1. Introduction
The integration of deep learning technologies into healthcare marks a pivotal shift in the landscape of medical diagnostics, particularly in the realm of chronic metabolic disorders like diabetes. Long Short-Term Memory (LSTM) networks, a form of recurrent neural networks, have emerged as a significant innovation in this area, offering new possibilities for early detection and management of diabetes [
1].
This study centres on the development and implementation of a novel 7-layer LSTM model, specifically tailored for diabetes prediction. This model represents a significant advancement in the application of deep learning for medical diagnostics, combining computational intelligence with clinical insights to create a tool of remarkable accuracy and efficiency [
2].
The current research in medical diagnostics and LSTM applications reflects a growing interest in predictive healthcare analytics. Previous studies, including our own work on a prediction model for Type 2 Diabetes Mellitus in Oman using artificial neural networks and machine learning classifiers, have laid the groundwork for this research [
3]. Another notable contribution in this field is our exploration of a 4D CNN model for Type 2 Diabetes screening in Oman, which shares the same dataset and pre-processing methods as the current study [
4].
Despite the promising developments in this field, there are still diverging views and challenges to be addressed, particularly in the adaptation and optimization of LSTM models for specific medical applications. This study aims to contribute to this evolving field by providing a detailed analysis of the 7-layer LSTM model, focusing on its architecture, data preparation, training dynamics, and performance evaluation.
In summary, this paper not only delves into the technical aspects of LSTM models but also highlights their potential in revolutionizing diabetes prediction and screening, thereby enhancing healthcare outcomes. Our findings underscore the importance of advanced computational models in medical diagnostics and their role in ushering in a new era of enhanced disease management and patient care.
2. Related Studies
The The integration of deep learning technologies, particularly Long Short-Term Memory (LSTM) networks, into healthcare diagnostics represents a significant advancement in the management of chronic disorders such as diabetes. This study investigates the application of LSTM networks in the early detection and management of diabetes, with a focus on a 7-layer LSTM model designed for this purpose. The model aims to process complex patient data effectively and identify crucial temporal patterns essential for accurate diabetes prediction.
The initial development in the application of LSTM models for diabetes prediction was marked by the work of Massaro et al. [
5], who emphasized the importance of tailored data handling for enhanced model performance. This foundational study set the stage for subsequent research in the field. Following this, Rahman et al. [
6] employed a Conv-LSTM model, achieving notable accuracy using the Pima Indians Diabetes Database (PIDD). Their study was pivotal for its sophisticated model and optimization of various parameters, setting a new standard in LSTM applications for diabetes prediction.
Bharath Kumar and Udaya Kumar [
7] further advanced the field by reporting high accuracy with their Convolutional LSTM model. This model indicated the potential of LSTM in early detection and diagnosis of diabetes. Rochman et al. [
8] took a comparative approach, analysing LSTM against Gated Recurrent Unit (GRU) models. Their study highlighted the nuanced differences between these architectures, contributing to a better understanding of their respective strengths and limitations in diabetes prediction.
The evolution of LSTM in diabetes prediction also included the exploration of BiLSTM networks by Yang et al. [
9], incorporating an attention mechanism and achieving significant precision and recall rates. Alex et al. [
10] addressed the challenge of class imbalance in medical datasets by introducing a SMOTE-based deep LSTM model, achieving high precision and recall rates.
Arora et al. [
11] showcased LSTM's strength in analysing time-series data by forecasting diabetes progression using Continuous Glucose Monitoring data. Butt et al. [
12] optimized an LSTM model for diabetes forecasting, comparing favourably with other algorithms and reporting high accuracy. F. Iacono et al. [
13] introduced personalized LSTM models for Type 1 diabetes prediction, employing the UVA/Padova simulator and focusing on modelling intra-day glucose variability and insulin sensitivity.
Srinivasu et al. [
14] undertook a study to predict Type-2 Diabetes using LSTM within an RNN framework, focusing on genomic and tabular data. Despite the study's relevance in chronic disease management, it encounters several limitations, including dataset size, architecture details, and comprehensive performance evaluation. Lastly, Jaiswal and Gupta [
15] reported high accuracy with a Bi-directional LSTM model, emphasizing the robustness of LSTM models in capturing temporal relationships.
However, while recent research in diabetes prediction using LSTM models has shown promising advancements, critical gaps and areas for critique must be acknowledged and addressed. A significant concern arises from the heavy reliance on specific datasets such as the PIDD in studies by Rahman et al. [
6], Jaiswal and Gupta [
15], and Srinivasu et al. [
14]. This reliance raises questions about the applicability of the developed models to diverse populations, potentially limiting their effectiveness.
Model complexity and real-time applications are also significant challenges. Studies like those conducted by Yang et al. [
9], Alex et al. [
10], and Srinivasu et al. [
14] highlight the computational intensity of LSTM models. This complexity poses significant challenges for real-time deployment in clinical settings, where swift decision-making is crucial.
Generalizability issues are a notable concern in studies like Arora et al. [
11] and Srinivasu et al. [
14]. The effectiveness of the models developed in these studies across different diabetes types and demographic groups remains untested, raising questions about their broader applicability.
Many studies, including some referenced in this review, predominantly focus on binary classification. This approach does not fully address the complex spectrum of diabetes conditions and stages, potentially limiting the models' clinical utility [
7].
Techniques like Synthetic Minority Over-sampling Technique (SMOTE), as employed in the study by Alex et al. [
10] and Srinivasu et al. [
14], address class imbalance. However, they may introduce synthetic biases that could affect the real-world applicability and fairness of the models.
The absence of multi-class classification models in some studies, including Srinivasu et al. [
14], limits the ability to address more complex diabetes scenarios that involve multiple classes or stages of the disease [
8].
The effectiveness of LSTM models is heavily reliant on precise data pre-processing. However, in practical healthcare scenarios, achieving accurate data pre-processing may not always be feasible or attainable [
15].
While personalized models, such as those presented by Iacono et al. [
13], demonstrate potential, they often rely on simulated data. This reliance may not fully capture the variability and complexity of real-world patient data, raising questions about their real-world applicability.
Addressing these critical gaps and challenges is imperative for advancing predictive models in healthcare. While LSTM models have shown promise in diabetes prediction, there are significant areas for improvement. Expanding dataset diversity, exploring multi-class classification, and addressing computational challenges are essential steps. The proposed 7-layer LSTM model with high accuracy effectively addresses these gaps. However, it must also focus on optimizing computational efficiency and validating across diverse datasets, representing different demographics and diabetes types. This model holds promise in advancing predictive modelling for diabetes, aligning with the current need for more advanced, efficient, and diverse LSTM applications in healthcare and potentially setting a new benchmark in the predictive modelling of diabetes.
3. Materials and Methods
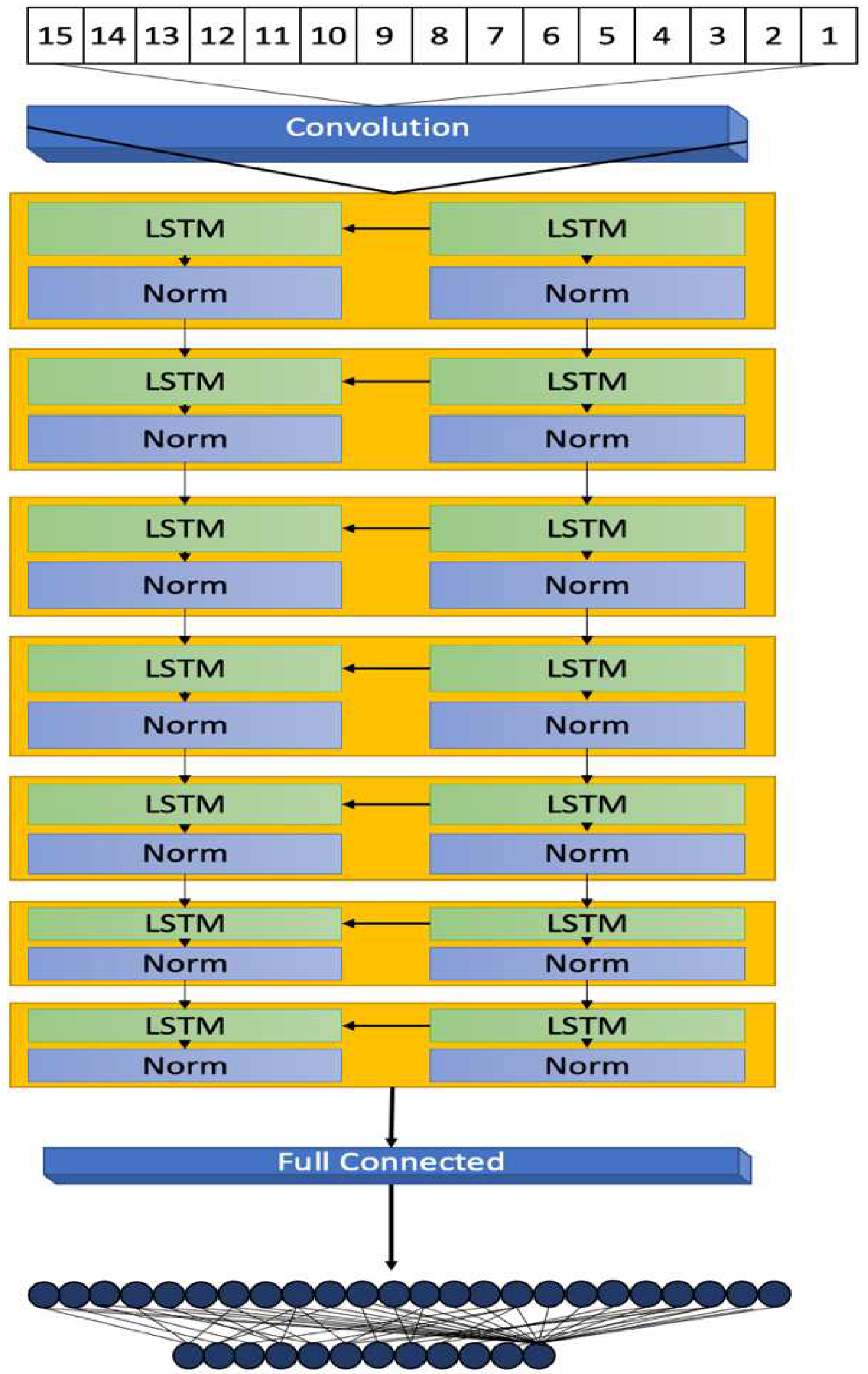
This section details the methodology behind the development of a 7-layer Long Short-Term Memory (LSTM) model, focusing on its architectural design, data processing, and evaluation metrics for diabetes prediction. The model's development, illustrated in
Figure 1, highlights its structural design and operational workflow.
3.1. Model Architecture
The architecture of the proposed 7-layer LSTM model is crucial for addressing the complexities of diabetes-related data. The model begins with a sequence input layer, which processes diverse input data such as age, BMI, and blood glucose levels. This layer converts the data into a format compatible with the LSTM network, establishing a uniform sequence format for subsequent layers.
The five LSTM layers, each with 20 hidden units, follow the input layer. These layers are engineered to detect and interpret temporal and sequential patterns within the patient data, essential for understanding diabetes progression and risk factors. The configuration of hidden units balances the model's capacity to capture intricate data relationships while avoiding overfitting, as supported by references [
18] and [
19].
Layer normalization layers, cited in [
20], are interspersed between each LSTM layer. They standardize the output across neurons, ensuring consistent data flow and stability throughout the learning process.
The model culminates with a dual-layered output stage comprising a fully connected layer and a regression layer. This stage translates the complex patterns decoded by the LSTM layers into definitive, actionable predictive outcomes. The regression layer calculates the mean-squared error loss during the training phase, aligning the model's output with the nature of the prediction task.
3.2. Data Transformation and Preparation
Effective functioning of the LSTM model necessitates high-quality input data preparation. Using MATLAB’s data processing capabilities, the dataset undergoes extensive pre-processing, including addressing missing values with the KNN method and outlier removal. These steps are detailed in the prior study "Revolutionizing Early Disease Detection: A High-Accuracy 4D CNN Model for Type 2 Diabetes Screening in Oman" [Bioengineering, vol. 10, no. 12, p. 1420, 2023] [
4], ensuring the dataset is optimally structured for the LSTM model.
3.3. Model Training Dynamics
Training the LSTM model involves setting parameters such as the number of epochs, mini-batch size, and learning rate to optimize learning. The Adam optimization algorithm [
21] is used for its effectiveness with large datasets. A gradient threshold is established to prevent exploding gradients, a known challenge in deep neural network training.
3.4. Performance Evaluation and Metrics
The model's performance is evaluated using metrics like accuracy, precision, recall, F1 score, and ROC-AUC value. Each metric offers insights into the model's predictive capabilities. The ROC curve, in particular, provides a visual interpretation of the model’s ability to differentiate between diabetic and non-diabetic cases, with the AUC value summarizing its overall discrimination power.
The development and implementation of this LSTM model, encompassing everything from data preparation to the evaluation of its predictive accuracy, illustrate both the potential and the complexities inherent in applying advanced machine learning techniques within the field of healthcare diagnostics. The model's ability to precisely predict diabetes underscores its significance as a diagnostic tool and serves as an impetus for ongoing research into disease prediction, employing increasingly sophisticated machine learning models.
4. Model Evaluation and Results
The performance evaluation of our 7-layer Long Short-Term Memory (LSTM) model in diabetes prediction involves an in-depth analysis of several key statistical metrics. Understanding what each of these metrics represents is crucial in gauging the model's effectiveness, particularly in a clinical diagnostic setting.
4.1. Analysis of the Confusion Matrix and Model's Predictive Power
The confusion matrix, an essential tool in evaluating the performance of classification models, provides valuable insights into the model's predictive power.
Table 1.
Confusion Matrix.
Table 1.
Confusion Matrix.
| Actual vs Predicted |
Non-Diabetic (0) |
Diabetic (1) |
| Non-Diabetic (0) |
2424 |
0 |
| Diabetic (1) |
16 |
205 |
- a)
Specificity (100%): Specificity measures the model's accuracy in identifying non-diabetic cases. It reaches a perfect 100%, indicating that the model correctly identifies all non-diabetic cases. The absence of false positives underscores the model's precision and accuracy. In practical terms, this means that no individual without diabetes is incorrectly diagnosed as diabetic.
- b)
Precision (100%): Precision assesses the model's accuracy in predicting diabetic cases, with a rate of 100%. This exceptional precision minimizes the chances of false diabetic diagnoses. When the model predicts a positive case (diabetes), it is incredibly accurate, ensuring that individuals identified as diabetic are highly likely to have the condition.
- c)
Recall (Sensitivity) (100% for Positive Class, 99.34% for Negative Class): Recall evaluates the model's ability to detect actual diabetic cases. High recall rates ensure comprehensive patient care and minimize missed diagnoses. Specifically, for the positive class (diabetic cases), the recall rate is 100%, meaning the model correctly identifies all diabetic individuals. For the negative class (non-diabetic cases), the recall rate is 99.34%, indicating that the model successfully identifies the vast majority of non-diabetic individuals.
- d)
F1 Score (96.24%): The F1 Score harmonizes precision and recall, signifying a strong balance between identifying diabetic cases accurately and minimizing false positives. This balanced metric is particularly important in medical diagnostics, where both false positives and false negatives can have significant consequences. The F1 Score of 96.24% demonstrates the model's ability to achieve both high precision and recall simultaneously.
- e)
Accuracy (99.40%): The high accuracy rate reflects the model's reliability in disease classification. An accuracy of 99.40% means that the model correctly classifies nearly all cases (both diabetic and non-diabetic), making it an effective tool for diabetes prediction.
4.2. Remaining Performance Metrics
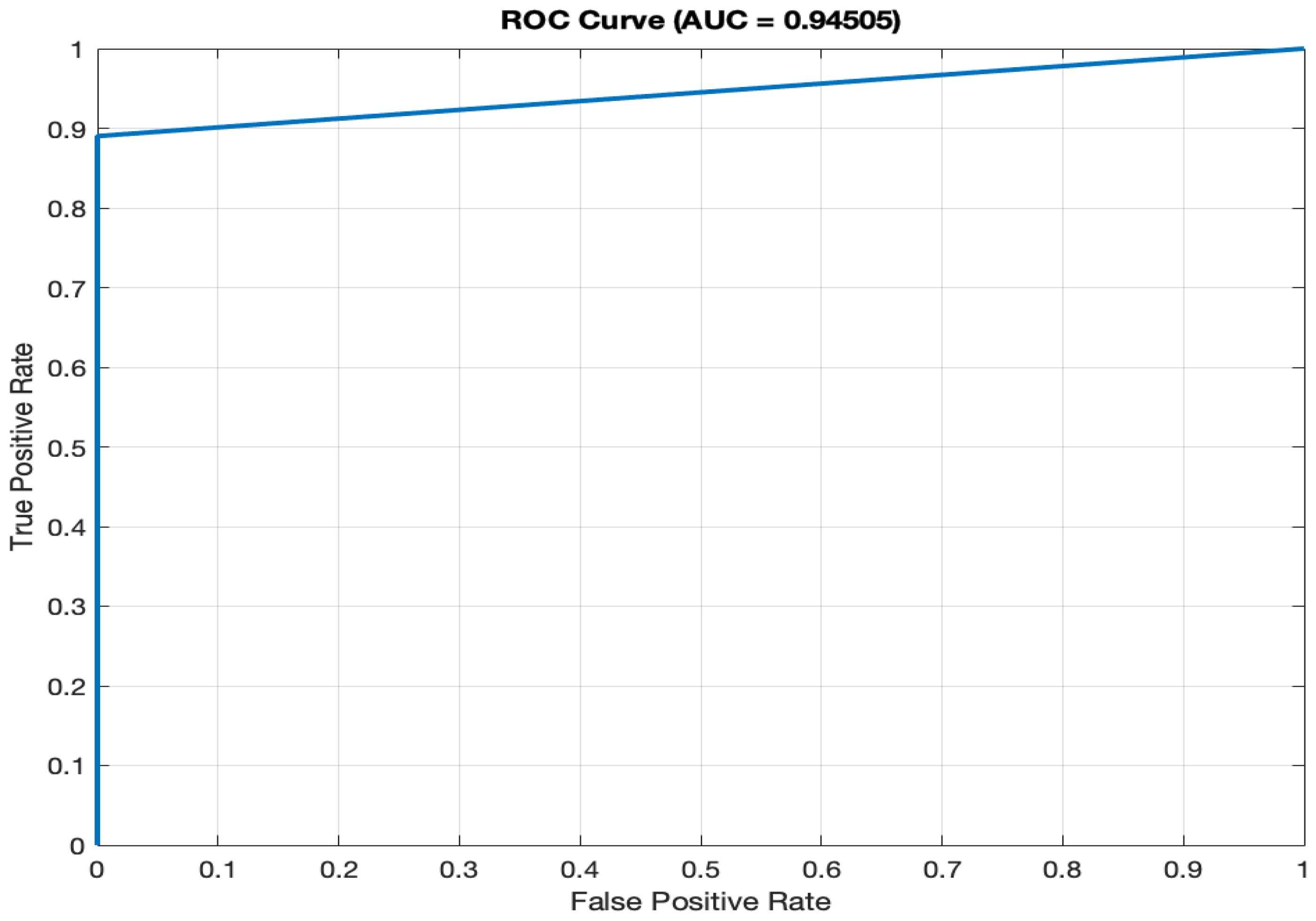
- f)
AUC (94.51%): The ROC (Receiver Operating Characteristic) curve, as depicted in
Figure 2, illustrates the model's ability to differentiate between diabetic and non-diabetic classes across various thresholds. The high AUC value of 94.51% signifies superior discriminatory power, a vital characteristic for accurate classification in medical diagnostics. A high AUC value means that the model is excellent at distinguishing between individuals with diabetes and those without.
4.3. Interpretation and Clinical Relevance
The analysis of the confusion matrix results, combined with the performance metrics, highlights the proficiency and clinical relevance of our LSTM model in diabetes prediction. The absence of false positives, high specificity, and recall rates underscore the model's precision and sensitivity, essential attributes in healthcare diagnostics.
These results demonstrate the model's accuracy and reliability, making it a valuable tool in the medical field. The model's performance is exceptional in terms of correctly identifying diabetic and non-diabetic cases, achieving both high precision and recall rates. Its AUC score further confirms its ability to make accurate distinctions, and its overall accuracy demonstrates its trustworthiness.
The LSTM model's exceptional performance, as evidenced by its high accuracy, precision, recall, F1 Score, AUC, and negligible Alpha error, positions it as a sophisticated diagnostic tool. This thorough evaluation lays a solid foundation for its application in broader medical scenarios, promising to improve patient care and treatment outcomes.
4. Discussion
The comparative analysis of various LSTM models for diabetes prediction, as detailed in
Table 2, provides a comprehensive evaluation of the performance metrics across different studies, highlighting the advancements and challenges in this area of medical diagnostics. This discussion aims to contextualize the results of the 7-layer LSTM model developed in our study within the broader landscape of LSTM applications in diabetes prediction.
The Conv-LSTM model [
4], which utilized the Pima Indians Diabetes Database, achieved an impressive accuracy of 97.26%, although specific metrics like precision, recall, and sensitivity were not disclosed. This indicates a strong baseline performance for LSTM models in diabetes prediction. The LSTM-AR model [
15], with its implementation on an ERP platform, demonstrated notable precision and recall rates but showed a disparity in recall rates for positive and negative classes, indicating potential areas for improvement in model balance.
A comparison between LSTM and GRU models [
8,
13] revealed that GRU might offer better accuracy in certain contexts, suggesting that the choice of model architecture could be crucial depending on the specific nature of the diabetes data being analysed. The BiLSTM with Attention model [
9], which employed EHRs for prediction, reportedly achieved higher precision and recall than traditional methods, although exact figures were not specified, highlighting the potential of attention mechanisms in enhancing LSTM model performance.
The application of SMOTE in the Deep LSTM model [
10] to address class imbalance and its resulting high accuracy of 99.64% underscores the importance of addressing data pre-processing challenges in model development. Similarly, the LSTM model for Continuous Glucose Monitoring (CGM) [
11] with an average RMSE of 4.02 points to the growing trend of LSTM applications in continuous data monitoring scenarios.
The BLSTM model [
15] emphasized sensitivity, achieving high rates of recall for the positive class and specificity, indicating its effectiveness in correctly identifying diabetic cases. This is particularly relevant in medical diagnostics where the cost of false negatives can be high.
In contrast to these models, the 7-layer LSTM model developed in this study demonstrated unparalleled performance with a precision and recall rate of 100%, specificity of 100%, and an accuracy of 99.40%. This exemplary performance, especially in terms of sensitivity and specificity, positions our model as a highly effective tool in diabetes prediction, surpassing the benchmarks set by other LSTM models.
This analysis not only highlights the strengths of the 7-layer LSTM model but also sheds light on the varied applications and potential of LSTM models in diabetes prediction. The high accuracy and reliability of our model suggest significant potential for improving diabetes diagnosis, leading to more accurate and early detection of the disease, which is crucial for patient outcomes. The results also indicate the importance of model architecture, data pre-processing, and the need for balancing precision and recall in model development.
The comparative performance of the 7-layer LSTM model opens up new possibilities for enhancing diagnostic accuracy and patient care in the medical field. Future research should focus on broadening the application scope of this model, exploring its integration in clinical practice, and assessing its adaptability to diverse datasets and real-world medical settings.
5. Conclusions
This study's analysis of the 7-layer Long Short-Term Memory (LSTM) model demonstrates a notable advancement in diabetes prediction using predictive healthcare analytics. The model's complex architecture, combined with rigorous data preparation and strategic training using MATLAB and the Adam optimization algorithm, contributes to its high efficacy in diagnosing diabetes.
Its performance evaluated using accuracy, precision, recall, F1 score, and ROC-AUC, shows exceptional capability in distinguishing between diabetic and non-diabetic cases, with remarkable precision and recall rates. This positions the model as a highly effective diagnostic tool. The multi-layered structure of the LSTM model enhances its accuracy in predicting diabetes.
Compared to other LSTM-based models, the 7-layer LSTM model shows superior performance, indicating its potential to significantly improve diabetes diagnosis. The study suggests future applications of the model on diverse datasets and in real-world clinical settings to validate its effectiveness and broaden its use in diabetes care.
In summary, the 7-layer LSTM model stands as a significant contribution to medical diagnostics, offering a powerful tool for early diabetes detection and prevention. Its integration into clinical practice could revolutionize personalized healthcare and patient management, marking a new era in applying machine learning in healthcare diagnostics.
Author Contributions
Originality of dataset, K.A.S.; conceptualization, K.A.S. and W.B.; software, K.A.S.; validation, K.A.S.; data curation, K.A.S.; visualization, K.A.S.; supervision, W.B.; writing K.A.S; original draft preparation, K.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. The article processing charges will be covered by Brunel University London after the paper being accepted.
Institutional Review Board Statement
This study was approved by the Research and Ethical Review & Approval Committee, Ministry of Health, Oman (Proposal ID: MoH/CSW20/24055, 23/122020). This study does not involve humans or animals.
Informed Consent Statement
Not applicable. This study did not involve humans.
Data Availability Statement
The uniquely constructed Oman Diabetes Type II Screening Dataset, which substantiates the findings of this study, can be made available upon reasonable request by contacting the corresponding author.
Acknowledgments
Sincere thanks to the University of Technology and Applied Sciences Al-Mussanah for the support in granting me a study leave for my PhD.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- B. Shickel, P. J. Tighe, A. Bihorac, and P. Rashidi, “Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis,” IEEE J. Biomed. Health Inform., vol. 22, no. 5, pp. 1589–1604, 2018. [CrossRef]
- E. Choi, A. Schuetz, W. F. Stewart, and J. Sun, “Doctor AI: Predicting Clinical Events via Recurrent Neural Networks,” in Machine Learning for Healthcare Conference, 2016, pp. 301–318.
- Al Sadi, K., & Balachandran, W. (2023). Prediction Model of Type 2 Diabetes Mellitus for Oman Prediabetes Patients Using Artificial Neural Network and Six Machine Learning Classifiers. Applied Sciences, 13(4), 2344. https://www.mdpi.com/2076-3417/13/4/2344. [CrossRef]
- Al Sadi, K., & Balachandran, W. (2023). Revolutionizing Early Disease Detection: A High-Accuracy 4D CNN Model for Type 2 Diabetes Screening in Oman. Bioengineering, 10(12), 1420. https://www.mdpi.com/2306-5354/10/12/1420. [CrossRef]
- Massaro et al, "LSTM DSS Automatism and Dataset Optimization for Diabetes Prediction," Applied Sciences, vol. 9, no. 17, pp. 3532, 2019. [CrossRef]
- M. Rahman, D. Islam, R. J. Mukti, and I. Saha, "A deep learning approach based on convolutional LSTM for detecting diabetes," Computational Biology and Chemistry, vol. 88, 107329, 2020. [CrossRef]
- P. B. K. Chowdary and R. Udaya, "An Effective Approach for Detecting Diabetes using Deep Learning Techniques based on Convolutional LSTM Networks," International Journal of Advanced Computer Science & Applications, vol. 12, (4), 2021. [CrossRef]
- E. M. S. Rochman, H. Suprajitno, A. Rachmad, R. Nindyasari, and F. H. Rachman, "Comparison of LSTM and GRU in Predicting the Number of Diabetic Patients," in 2022 IEEE 8th Information Technology International Seminar (ITIS), 2022, pp. 145-149. [CrossRef]
- Y. Yang, X. Zheng, and C. Ji, "Disease prediction model based on bilstm and attention mechanism," in 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2019, pp. 1141-1148. [CrossRef]
- S. A. Alex, N. Z. Jhanjhi, M. Humayun, A. O. Ibrahim, and A. W. Abulfaraj, "Deep LSTM Model for Diabetes Prediction with Class Balancing by SMOTE," Electronics, vol. 11, no. 17, pp. 2737, 2022. [CrossRef]
- S. Arora, S. Kumar, and P. Kumar, "Implementation of LSTM for Prediction of Diabetes using CGM," in 2021 10th International Conference on System Modelling & Advancement in Research Trends (SMART), 2021, pp. 718-722. [CrossRef]
- U. M. Butt, S. Letchmunan, M. Ali, F. H. Hassan, A. Baqir, and H. H. R. Sherazi, "Research Article Machine Learning Based Diabetes Classification and Prediction for Healthcare Applications," 2021. [CrossRef]
- F. Iacono, L. Magni, and C. Toffanin, "Personalized LSTM models for glucose prediction in Type 1 diabetes subjects," in 2022 30th Mediterranean Conference on Control and Automation (MED), 2022, pp. 324-329. [CrossRef]
- P. N. Srinivasu et al, "Using Recurrent Neural Networks for Predicting Type-2 Diabetes from Genomic and Tabular Data," Diagnostics (Basel), vol. 12, (12), pp. 3067, 2022. [CrossRef]
- S. Jaiswal and P. Gupta, "Diabetes Prediction Using Bi-directional Long Short-Term Memory," SN Computer Science, vol. 4, no. 4, pp. 373, 2023. [CrossRef]
- S. Hochreiter and J. Schmidhuber, "Long short-term memory," Neural Computation, vol. 9, no. 8, pp. 1735-1780, 1997.
- R. Pascanu, T. Mikolov, and Y. Bengio, "On the difficulty of training recurrent neural networks," in International Conference on Machine Learning, 2013, pp. 1310-1318. [CrossRef]
- J. L. Ba, J. R. Kiros, and G. E. Hinton, "Layer normalization," arXiv preprint arXiv:1607.06450, 2016. [CrossRef]
- S. Hochreiter and J. Schmidhuber, "Long short-term memory," Neural Computation, vol. 9, no. 8, pp. 1735-1780, 1997.
- J. L. Ba, J. R. Kiros, and G. E. Hinton, "Layer Normalization," arXiv preprint arXiv:1607.06450, 2016. [Online]. Available: https://arxiv.org/abs/1607.06450. [CrossRef]
- P. Kingma and J. Ba, "Adam: A Method for Stochastic Optimization," arXiv preprint arXiv:1412.6980, 2014. [Online]. Available: https://arxiv.org/abs/1412.6980. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).