Lincrna 1281 Is Critically Required for Mesc Differentiation Potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xu, T.; Sun, K. N6-Methyladenosine Rna Modification in Inflammation: Roles, Mechanisms, and Applications. Front Cell Dev Biol 2021, 9, 670711. [Google Scholar] [CrossRef]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; F. Yu, G. Yuan, and J. Jia. Update on N6-Methyladenosine Methylation in Obesity-Related Diseases. Obesity (Silver Spring) (2023). [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-Methyladenosine and Its Role in Cancer. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Zhou, M.; Yu, L.; Si, Z. M6a Modified Bace1-as Contributes to Liver Metastasis and Stemness-Like Properties in Colorectal Cancer through Tuft1 Dependent Activation of Wnt Signaling. J. Exp. Clin. Cancer Res. 2023, 42, 1–20. [Google Scholar] [CrossRef]
- Zhu, Z.-M.; Huo, F.-C.; Pei, D.-S. Function and Evolution of Rna N6-Methyladenosine Modification. Int. J. Biol. Sci. 2020, 16, 1929–1940. [Google Scholar] [CrossRef]
- Śledź, P.; Jinek, M. Structural Insights into the Molecular Mechanism of the M(6)a Writer Complex. eLife 2016, 5. [Google Scholar] [CrossRef]
- Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Methyltransferase-Like Protein 16 Binds the 3'-Terminal Triple Helix of Malat1 Long Noncoding Rna. Proc. Natl. Acad. Sci. 2016, 113, 14013–14018. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; He, P.C.; Liu, W.; Wei, J.; Zhao, Z.; Gao, L.; Han, L.; et al. Mettl16 Exerts an M(6)a-Independent Function to Facilitate Translation and Tumorigenesis. Nature 2022, 24, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dai, X.-Y.; Qian, J.-Y.; Xu, F.; Wang, Z.-W.; Xia, T.; Zhou, X.-J.; Li, X.-X.; Shi, L.; Wei, J.-F.; et al. SMC1A regulated by KIAA1429 in m6A-independent manner promotes EMT progress in breast cancer. Mol. Ther. - Nucleic Acids 2022, 27, 133–146. [Google Scholar] [CrossRef]
- Zeng, X.; Chen, K.; Li, L.; Tian, J.; Ruan, W.; Hu, Z.; Peng, D.; Chen, Z. Epigenetic Activation of Rbm15 Promotes Clear Cell Renal Cell Carcinoma Growth, Metastasis and Macrophage Infiltration by Regulating the M6a Modification of Cxcl11. Free. Radic. Biol. Med. 2022, 184, 135–147. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Li, H.B.; Yin, Z.; Flavell, R.A. Recent Advances in Dynamic M6a Rna Modification. Open Biol 2016, 6, 160003. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-Y.; Lu, A.-Q. The Biological Function of M6a Reader Ythdf2 and Its Role in Human Disease. Cancer Cell Int. 2021, 21, 109. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-Methyladenosine Modulates Messenger Rna Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. Ythdc1 Mediates Nuclear Export of N(6)-Methyladenosine Methylated Mrnas. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; Wang, X.; Ma, H.L.; Huang, C.M.; Yang, Y.; Huang, N.; Jiang, G.B.; Wang, H.L.; Zhou, Q.; Wang, X.J.; Zhao, Y.L.; Yang, Y.G. Nuclear M(6)a Reader Ythdc1 Regulates Mrna Splicing. Mol Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Zeng, M.; Dai, X.; Liang, Z.; Sun, R.; Huang, S.; Luo, L.; Li, Z. Critical Roles of Mrna M(6)a Modification and Ythdc2 Expression for Meiotic Initiation and Progression in Female Germ Cells. Gene 2020, 753, 144810. [Google Scholar] [CrossRef]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.-W. Cancer and Radiation Therapy: Current Advances and Future Directions. Int. J. Med Sci. 2012, 9, 193–199. [Google Scholar] [CrossRef]
- Ladbury, C.; Eustace, N.; Amini, A.; Dandapani, S.; Williams, T. Biology-Guided Radiation Therapy: An Evolving Treatment Paradigm. Surg Oncol Clin N Am 2023, 32, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-X.; Zhou, P.-K. DNA Damage Response Signaling Pathways and Targets for Radiotherapy Sensitization in Cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, Y.; Hu, S.; Ren, G.; Cui, F.; Zhou, P.-K. Radiotherapy Exposure in Cancer Patients and Subsequent Risk of Stroke: A Systematic Review and Meta-Analysis. Front. Neurol. 2019, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-Y.; Li, H.-Z.; Xie, D.-F.; Gao, S.-S.; Huang, X.; Guan, H.; Bai, C.-J.; Zhou, P.-K. Lpar5 Confers Radioresistance to Cancer Cells Associated with Emt Activation Via the Erk/Snail Pathway. J. Transl. Med. 2022, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-T.; Li, J.-H.; Zhu, X.-X.; Huang, C.-S.; Gao, Z.-X.; Xu, Q.-C.; Zhao, W.; Yin, X.-Y. Hnrnpc Impedes M(6)a-Dependent Anti-Metastatic Alternative Splicing Events in Pancreatic Ductal Adenocarcinoma. Cancer Lett. 2021, 518, 196–206. [Google Scholar] [CrossRef]
- Lin, W.; Liu, Y.; Zhou, Y.; Lin, M.; Liu, C.; Tang, Y.; Wu, B.; Lin, C. Methyltransferase-Like 3 Modulates Visceral Hypersensitivity through Regulating the Nuclear Export of Circkcnk9 in Ythdc1-Dependent Manner. Mol. Pain 2022, 18. [Google Scholar] [CrossRef]
- Li, D.; Cai, L.; Meng, R.; Feng, Z.; Xu, Q. Mettl3 Modulates Osteoclast Differentiation and Function by Controlling Rna Stability and Nuclear Export. Int. J. Mol. Sci. 2020, 21, 1660. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. Ythdf2 Destabilizes M(6)a-Containing Rna through Direct Recruitment of the Ccr4-Not Deadenylase Complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Hou, G.; Zhao, X.; Li, L.; Yang, Q.; Liu, X.; Huang, C.; Lu, R.; Chen, R.; Wang, Y.; Jiang, B.; et al. Sumoylation of Ythdf2 Promotes Mrna Degradation and Cancer Progression by Increasing Its Binding Affinity with M6a-Modified Mrnas. Nucleic Acids Res. 2021, 49, 2859–2877. [Google Scholar] [CrossRef]
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J.; Si, Y.; et al. The M6a Reader Ythdf1 Promotes Ovarian Cancer Progression Via Augmenting Eif3c Translation. Nucleic Acids Res. 2020, 48, 3816–3831. [Google Scholar] [CrossRef]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. Mrna Circularization by Mettl3-Eif3h Enhances Translation and Promotes Oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of M(6)a-Containing Rnas by Rnase P/Mrp Complex. Mol. Cell 2019, 74, 494–507. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive Translation of Circular Rnas Driven by N(6)-Methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, Z.; Yan, Z.; Liang, X.; Liu, X.; Liu, Y.; Wang, P.; Bai, C.; Gu, Y.; Zhou, P.K. Mirna-155-5p Inhibits Epithelium-to-Mesenchymal Transition (Emt) by Targeting Gsk-3beta During Radiation-Induced Pulmonary Fibrosis. Arch Biochem Biophys 2021, 697, 108699. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Cao, Y.; Li, X.; Yu, F.; Han, X. E2f1 Regulates Mir-215-5p to Aggravate Paraquat-Induced Pulmonary Fibrosis Via Repressing Bmpr2 Expression. Toxicol. Res. 2022, 11, 940–950. [Google Scholar] [CrossRef]
- Wang, K.; Chen, Z.; Qiao, X.; Zheng, J. Lncrna Norad Regulates the Mechanism of the Mir-532-3p/Nectin-4 Axis in Pancreatic Cancer Cell Proliferation and Angiogenesis. Toxicol. Res. 2023, 12, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Das Mandal, S.; Ray, P.S. Transcriptome-Wide Analysis Reveals Spatial Correlation between N6-Methyladenosine and Binding Sites of Micrornas and Rna-Binding Proteins. Genomics 2020, 113, 205–216. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of Microrna Biogenesis and Its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-Methyladenosine Marks Primary Micrornas for Processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-Methyladenosine-Dependent Rna Structural Switches Regulate Rna–Protein Interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Zhou, K.I.; Parisien, M.; Dai, Q.; Liu, N.; Diatchenko, L.; Sachleben, J.R.; Pan, T. N6-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding. J. Mol. Biol. 2016, 428, 822–833. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, M.; He, X.; Cao, Y.; Liu, P.; Li, F.; Zou, S.; Wen, C.; Zhan, Q.; Xu, Z.; Wang, J.; Sun, B.; Shen, B. Lncrna-Pacerr Induces Pro-Tumour Macrophages Via Interacting with Mir-671-3p and M6a-Reader Igf2bp2 in Pancreatic Ductal Adenocarcinoma. J Hematol Oncol 2022, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-C.; Zhou, P.-K. Tissue Reactions and Mechanism in Cardiovascular Diseases Induced by Radiation. Int. J. Mol. Sci. 2022, 23, 14786. [Google Scholar] [CrossRef]
- McBride, W. H.; Schaue, D. Radiation-Induced Tissue Damage and Response. J Pathol 2020, 250, 647–655. [Google Scholar] [CrossRef]
- Nepon, H.; Safran, T.; Reece, E.M.; Murphy, A.M.; Vorstenbosch, J.; Davison, P.G. Radiation-Induced Tissue Damage: Clinical Consequences and Current Treatment Options. Semin. Plast. Surg. 2021, 35, 181–188. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. Rna M(6)a Methylation Regulates the Ultraviolet-Induced DNA Damage Response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, S.; Cui, Y.-H.; Wei, J.; Shah, P.; Park, G.; Cui, X.; He, C.; He, Y.-Y. Mettl14 Facilitates Global Genome Repair and Suppresses Skin Tumorigenesis. Proc. Natl. Acad. Sci. 2021, 118. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, L.; Peng, D.; Jiang, A.; He, Y.; Zeng, Y.; Xie, C.; Zhou, H.; Luo, X.; Liu, H.; et al. Mettl3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of Dsbs by Modulating DNA-Rna Hybrid Accumulation. Mol. Cell 2020, 79, 425–442. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, X.; Yang, W.; Zhang, Q.; Hao, R.; Jiang, S.; Han, H.; Yu, Z.; Xing, S.; Feng, C.; et al. Rna N6-Methyladenosine Modification-Based Biomarkers for Absorbed Ionizing Radiation Dose Estimation. Nat. Commun. 2023, 14, 1–16. [Google Scholar] [CrossRef]
- Jin, Z.; Dong, Z.; Zhao, X.; Hang, X.; Lu, Y.; Zhang, Q.; Chen, H.; Huang, Z.; Wang, Y.; Zhou, G.; et al. Sensitive, Rapid Detection of Ncoa4-M6a Towards Precisely Quantifying Radiation Dosage on a Cas13a-Microdroplet Platform. Biosens. Bioelectron. 2023, 242, 115753. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Bai, C.; Li, H.; Xie, D.; Chen, S.; Han, Y.; Luo, J.; Li, Y.; Ye, Y.; Jia, J.; et al. Parp1 Modulates Mettl3 Promoter Chromatin Accessibility and Associated Lpar5 Rna M(6)a Methylation to Control Cancer Cell Radiosensitivity. Mol. Ther. 2023, 31, 2633–2650. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential Role of Mettl3-Mediated M(6)a Modification in Glioma Stem-Like Cells Maintenance and Radioresistance. Oncogene 2017, 37, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The Epitranscriptome M6a Writer Mettl3 Promotes Chemo- and Radioresistance in Pancreatic Cancer Cells. Int. J. Oncol. 2017, 52, 621–629. [Google Scholar] [CrossRef]
- Shi, J.; Rui, X.; Han, C.; Wang, C.; Xu, L.; Jiang, X. Circrnf13, a Novel N(6)-Methyladenosine-Modified Circular Rna, Enhances Radioresistance in Cervical Cancer by Increasing Cxcl1 Mrna Stability. Cell Death Discov. 2023, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Cheon, N.Y.; Park, H.; Jeong, G.W.; Ye, B.J.; Yoo, E.J.; Lee, J.H.; Hur, J.-H.; Lee, E.-A.; Kim, H.; et al. Tonebp Recognizes R-Loops and Initiates M6a Rna Methylation for R-Loop Resolution. Nucleic Acids Res. 2020, 49, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Horton, J.R.; Yang, J.; Hajian, T.; Vedadi, M.; A Sagum, C.; Bedford, M.T.; Blumenthal, R.M.; Zhang, X.; Cheng, X. Human Mettl3-Mettl14 Rna Adenine Methyltransferase Complex Is Active on Double-Stranded DNA Containing Lesions. Nucleic Acids Res. 2021, 49, 11629–11642. [Google Scholar] [CrossRef]
- Svobodova Kovarikova, A.; Stixova, L.; Kovarik, A.; Komurkova, D.; Legartova, S.; Fagherazzi, P.; Bartova, E. N(6)-Adenosine Methylation in Rna and a Reduced M(3)G/Tmg Level in Non-Coding Rnas Appear at Microirradiation-Induced DNA Lesions. Cells 2020, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-M.; Li, Z.-X.; Wu, Y.-H.; Shi, Z.-L.; Mi, J.-L.; Hu, K.; Wang, R.-S. M6a Demethylase Fto Renders Radioresistance of Nasopharyngeal Carcinoma Via Promoting Otub1-Mediated Anti-Ferroptosis. Transl. Oncol. 2022, 27, 101576. [Google Scholar] [CrossRef]
- Hu, X.; Wu, J.; Feng, Y.; Ma, H.; Zhang, E.; Zhang, C.; Sun, Q.; Wang, T.; Ge, Y.; Zong, D.; Chen, W.; He, X. Mettl3-Stabilized Super Enhancers-Lncrna Suclg2-As1 Mediates the Formation of a Long-Range Chromatin Loop between Enhancers and Promoters of Sox2 in Metastasis and Radiosensitivity of Nasopharyngeal Carcinoma. Clin Transl Med 2023, 13, e1361. [Google Scholar] [CrossRef]
- Tatekawa, S.; Tamari, K.; Chijimatsu, R.; Konno, M.; Motooka, D.; Mitsufuji, S.; Akita, H.; Kobayashi, S.; Murakumo, Y.; Doki, Y.; et al. N(6)-Methyladenosine Methylation-Regulated Polo-Like Kinase 1 Cell Cycle Homeostasis as a Potential Target of Radiotherapy in Pancreatic Adenocarcinoma. Sci. Rep. 2022, 12, 1–20. [Google Scholar] [CrossRef]
- Wang, L.; Dou, X.; Chen, S.; Yu, X.; Huang, X.; Zhang, L.; Chen, Y.; Wang, J.; Yang, K.; Bugno, J.; et al. Ythdf2 Inhibition Potentiates Radiotherapy Antitumor Efficacy. Cancer Cell 2023, 41, 1294–1308. [Google Scholar] [CrossRef] [PubMed]
- Henry, E.; Souissi-Sahraoui, I.; Deynoux, M.; Lefèvre, A.; Barroca, V.; Campalans, A.; Ménard, V.; Calvo, J.; Pflumio, F.; Arcangeli, M.-L. Human Hematopoietic Stem/Progenitor Cells Display Reactive Oxygen Species-Dependent Long-Term Hematopoietic Defects after Exposure to Low Doses of Ionizing Radiations. Haematologica 2019, 105, 2044–2055. [Google Scholar] [CrossRef]
- Yu, F.; Wei, J.; Cui, X.; Yu, C.; Ni, W.; Bungert, J.; Wu, L.; He, C.; Qian, Z. Post-Translational Modification of Rna M6a Demethylase Alkbh5 Regulates Ros-Induced DNA Damage Response. Nucleic Acids Res. 2021, 49, 5779–5797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dong, J.; Li, Y.; Xiao, H.; Shang, Y.; Wang, B.; Chen, Z.; Zhang, M.; Fan, S.; Cui, M. Gamma-Irradiation Fluctuates the Mrna N(6)-Methyladenosine (M(6)a) Spectrum of Bone Marrow in Hematopoietic Injury. Environ. Pollut. 2021, 285, 117509. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Y.; Niu, H.; Zhao, X.; Chen, G.; Zhao, Q.; Ma, G.; Du, S.; Zeng, Z. Mettl3-Mediated Sting Upregulation and Activation in Kupffer Cells Contribute to Radiation-Induced Liver Disease Via Pyroptosis. Endocrine 2023. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhao, Q.; Yuan, B.; Wang, B.; Zhang, Y.; Li, Z.; Du, S.; Zeng, Z. Alkbh5-Modified Hmgb1-Sting Activation Contributes to Radiation Induced Liver Disease Via Innate Immune Response. Endocrine 2021, 111, 491–501. [Google Scholar] [CrossRef]
- Feng, Y.; Yuan, P.; Guo, H.; Gu, L.; Yang, Z.; Wang, J.; Zhu, W.; Zhang, Q.; Cao, J.; Wang, L.; Jiao, Y. Mettl3 Mediates Epithelial-Mesenchymal Transition by Modulating Foxo1 Mrna N(6) -Methyladenosine-Dependent Ythdf2 Binding: A Novel Mechanism of Radiation-Induced Lung Injury. Adv Sci (Weinh) 2023, 10, e2204784. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Han, D.-X.; Wang, C.-B.; Wang, X.-L. Zbtb7b Suppresses Aseptic Inflammation by Regulating M(6)a Modification of Il6 Mrna. Biochem. Biophys. Res. Commun. 2020, 530, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, Z.; Liu, Y.; Wu, H.; Zhang, Q.; Han, J.; Liu, J.; Zhang, C. Carbon Dots from Lycium Barbarum Attenuate Radiation-Induced Bone Injury by Inhibiting Senescence Via Mettl3/Clip3 in an M(6)a-Dependent Manner. ACS Appl Mater Interfaces 2023, 15, 20726–20741. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-D.; Sun, G.; Li, J.; Xu, J.; Wang, X. Mechanisms and Therapeutic Potentials of Cancer Immunotherapy in Combination with Radiotherapy and/or Chemotherapy. Cancer Lett. 2019, 452, 66–70. [Google Scholar] [CrossRef]
- Zhou, M.; He, J.; Li, Y.; Jiang, L.; Ran, J.; Wang, C.; Ju, C.; Du, D.; Xu, X.; Wang, X.; Li, H.; He, F.; Wen, H. N(6)-Methyladenosine Modification of Reg1alpha Facilitates Colorectal Cancer Progression Via Beta-Catenin/Myc/Ldha Axis Mediated Glycolytic Reprogramming. Cell Death Dis 2023, 14, 557. [Google Scholar] [CrossRef]
- Liu, R.; Yin, G.; Tuo, H.; Guo, Y.; Zhu, Y.; Zhang, L.; Yang, W.; Liu, Q.; Wang, Y. Mettl3-Induced Lncrna Gbap1 Promotes Hepatocellular Carcinoma Progression by Activating Bmp/Smad Pathway. Biol. Direct 2023, 18, 1–16. [Google Scholar] [CrossRef]
- Chen, M.; Tian, B.; Hu, G.; Guo, Y. Mettl3-Modulated Circuhrf2 Promotes Colorectal Cancer Stemness and Metastasis through Increasing Ddx27 Mrna Stability by Recruiting Igf2bp1. Cancers 2023, 15, 3148. [Google Scholar] [CrossRef]
- Zhu, C.; Wu, Q.; Xu, Y.; Ma, J.; Hu, Y.; Wang, J.; Gao, Z.; Da, M. Prognostic Significance of N6-Methyladenosine-Modified Related Chemotransferase Mettl3 in Gastric Carcinoma: Evidence from Meta-Analysis. Int. J. Biol. Markers 2023, 38, 185–193. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, L.; Qi, P.; Pang, R.; Wang, Z.; Liu, X.; Shi, Q.; Zhang, Q. Potential Role of Lpar5 Gene in Prognosis and Immunity of Thyroid Papillary Carcinoma and Pan-Cancer. Sci. Rep. 2023, 13, 1–14. [Google Scholar] [CrossRef]
- Sun, X.; Bai, C.; Li, H.; Xie, D.; Chen, S.; Han, Y.; Luo, J.; Li, Y.; Ye, Y.; Jia, J.; et al. Parp1 Modulates Mettl3 Promoter Chromatin Accessibility and Associated Lpar5 Rna M(6)a Methylation to Control Cancer Cell Radiosensitivity. Mol. Ther. 2023, 31, 2633–2650. [Google Scholar] [CrossRef] [PubMed]
- Kowalski-Chauvel, A.; Lacore, M.G.; Arnauduc, F.; Delmas, C.; Toulas, C.; Cohen-Jonathan-Moyal, E.; Seva, C. The M6a Rna Demethylase Alkbh5 Promotes Radioresistance and Invasion Capability of Glioma Stem Cells. Cancers 2020, 13, 40. [Google Scholar] [CrossRef]
- Wang, L.; Dou, X.; Chen, S.; Yu, X.; Huang, X.; Zhang, L.; Chen, Y.; Wang, J.; Yang, K.; Bugno, J.; et al. Ythdf2 Inhibition Potentiates Radiotherapy Antitumor Efficacy. Cancer Cell 2023, 41, 1294–1308. [Google Scholar] [CrossRef] [PubMed]
- He, J.-J.; Li, Z.; Rong, Z.-X.; Gao, J.; Mu, Y.; Guan, Y.-D.; Ren, X.-X.; Zi, Y.-Y.; Liu, L.-Y.; Fan, Q.; et al. M(6)a Reader Ythdc2 Promotes Radiotherapy Resistance of Nasopharyngeal Carcinoma Via Activating Igf1r/Akt/S6 Signaling Axis. Front. Oncol. 2020, 10, 1166. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.-C.; Xu, C.-Y.; Zhao, X.-Y.; Luo, J.-N.; Wang, G.; Zhao, L.-H.; Ge, X.-F. Up-Regulation of Vangl1 by Igf2bps and Mir-29b-3p Attenuates the Detrimental Effect of Irradiation on Lung Adenocarcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6a Rna Methylomes Revealed by M6a-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Luo, G.Z.; He, C. High-Resolution Mapping of N(6)-Methyladenosine in Transcriptome and Genome Using a Photo-Crosslinking-Assisted Strategy. Methods Enzymol 2015, 560, 161–185. [Google Scholar]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-Nucleotide-Resolution Mapping of M6a and M6am Throughout the Transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Molinie, B.; Wang, J.; Lim, K.S.; Hillebrand, R.; Lu, Z.-X.; Van Wittenberghe, N.; Howard, B.D.; Daneshvar, K.; Mullen, A.C.; Dedon, P.; et al. M(6)a-Laic-Seq Reveals the Census and Complexity of the M(6)a Epitranscriptome. Nat. Methods 2016, 13, 692–698. [Google Scholar] [CrossRef]
- McIntyre, A.B.R.; Gokhale, N.S.; Cerchietti, L.; Jaffrey, S.R.; Horner, S.M.; Mason, C.E. Limits in the Detection of M(6)a Changes Using Merip/M(6)a-Seq. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for Snornas and Snorna-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding Rna as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Marusic, M.; Toplishek, M.; Plavec, J. Nmr of Rna - Structure and Interactions. Curr Opin Struct Biol 2023, 79, 102532. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



