Submitted:
18 January 2024
Posted:
18 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. General methods
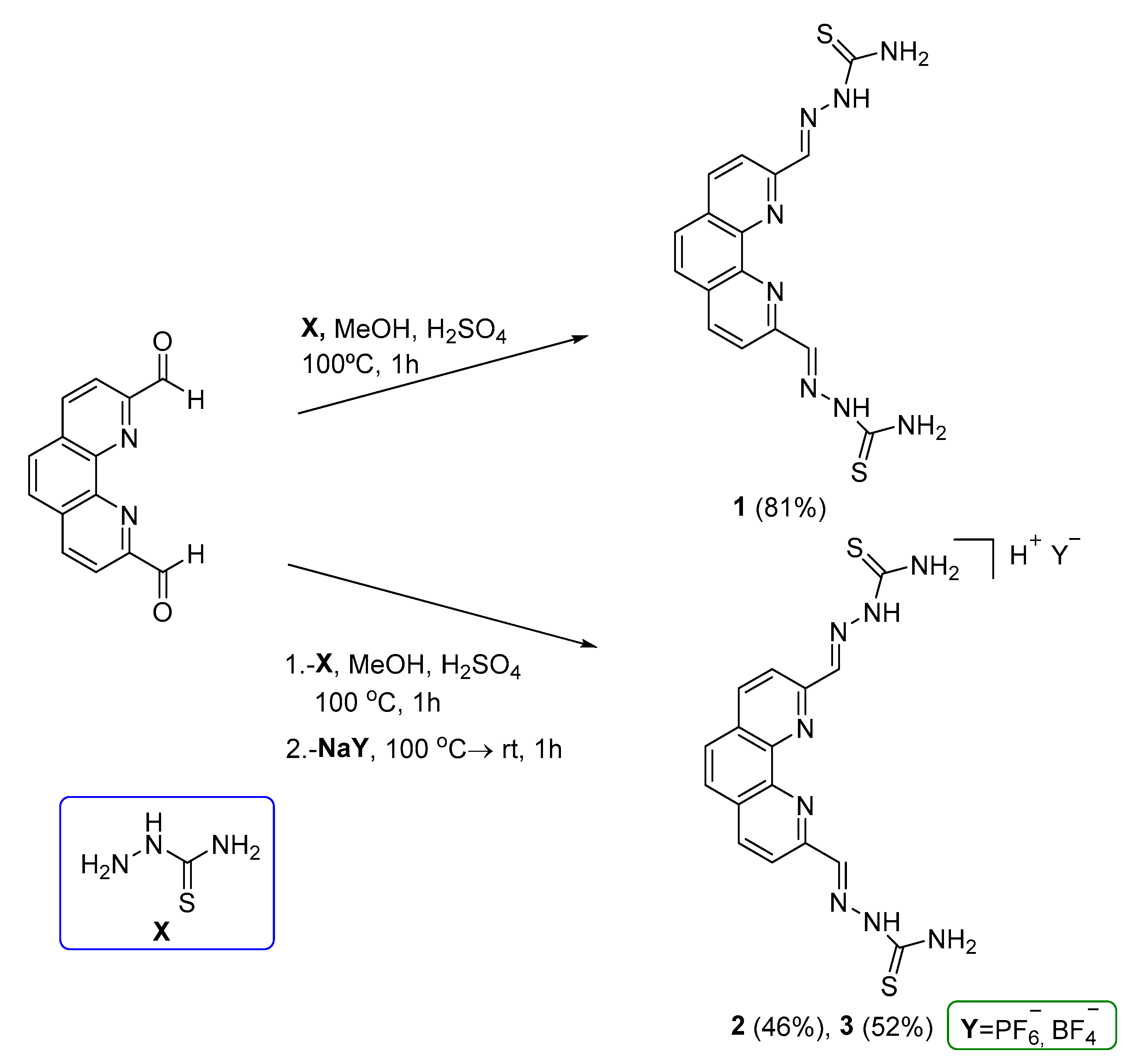
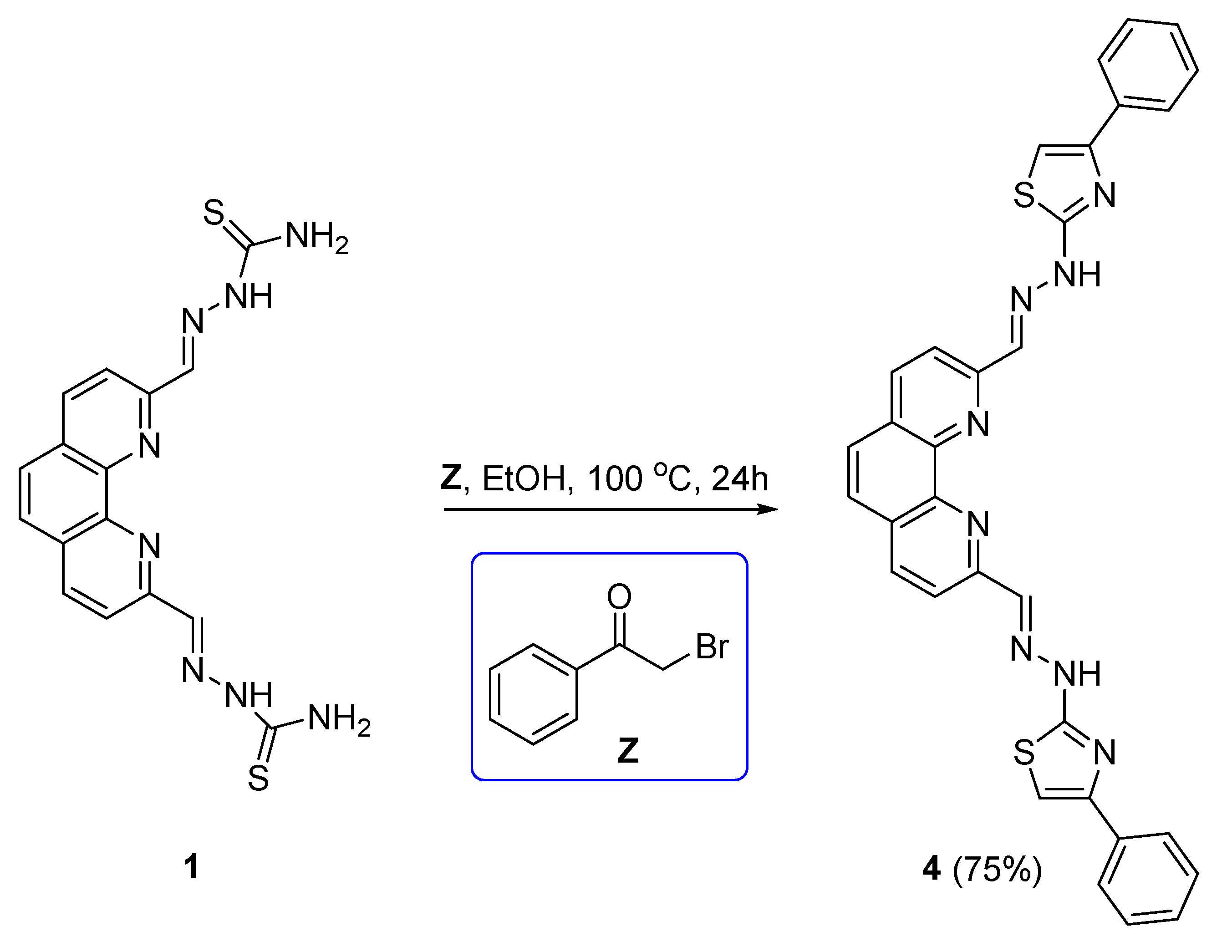
2.2. Synthesis and characterization of chemical compounds
2. ,2’-(1,10-phenanthroline-2,9-diyl)-bis(methylene)-bis(hydrazine-1-carbothioamide) (1)
2. ,2’-(1,10-phenanthroline-2,9-diyl)-bis(methylene)-bis(hydrazine-1-carbothioamide) hexafluorophosphate salt (2)
2. ,2’-(1,10-phenanthroline-2,9-diyl)-bis(methylene)-bis(hydrazine-1-carbothioamide) tetrafluoroborate salt (3)
2. ,9-bis((E)-(2-(4-phenylthiazol-2-yl)hydrazineylidene)methyl)-1,10-phenanthroline (4)
2.3. DNA interaction studies and biological activity
2.3.1. Fret-based DNA melting assays
2.3.2. Tel22 CD titration spectra
2.3.3. Non-competition and competition equilibrium dialysis experiments
2.3.4. CT viscosity assay
2.3.5. Cell culture conditions and MTT viability assay
2.3.6. Cell cycle and Annexin-V apoptosis assays
3. Results
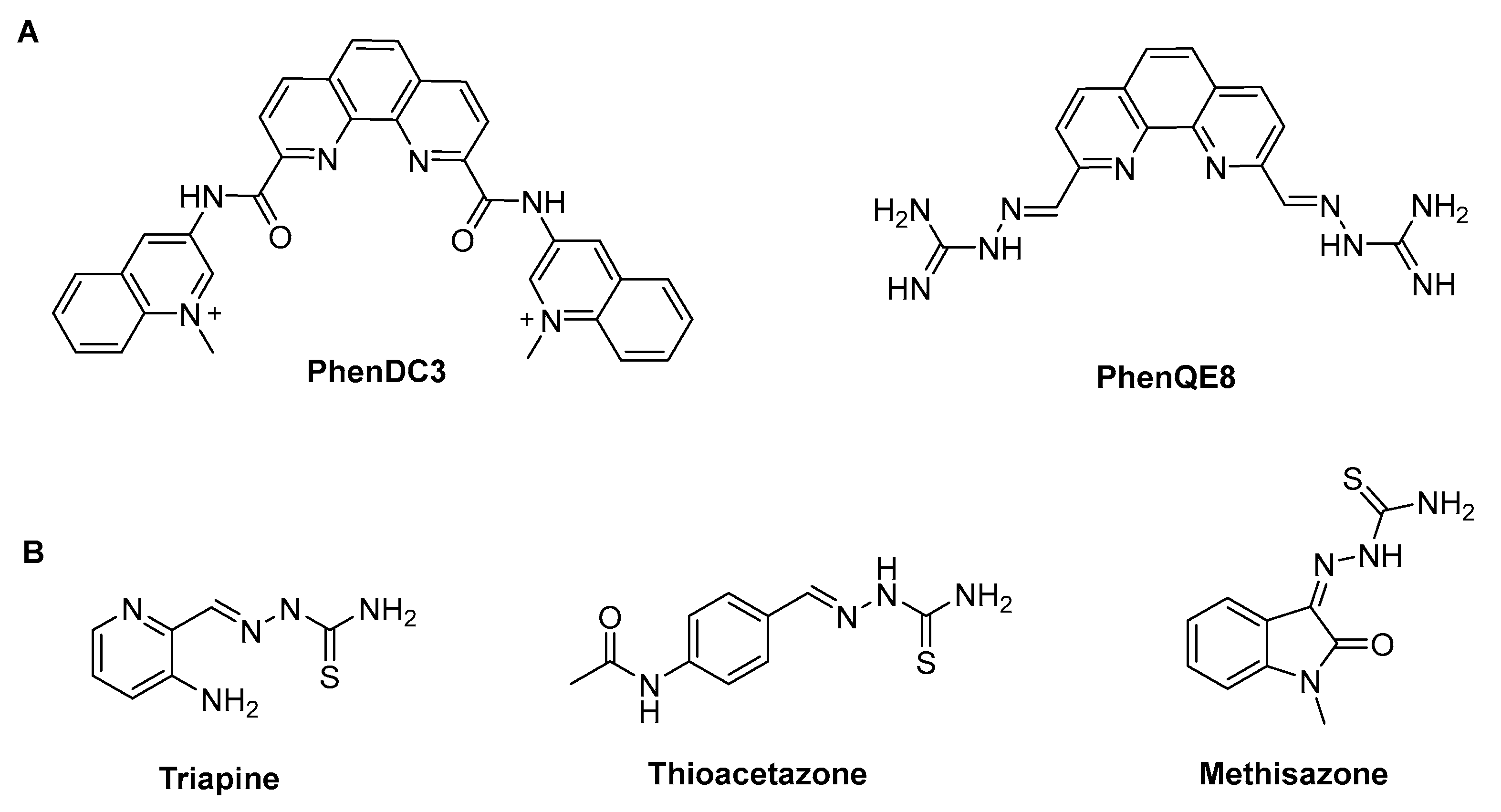
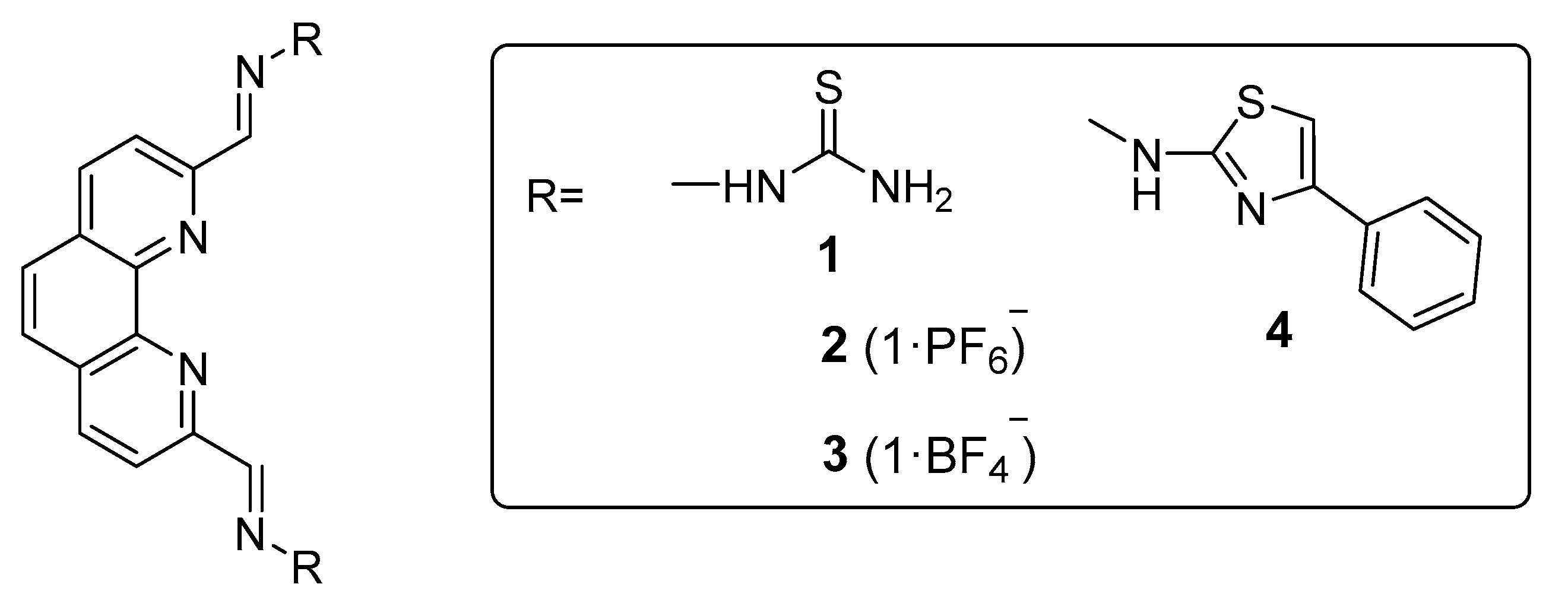
3.1. Synthesis and characterization of 1,10-phenanthroline TSCs and derived 4-phenylthiazole analogue.
3.2. DNA interactions with telomeric G4
3.2.1. FRET-based DNA melting experiments
3.2.2. Circular dichroism
3.2.3. Non-competition and competition equilibrium dialysis
3.2.4. CT DNA viscosity titration
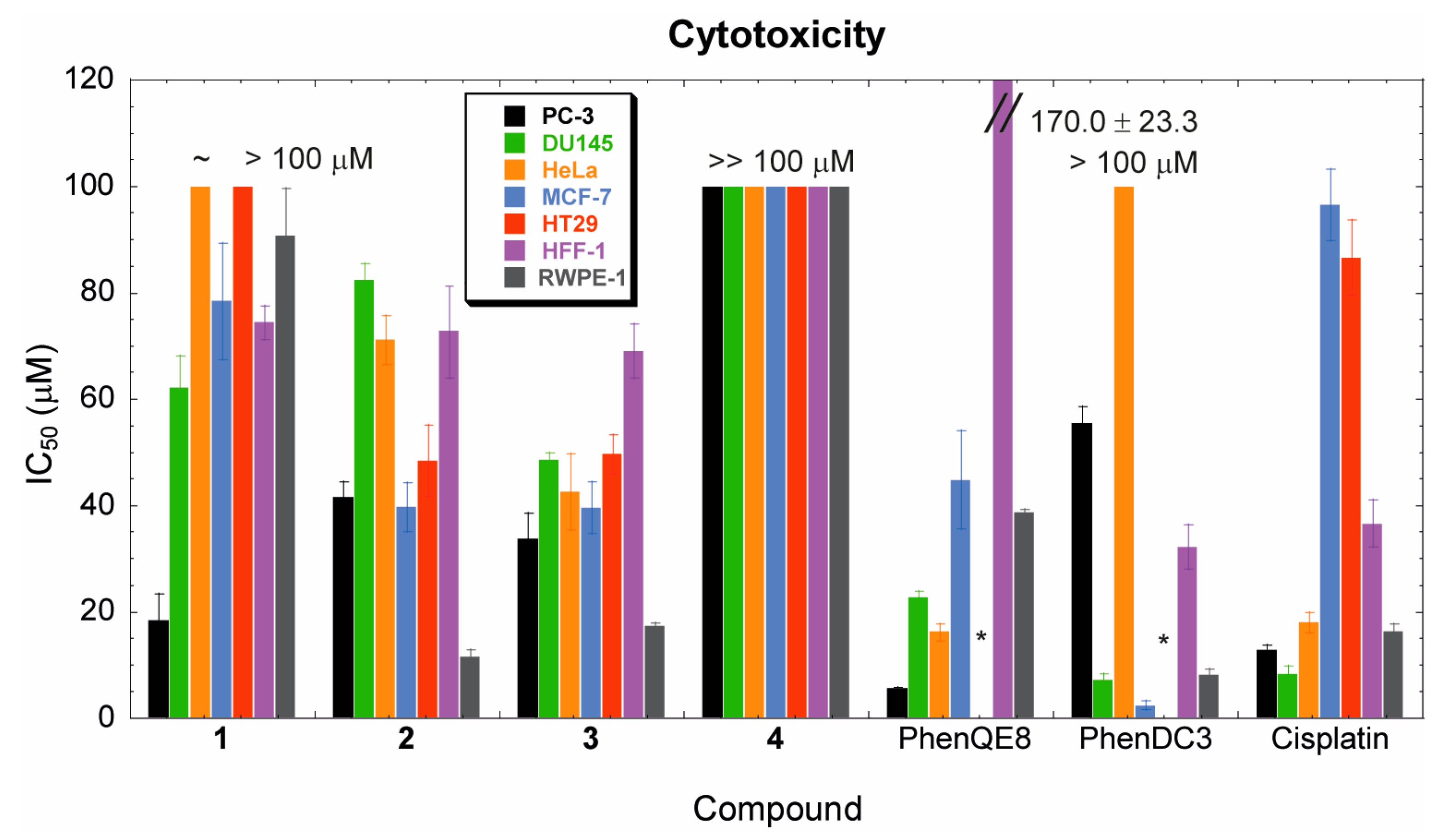
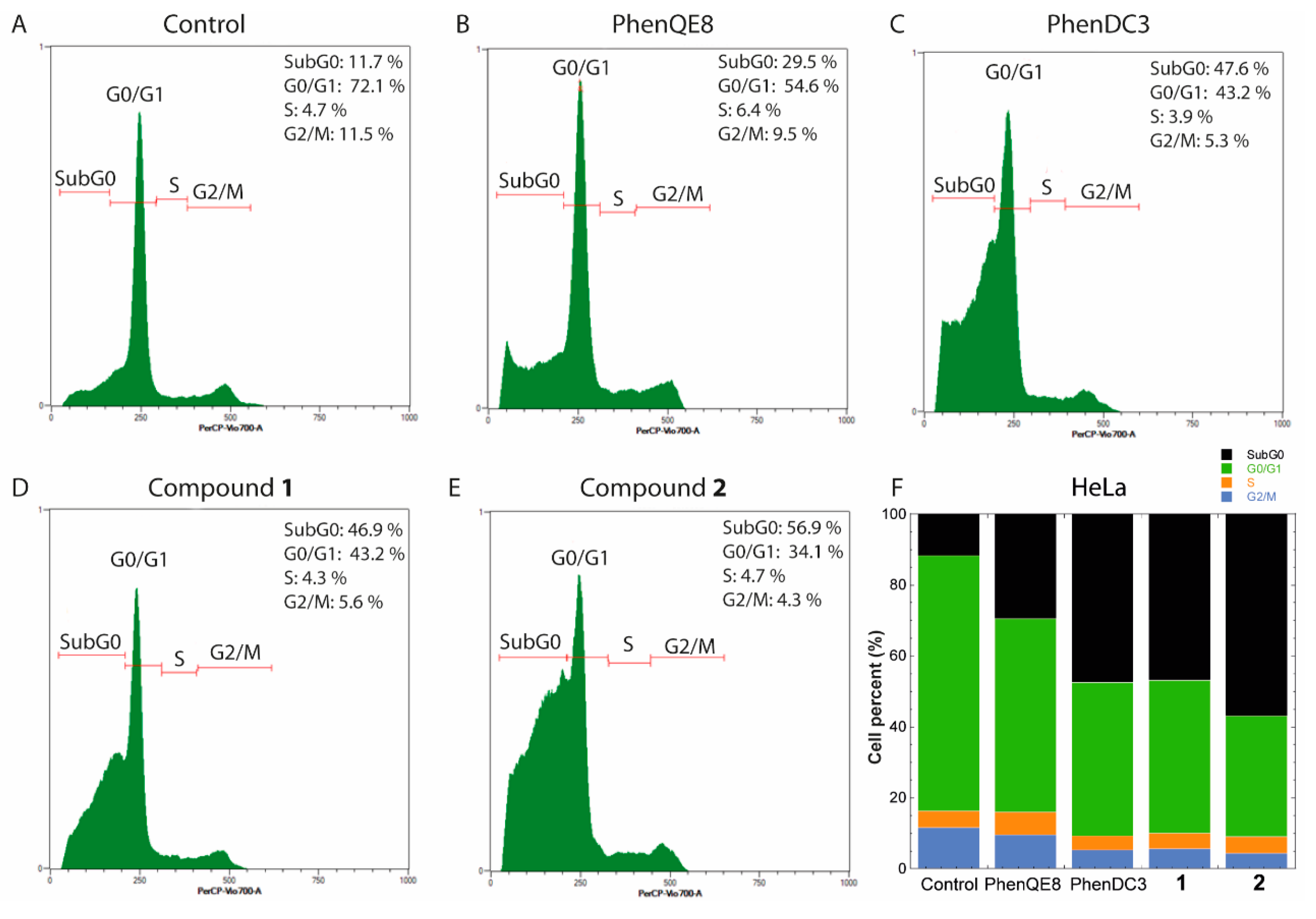
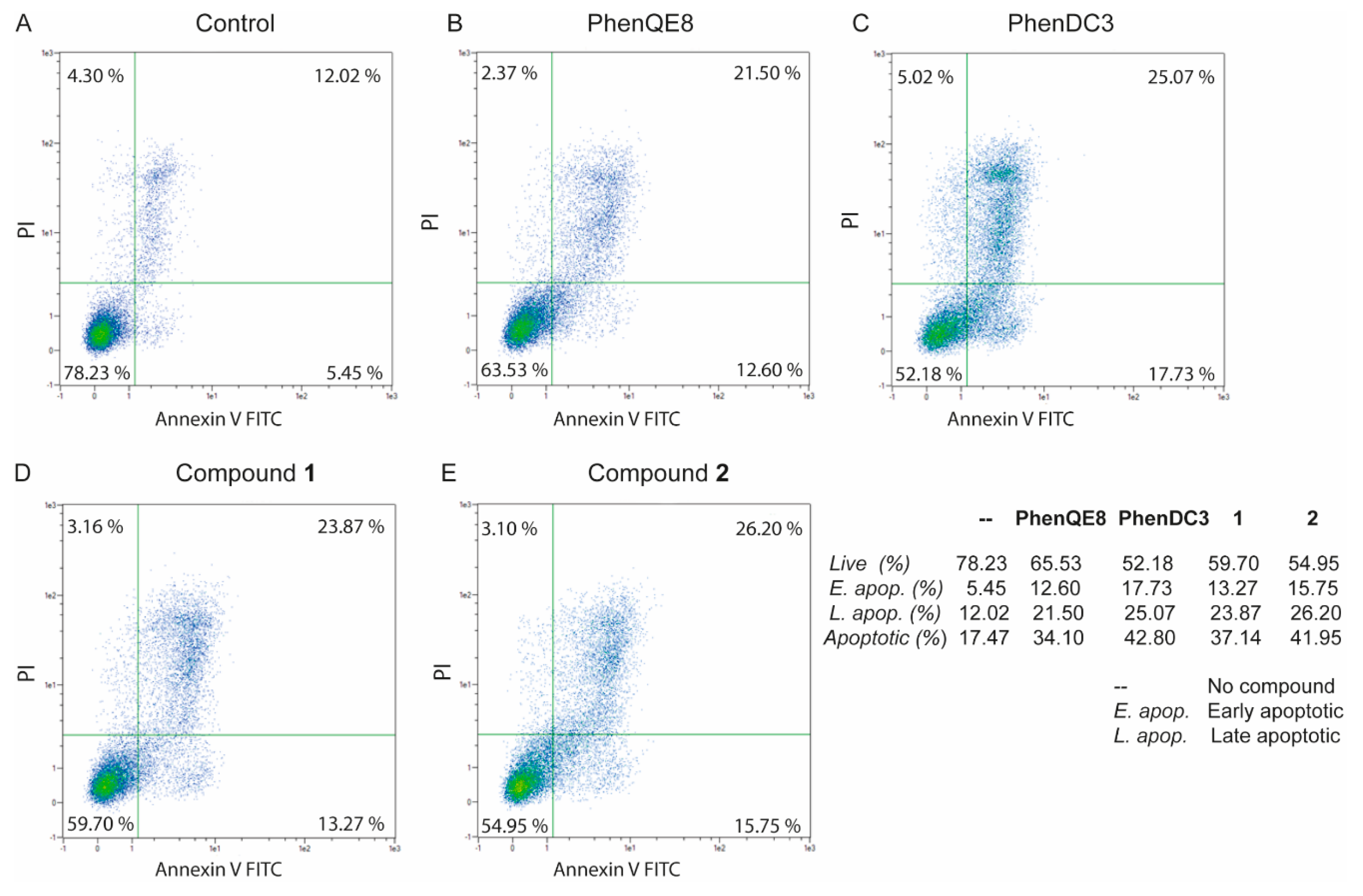
3.3. Biological activity. MTT, cell cycle and apoptosis assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflict of Interest
References
- Watson, J.D.; Crick, F.H. Molecular of Nucleic Acids; A Structure for deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Franklin, R.; Gosling, R. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, D. Sequence, Stability, and Structure of G-Quadruplexes and Their Interactions with Drugs. Curr Protoc Nucleic Acid Chem 2012. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Quadruplex Nucleic Acids as Targets for Anticancer Therapeutics. Nat Rev Chem 2017, 1. [Google Scholar] [CrossRef]
- Wang, H.; Li, P.; Sun, M.; Wei, J. Not Unusual, Just Different! Chemistry, Biology and Applications of G-Quadruplex Nucleic Acids. Acta Astronaut 2017, 137, 214–221. [Google Scholar] [CrossRef]
- Falanga, A.P.; Terracciano, M.; Oliviero, G.; Roviello, G.N.; Borbone, N. Exploring the Relationship between G-Quadruplex Nucleic Acids and Plants: From Plant G-Quadruplex Function to Phytochemical G4 Ligands with Pharmaceutic Potential. Pharmaceutics 2022, 14, 2377. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Dipankar, S. Chem Rev 2019, 119, 6290–6325. [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin, 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gellert, I.; Lipsett, M.N.; Davies, D.R. Helix Formation By Guanylic Acid. Proc Natl Acad Sci U S A. 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Bang, I.; Untersuchungen Über Die Guanylsäure. Biochem Z 1910, 26, 293.
- Sen, D.; Gilbert, W. Formation of Parallel Four-Stranded Complexes by Guanine-Rich Motifs in DNA and Its Implications for Meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Frasson, I.; Pirota, V.; Richter, S.N.; Doria, F. Multimeric G-Quadruplexes: A Review on Their Biological Roles and Targeting. Int J Biol Macromol 2022, 204, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Gilbert, W. A Sodium-Potassium Switch in the Formation of Four-Stranded G4-DNA. Nature 1990, 344, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Iida, K.; Nagasawa, K. Topologies of G-Quadruplex: Biological Functions and Regulation by Ligands. Biochem Biophys Res Commun 2020, 531, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA Secondary Structures: Stability and Function of G-Quadruplex Structures. Nat Rev Genet 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Therapeutic Applications of Quadruplex Nucleic Acids; 1st ed.; Academic Press, 2012.
- Andrews, L.G.; Tollefsbol, T.O. Methods of Telomerase Inhibition. Methods Mol Biol. 2007, 405, 1–7. [Google Scholar] [CrossRef]
- Fragkiadaki, P.; Renieri, E.; Kalliantasi, K.; Kouvidi, E.; Apalaki, E.; Vakonaki, E.; Mamoulakis, C.; Spandidos, D.A.; Tsatsakis, A. Τelomerase Inhibitors and Activators in Aging and Cancer: A Systematic Review. Mol Med Rep 2022, 25. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chemistry - A European Journal 2019, 25, 417–430. [Google Scholar] [CrossRef]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-Quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. A Hitchhiker’s Guide to G-Quadruplex Ligands. Org Biomol Chem 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Neidle, S. Design Principles for Quadruplex-Binding Small Molecules. In Therapeutic Applications of Quadruplex Nucleic Acids; Academic Press: London, UK, 2012. [Google Scholar]
- Brassart, B.; Gomez, D.; De Cian, A.; Paterski, R.; Montagnac, A.; Qui, K.H.; Temime-Smaali, N.; Trentesaux, C.; Mergny, J.L.; Gueritte, F.; et al. A New Steroid Derivative Stabilizes G-Quadruplexes and Induces Telomere Uncapping in Human Tumor Cells. Mol Pharmacol 2007, 72, 631–640. [Google Scholar] [CrossRef]
- Ohnmacht, S.A.; Neidle, S. Small-Molecule Quadruplex-Targeted Drug Discovery. Bioorg Med Chem Lett 2014, 24, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Hemalatha, C.N.; Vijey Aanandhi, M. G-Quadruplex Ligands as Stabilizer Targeting Telomerase Enzyme as Anti Cancer Agents. Asian Journal of Pharmaceutical and Clinical Research 2017, 10, 50–53. [Google Scholar]
- Carella, A.; Roviello, V.; Iannitti, R.; Palumbo, R.; La Manna, S.; Marasco, D.; Trifuoggi, M.; Diana, R.; Roviello, G.N. Evaluating the biological properties of synthetic 4-nitrophenyl functionalized benzofuran derivatives with telomeric DNA binding and antiproliferative activities. Int J Biol Macromol 2019, 121, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Hein, N.; Hannan, K.M.; George, A.J.; Sanij, E.; Hannan, R.D. The Nucleolus: An Emerging Target for Cancer Therapy. Trends Mol Med 2013, 19, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Hurley, L.H. A First-in-Class Clinical G-Quadruplex-Targeting Drug. The Bench-to-Bedside Translation of the Fluoroquinolone QQ58 to CX-5461 (Pidnarulex). Bioorg Med Chem Lett 2022, 77. [Google Scholar] [CrossRef] [PubMed]
- Gratal, P.B.; Quero, J.G.; Pérez-Redondo, A.; Gándara, Z.; Gude, L. PhenQE8, a Novel Ligand of the Human Telomeric Quadruplex. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Bollenbach, M.; Nemska, S.; Wagner, P.; Camelin, G.; Daubeuf, F.; Obrecht, A.; Villa, P.; Rognan, D.; Bihel, F.; Bourguignon, J.-J.; et al. Design, Synthesis and Biological Evaluation of Arylpyridin-2-Yl Guanidine Derivatives and Cyclic Mimetics as Novel MSK1 Inhibitors. An Application in an Asthma Model. Molecules 2021, 26(2):391. [CrossRef]
- Rahbari, M.; Rahlfs, S.; Jortzik, E.; Bogeski, I.; Becker, K. Exploring the Anti-Cancer Activity of Novel Thiosemicarbazones Generated through the Combination of Retro-Fragments: Dissection of Critical Structure-Activity Relationships. PLoS One 2014, 9, 1–15. [Google Scholar] [CrossRef]
- Dincel, E.D.; Akdağ, Ç.; Kayra, T.; Coşar, E.D.; Aksoy, M.O.; Akalın-Çiftçi, G.; Ulusoy-Güzeldemirci, N. Design, Synthesis, Characterization, Molecular Docking Studies and Anticancer Activity Evaluation of Novel Hydrazinecarbothioamide, 1,2,4-Triazole-3-Thione, 4-Thiazolidinone and 1,3,4-Oxadiazole Derivatives. J Mol Struct 2022, 1268. [Google Scholar] [CrossRef]
- Sun, Y.; Lu, Y.; Bian, M.; Yang, Z.; Ma, X.; Liu, W. Pt(II) and Au(III) Complexes Containing Schiff-Base Ligands: A Promising Source for Antitumor Treatment. Eur J Med Chem 2021, 211. [Google Scholar] [CrossRef]
- Arifuzzaman, Md.; Karim, M.R.; Siddiquee, T.A.; Mirza, A.H.; Ali, M.A. Synthesis and Characterization of New Schiff Bases Formed by Condensation of 2,9-Phenathroline-1,10-Dialdehyde with Sulfur-Containing Amines. Int J Org Chem (Irvine) 2013, 03, 81–86. [Google Scholar] [CrossRef]
- Miglietta, G.; Russo, M.; Duardo, R.C.; Capranico, G. G-Quadruplex Binders as Cytostatic Modulators of Innate Immune Genes in Cancer Cells. Nucleic Acids Res 2021, 49, 6673–6686. [Google Scholar] [CrossRef]
- Chaires, J.B. Structural Selectivity of Drug-Nucleic Acid Interactions Probed by Competition Dialysis. Top Curr Chem 2005, 253. [Google Scholar] [CrossRef]
- van Engeland, M.; Nieland, L.J.; Ramaekers, F.C.; Schutte, B., Reutelingsperger, C.P. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry 1998, 31, 1–9. [CrossRef]
- Lakshmanan, I.; Batra, S.K. Protocol for Apoptosis Assay by Flow Cytometry Using Annexin V Staining Method. Bio Protoc. 2013, 3, e374. [Google Scholar] [CrossRef]
- Loughrey, B.T.; Healy, P.C.; Parsons, P.G.; Williams, M.L. Selective Cytotoxic Ru(II) Arene Cp* Complex Salts [R-PhRuCp*]+X- for X = BF4¯, PF6¯, and BPh4¯. Inorg Chem 2008, 47, 8589–8591. [Google Scholar] [CrossRef] [PubMed]
- Porchia, M.; Benetollo, F.; Refosco, F.; Tisato, F.; Marzano, C.; Gandin, V. Synthesis and Structural Characterization of Copper(I) Complexes Bearing N-Methyl-1,3,5-Triaza-7-Phosphaadamantane (MPTA). Cytotoxic Activity Evaluation of a Series of Water Soluble Cu(I) Derivatives Containing PTA, PTAH and MPTA Ligands. J Inorg Biochem 2009, 103, 1644–1651. [Google Scholar] [CrossRef]
- Healy, P.C.; Loughrey, B.T.; Williams, M.L.; Parsons, P.G. Synthesis, Structure and Cytotoxicity Studies of Four-Coordinate Bis (Cis-Bis(Diphenylphosphino)Ethene) Gold(I) Complexes, [Au(Dppey)2]X. J Inorg Biochem 2010, 104, 625–631. [Google Scholar] [CrossRef]
- Yurttaş, L.; Çavuşoğlu, B.K.; Cantürk, Z. Novel 2-(2-Hydrazinyl)Thiazole Derivatives as Chemotherapeutic Agents. Synth Commun 2020, 50, 3072–3079. [Google Scholar] [CrossRef]
- De Cian, A.; Guittat, L.; Kaiser, M.; Saccà, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.P.; Lacroix, L.; Mergny, J.L. Fluorescence-Based Melting Assays for Studying Quadruplex Ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Kieltyka, R.; Englebienne, P.; Fakhoury, J.; Autexier, C.; Moitessier, N.; Sleiman, H.F. A Platinum Supramolecular Square as an Effective G-Quadruplex Binder and Telomerase Inhibitor. J Am Chem Soc 2008, 130, 10040–10041. [Google Scholar] [CrossRef]
- del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angewandte Chemie 2018, 130, 7289–7293. [Google Scholar] [CrossRef]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular Dichroism of Quadruplex DNAs: Applications to Structure, Cation Effects and Ligand Binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, A.S.G.P.; D. S.M.W. Circular Dichroism of Quadruplex Structures. Top. Curr. Chem 2013, 330, 67–86. [Google Scholar] [PubMed]
- Neidle, S. Human Telomeric G-Quadruplex: The Current Status of Telomeric G-Quadruplexes as Therapeutic Targets in Human Cancer. FEBS Journal 2010, 277, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Teulade-Fichou, M.P. G4-FID: A Fluorescent DNA Probe Displacement Assay for Rapid Evaluation of Quadruplex Ligands. Methods Mol Biol 2010, 608. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.; Chaires, J.B. Criteria for the Mode of Binding of DNA Binding Agents. Bioorg Med Chem 1995, 3. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.; Eisenberg, H. Viscosity and Sedimentation Study of Sonicated DNA–Proflavine Complexes. Biopolymers 1969, 8. [Google Scholar] [CrossRef]
- Fairley, T.A.; Tidwell, R.R.; Donkor, I.; Naiman, N.A.; Ohemeng, K.A.; Lombardy, R.J.; Bentley, J.A.; Cory, M. Structure, DNA Minor Groove Binding, and Base Pair Specificity of Alkyl- and Aryl-Linked Bis(Amidinobenzimidazoles) and Bis(Amidinoindoles). J Med Chem 1993, 36. [Google Scholar] [CrossRef]
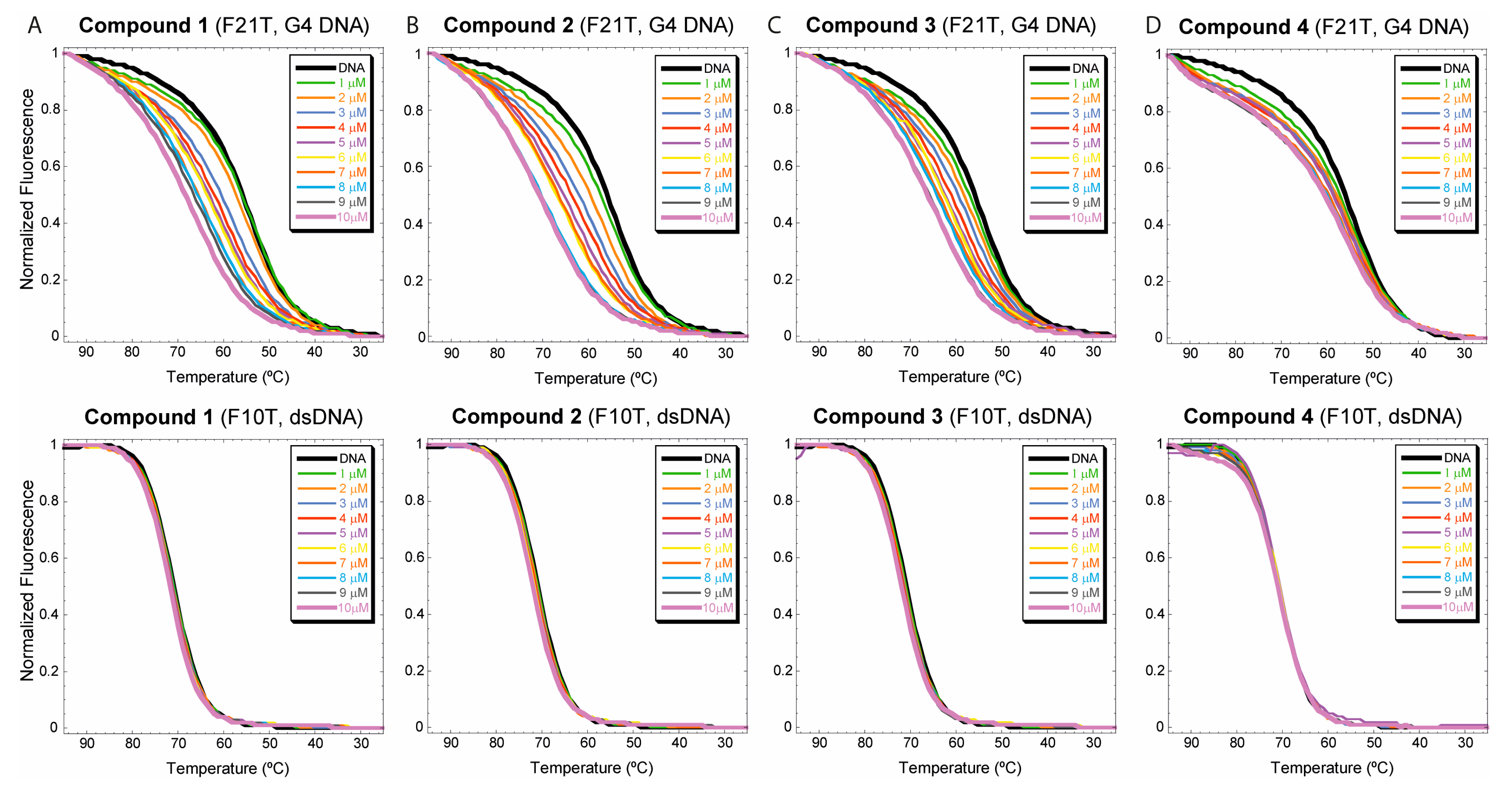
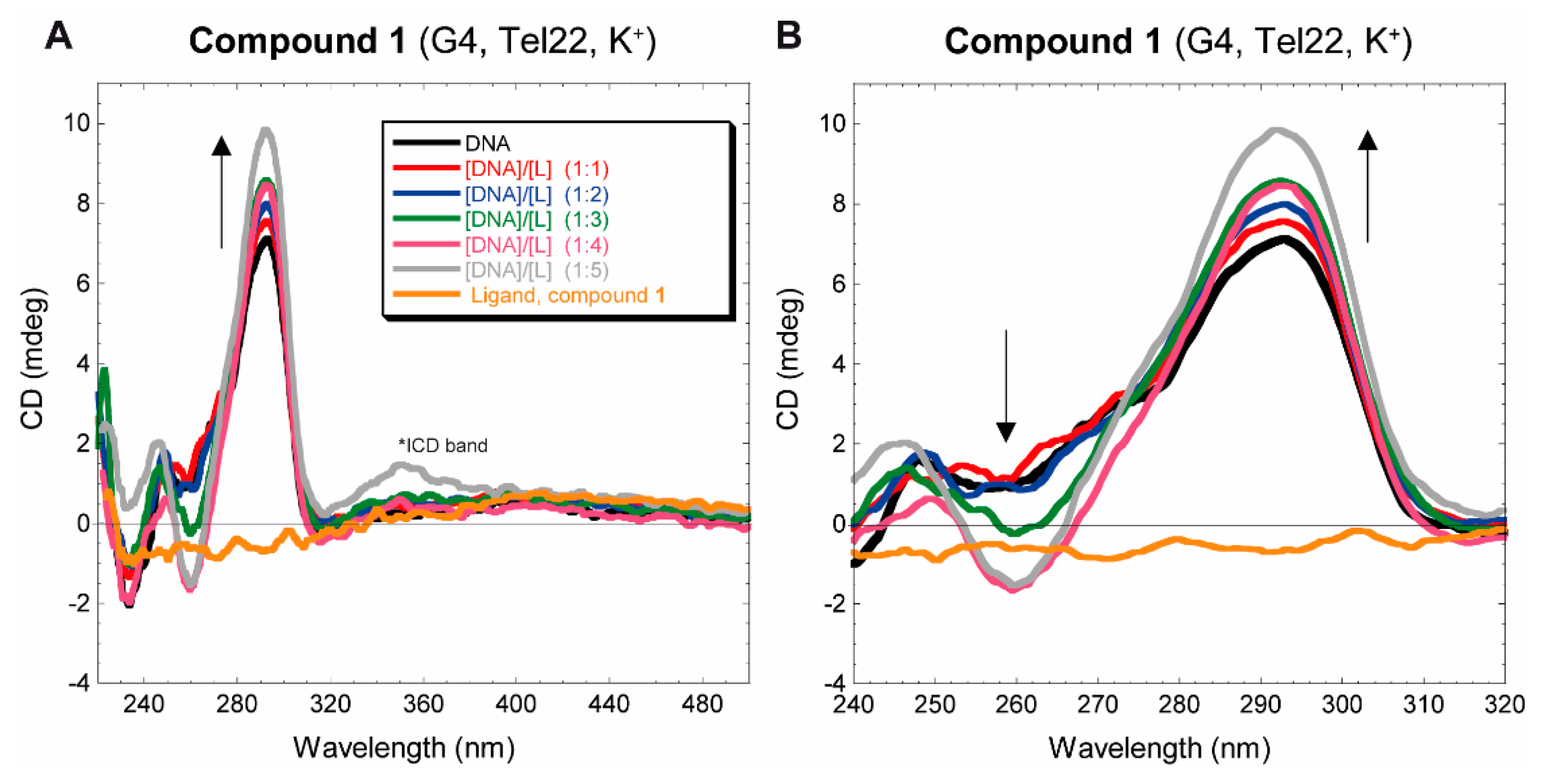
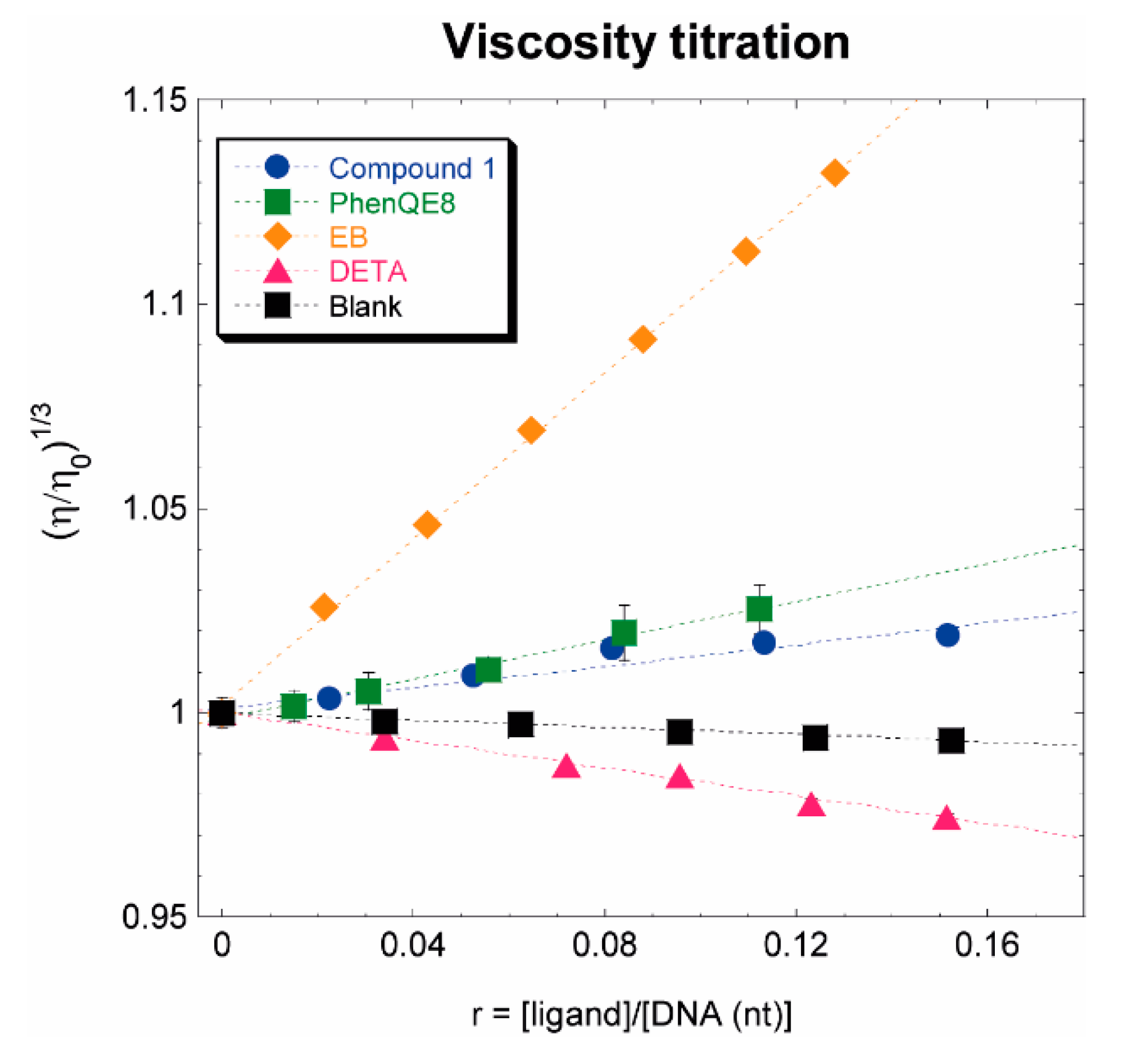
| 1 + Tel22 | 1 + ds17 |
1 + Tel22 + ds17 |
|
|---|---|---|---|
| DNA | Kapp (M-1) x10-5 | Kapp (M-1) x10-5 | Kapp (M-1) x10-5 |
| Tel22 (Q) | 1.1 ± 0.1 | -- | 1.2 ± 0.1 |
| ds17 (D) | -- | 0.06 ± 0.01 | 0.05 ± 0.01 |
| PC-3 | DU145 | HeLa | MCF-7 | HT29 | HFF-1 | RWPE-1 | |
|---|---|---|---|---|---|---|---|
| Ligand | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) |
| 1 | 18.4 ± 4.9 | 62.2 ± 5.9 | ~ 100 | 78.4 ± 11.0 | > 100 | 74.4 ± 3.1 | 90.8 ± 8.8 |
| 2 | 41.6 ± 2.8 | 82.5 ± 3.0 | 71.2 ± 4.6 | 39.7 ± 4.6 | 48.5 ± 6.7 | 72.9 ± 8.8 | 11.5 ± 1.3 |
| 3 | 33.8 ± 4.7 | 48.6 ± 1.4 | 42.6 ± 7.1 | 39.6 ± 4.8 | 49.7 ± 3.7 | 69.1 ± 5.0 | 17.3 ± 0.7 |
| 4 | >> 100 | >> 100 | >> 100 | >> 100 | >> 100 | >> 100 | >> 100 |
| PhenQE8 | 5.6 ± 0.3 | 22.7 ± 1.1 | 16.2 ± 1.6 | 44.9 ± 9.2 | N.D. | 170.0 ± 23.3 | 38.8 ± 0.5 |
| PhenDC3 | 55.5 ± 3.1 | 7.3 ± 1.0 | > 100 | 2.4 ± 0.8 | N.D. | 32.2 ± 4.2 | 8.2 ± 1.1 |
| cisplatin | 12.8 ± 0.9 | 8.4 ± 1.5 | 18.0 ± 1.9 | 96.5 ± 6.7 | 86.6 ± 7.0 | 36.6 ± 4.4 | 16.4 ± 1.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





