1. Introduction
Dairy cow farming stands as a pivotal component of modern agriculture, playing a multifaceted role in both sustenance and societal fabric [
1]. This agricultural pursuit centers around the husbandry of dairy cattle primarily for the production of milk and its derivatives, constituting an integral part of the global food supply chain [
2]. The significance of dairy cow farming reverberates through its contributions to nutrition, economic stability, and rural livelihoods. Milk, a primary output of these farms, serves as a rich source of essential nutrients, fostering public health and well-being [
3]. Economically, the dairy industry encompasses a vast network of producers, processors, and distributors, generating substantial revenue and employment opportunities worldwide.
Body Condition Score (BCS) is a crucial metric in dairy cow farming, serving as an indicator of the animal’s nutritional status and overall well-being [
4]. The assessment of body BCS during calving, along with the lowest BCS point and the overall loss in BCS throughout early lactation, has been linked to milk production, reproductive outcomes, and the occurrence of postparturient diseases [
5]. This underscores the significance of precisely monitoring nutrient reserves and their utilization during the periparturient phase [
6].
Traditionally, BCS assessment has been a manual process conducted by skilled experts or trained assessor [
7,
8,
9,
10,
11]. This subjective evaluation involves palpating and visually inspecting specific anatomical areas such as the tail-head, backbone, and ribs to assign a numerical score that reflects the cow’s body fat and muscle condition. Experts rely on their experience and tactile judgment to gauge the adequacy of the cow’s nutrition, influencing subsequent feeding and management decisions.
While manual BCS assessment has been a longstanding practice, its subjectivity and dependency on human expertise can introduce variability. The advent of technology, such as 3D imaging and artificial intelligence, has opened avenues for automated BCS scoring, offering potential advancements in accuracy, efficiency, and objectivity compared to traditional manual methods.
Deep Learning (DL), a subset of Machine Learning (ML), has emerged as a transformative paradigm in the field of artificial intelligence, showcasing unprecedented capabilities in processing complex data. Characterized by its utilization of deep neural networks (DNNs), DL excels in automatically learning hierarchical representations from data, enabling the extraction of intricate patterns and features. As articulated by LeCun et al. [
12], DNNs excel at feature abstraction, enabling the discovery of hierarchical representations that contribute to the model’s capacity to discern intricate patterns. This capacity has proven particularly advantageous in various domains, including image and speech recognition, natural language processing, and medical diagnostics. Moreover, the flexibility and adaptability of DNNs, as highlighted by Goodfellow et al. [
13], make them well-suited for diverse applications, allowing the models to learn complex representations directly from raw data. The potential of deep learning extends beyond traditional machine learning techniques, offering unprecedented opportunities for innovation and automation in numerous fields.
Recently, dairy cow farming has been revolutionized by the integration of smart technology, including artificial intelligence (AI), 3D imaging, computer vision, and sensors [
14,
15]. The assessment of BCS in dairy cows has seen transformative advancements with the integration of ML, DL, and computer vision techniques. Numerous studies have explored the potential of these technologies to automate and enhance the accuracy of BCS evaluations. Notably, researchers have employed DL models in the analysis of point cloud data derived from 3D scans of cow bodies. W. Shi et al. demonstrated the effectiveness of DNN models in capturing complex spatial relationships, providing a foundation for automated BCS assessments [
16]. Additionally, computer vision approaches utilizing depth maps have been investigated. Yukun et al. explored the application of Convolutional Neural Network (CNN) architectures in BCS prediction, showcasing the capability of these models to extract meaningful features from depth information [
17].
In this study, we present a novel approach to BCS assessment in dairy cows by introducing Point of View (POV) features and leveraging an enhanced PointNet model. Our proposed method capitalizes on the unique information embedded in specific anatomical viewpoints, allowing for a more nuanced analysis of cow body conditions. By integrating these POV features into the PointNet architecture, we enhance the model’s capacity to capture intricate spatial relationships within point cloud data. This augmentation, coupled with the robust capabilities of the enhanced PointNet model, aims to significantly improve the accuracy and efficiency of BCS assessments. The incorporation of POV features not only contributes to a more detailed understanding of cow anatomy but also aligns with the broader objective of advancing automated techniques for precise and reliable health evaluations in dairy farming practices.
The remainder of this article is organized as follows:
Section 2 delves into an exploration of the general task of Body Condition Scoring. Following this,
Section 3 elucidates the intricacies of the proposed method.
Section 4, presents a comprehensive overview of the experimental results and engages in a thorough discussion. Finally,
Section 5 concludes this work.
2. Problem Formulation
BCS scales are not universally standardized across countries. Each region may develop its own system or adopt a scale based on specific guidelines. For instance, the United States and typically uses a BCS scale ranging from 1 to 5 [
18], whereas some European Union countries may opt for a scale of 1 to 10 [
19]. In this work, a scale of 1 to 9 is employed. This variability underscores the importance of understanding and adhering to the specific BCS scale relevant to a particular country or region, emphasizing the need for consistency in assessing and managing the body condition of livestock.
A point cloud data sample can be represented mathematically as a set of points in a three-dimensional space. Each point in the point cloud corresponds to a specific position in the 3D coordinate system. Mathematically, a single point
in 3D space can be denoted as:
Here,
represents the i-th point in the point cloud, and
are the Cartesian coordinates of that point. For a point cloud with
N points, the entire point cloud can be represented as a set:
In practical terms, point cloud data is often stored as arrays or matrices where each row corresponds to a point, and the columns represent the
and
z coordinates. The point cloud can then be denoted as a matrix:
In this work, the task of BCS assessment is framed as a regression problem, seeking to quantify the relative amount of subcutaneous body fat or energy reserves in dairy cows. The regression-based approach employs a DNN with point cloud data as inputs. The core equation for the BCS regression task can be represented as follows:
where
is the predicted score,
f denotes the mapping function parameterized by
, and
P represents the three-dimensional structural information derived from the dairy cow’s anatomy, i.e., the corresponding point cloud data of that cow individual. The DNN learns the optimal parameters
through the training process, enabling it to capture intricate patterns within the point cloud data and produce a continuous prediction of the dairy cow’s body condition.
3. Materials and Methods
The proposed method for automatically scoring the BCS of dairy cows encompasses a systematic approach involving distinct steps as follows:
Data acquisition
Data pre-processing
Feature computation
BCS assessment by DNN
Initially, the process commences with the acquisition of data through a 3D scan system, capturing comprehensive information about the physical structure of the cows. Subsequently, a crucial step involves Data Alignment, wherein all point cloud data obtained from the scanned cows are precisely aligned in a uniform direction. Following this, the method incorporates a manual extraction phase, specifically targeting the rump part from the point cloud data. This meticulous extraction ensures that the subsequent analysis focuses on a standardized and consistent region of interest. Finally, the core of the proposed method involves the application of a DNN for the automated scoring of BCS. Leveraging the extracted rump part as inputs, the DNN employs sophisticated algorithms to discern and quantify the body condition of the dairy cows. This integrated approach aims to streamline and enhance the accuracy of BCS assessment, offering a technologically advanced and efficient solution for the dairy industry.
3.1. Data Acquisition
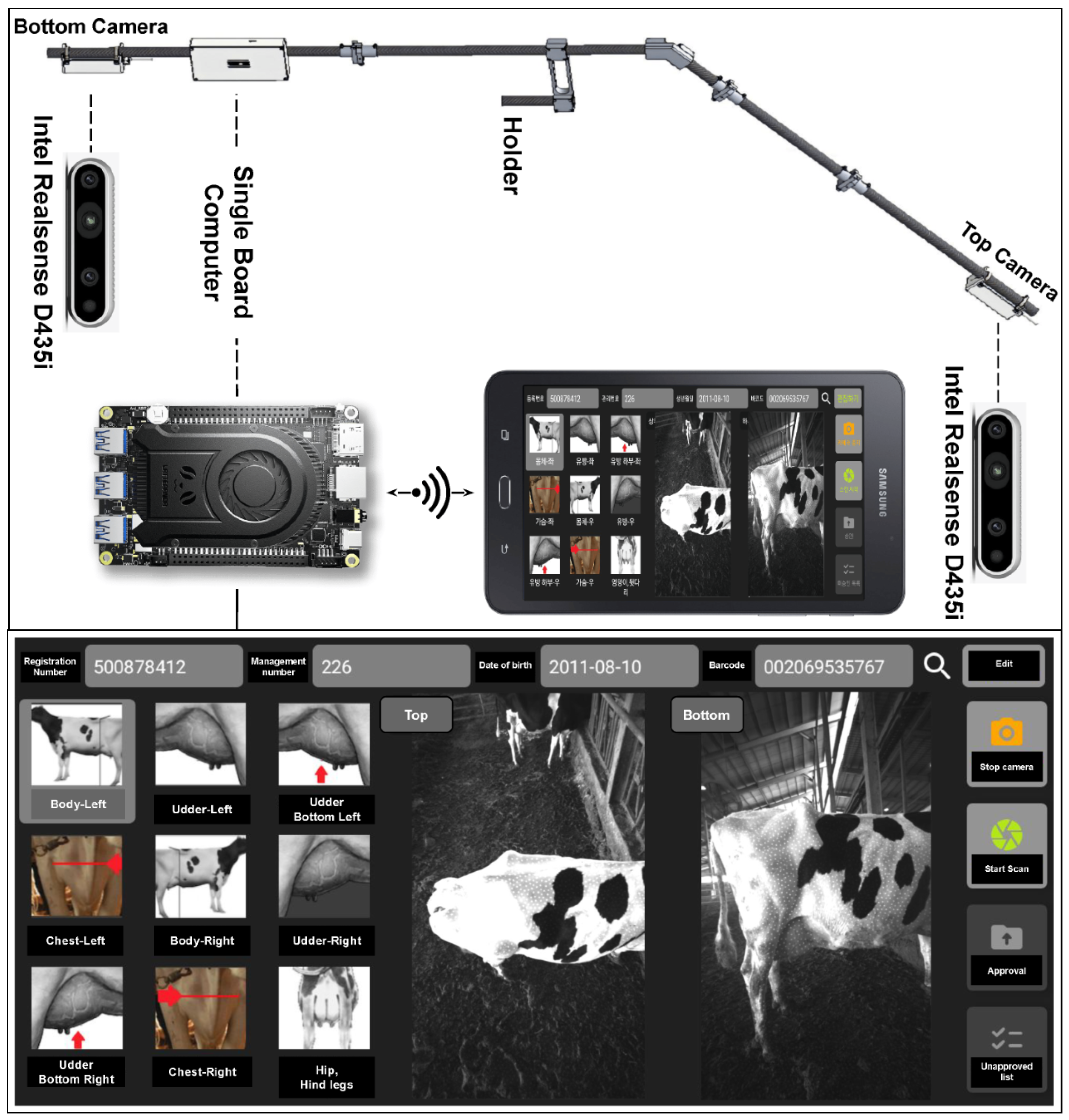
In our prior publication [
20], we introduced a specialized 3D scanning system designed specifically for capturing detailed scans of cow bodies. This technology was employed to scan a total of 403 individual dairy cows, generating a corresponding dataset of 403 point cloud samples. Concurrently, each cow underwent a BCS assessment, conducted by dairy farming experts with specialized knowledge. This meticulous assessment process by domain professionals yielded a dataset of 403 labeled dairy cows, providing a comprehensive foundation for our subsequent analyses and investigations.
Figure 1.
Dairy Cow 3D Scanning System.
Figure 1.
Dairy Cow 3D Scanning System.
3.2. Data Pre-Processing
3.2.1. Global Coordinate Alignment
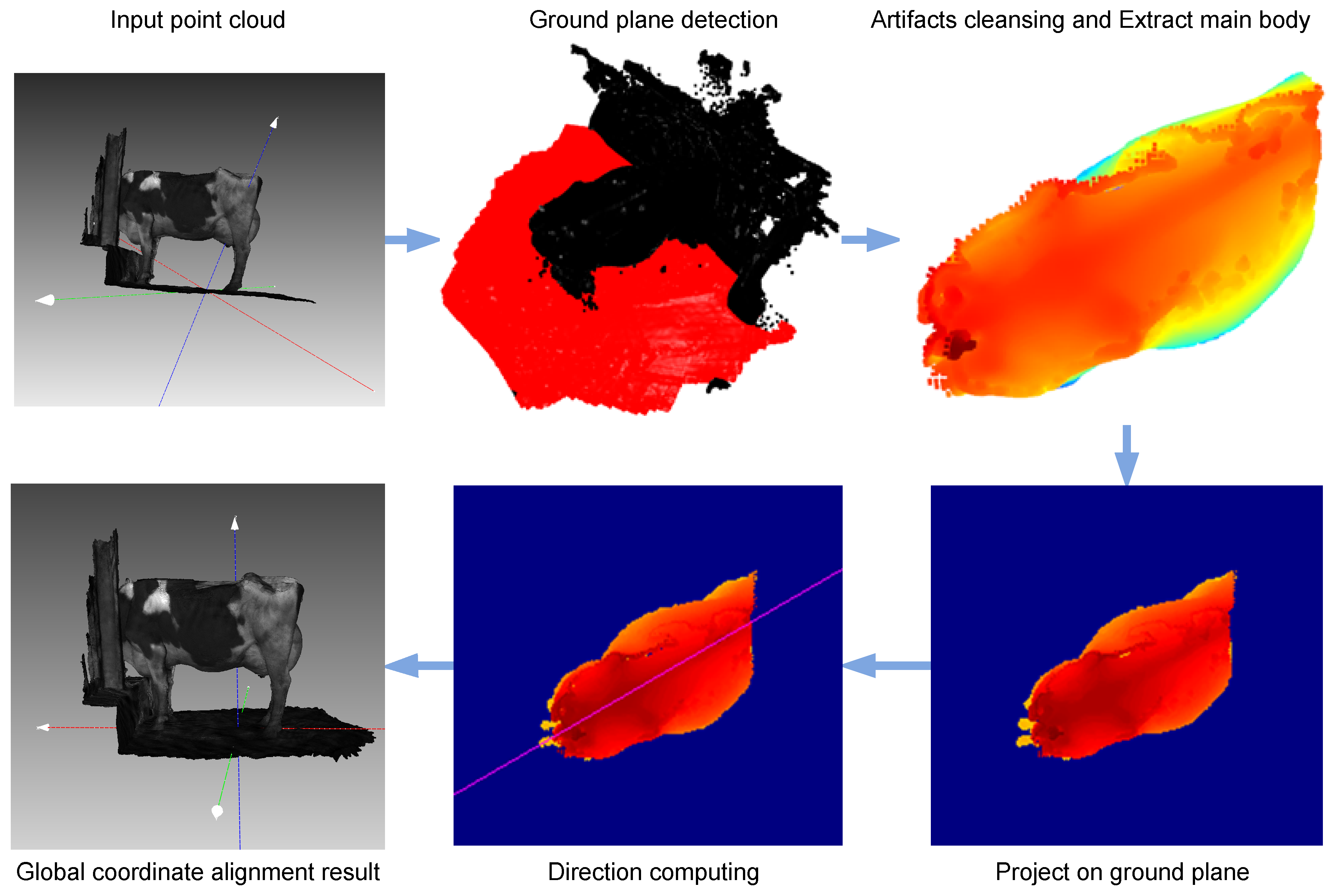
The Data Alignment step within the proposed methodology holds paramount importance due to the inherent variability in the output point cloud data obtained from the initial 3D scan of cows. Given that these scans may exhibit divergent orientations and directions, the alignment process becomes essential to ensure uniformity and consistency in subsequent analyses. The primary objective of this alignment step is to bring coherence to the dataset by aligning all scanned cow bodies effectively to the 3D Global Cartesian Coordinate System (Oxyz). By aligning the scans to a standardized coordinate system, variations in directionality are mitigated, facilitating a more accurate and coherent representation of the cows’ anatomical structures. This meticulous alignment not only establishes a common reference frame but also streamlines the subsequent stages of data processing, particularly the manual extraction of the rump part and the application of the Deep Neural Network (DNN) for automated Body Condition Score (BCS) assessment. The alignment step thus serves as a crucial precursor, ensuring the reliability and consistency of the entire methodology for automated BCS scoring in dairy cows.
Illustrated in
Figure 2, the initial step in the alignment process involves extracting the ground plane from the raw input point cloud. This task is accomplished through the application of the Random Sample Consensus (RANSAC) algorithm [
21], which aids in the identification of the two ground planes. Subsequently, the main body of the cow is isolated and projected onto the ground plane, utilizing the Principle Component Analysis (PCA) [
22] method to calculate the body’s orientation. Integrating information about both the ground plane and the direction of the cow’s body, the input point cloud data undergo alignment to a fixed coordinate system. This comprehensive process results in the harmonization of all input point cloud data of cows to the global coordinate, ensuring a standardized and consistent reference frame for subsequent analyses.
3.2.2. Rump Part Extraction
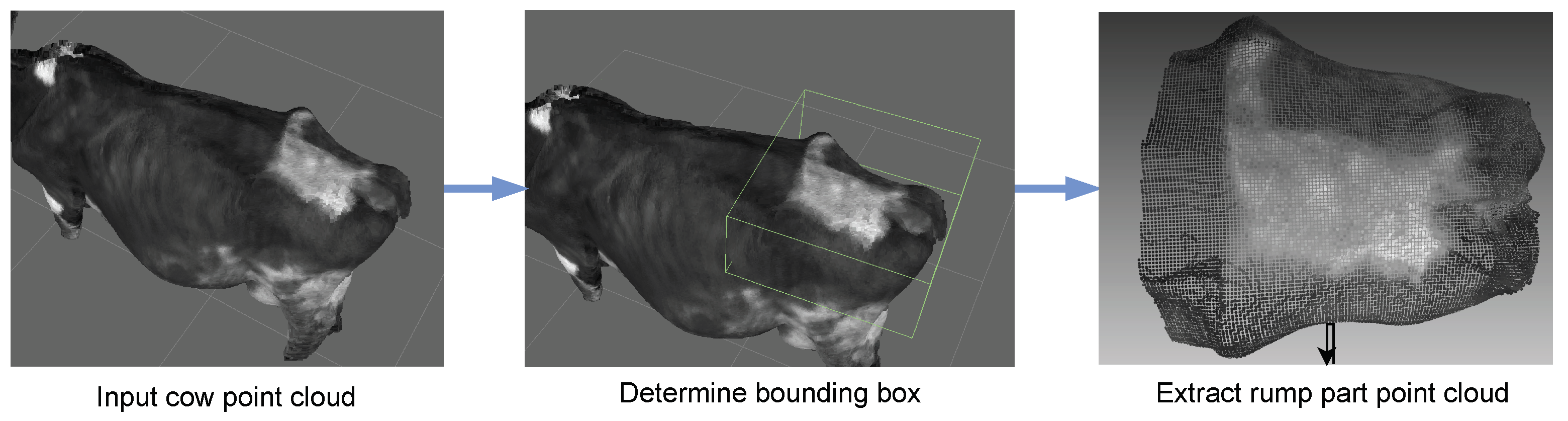
By employing a point cloud labelling tool [
23], the manual extraction of the rump part from the point cloud data of dairy cows is undertaken with precision and purpose. This meticulous process involves human intervention to isolate and delineate the specific anatomical region corresponding to the rump in each scanned cow. The rationale behind this manual extraction lies in the nuanced complexity of identifying and capturing the rump accurately, as automated algorithms may face challenges in discerning subtle variations in anatomy and ensuring consistent extraction across diverse datasets. By employing human expertise, potential variations in cow positioning and body structures can be accounted for, enhancing the robustness of subsequent analyses. The extracted rump part serves as a focused and standardized input for the subsequent application of the DNN in the BCS scoring process.
Figure 3.
Rump part extraction.
Figure 3.
Rump part extraction.
3.3. Features Computation
Given a point
, for the point cloud
, its
Point of View (POV) feature corresponding to
A is defined as follows:
where
is the Euclidean distance between point
and point
of the point cloud P. Explicitly, the Euclidean distance is computed by the following formula:
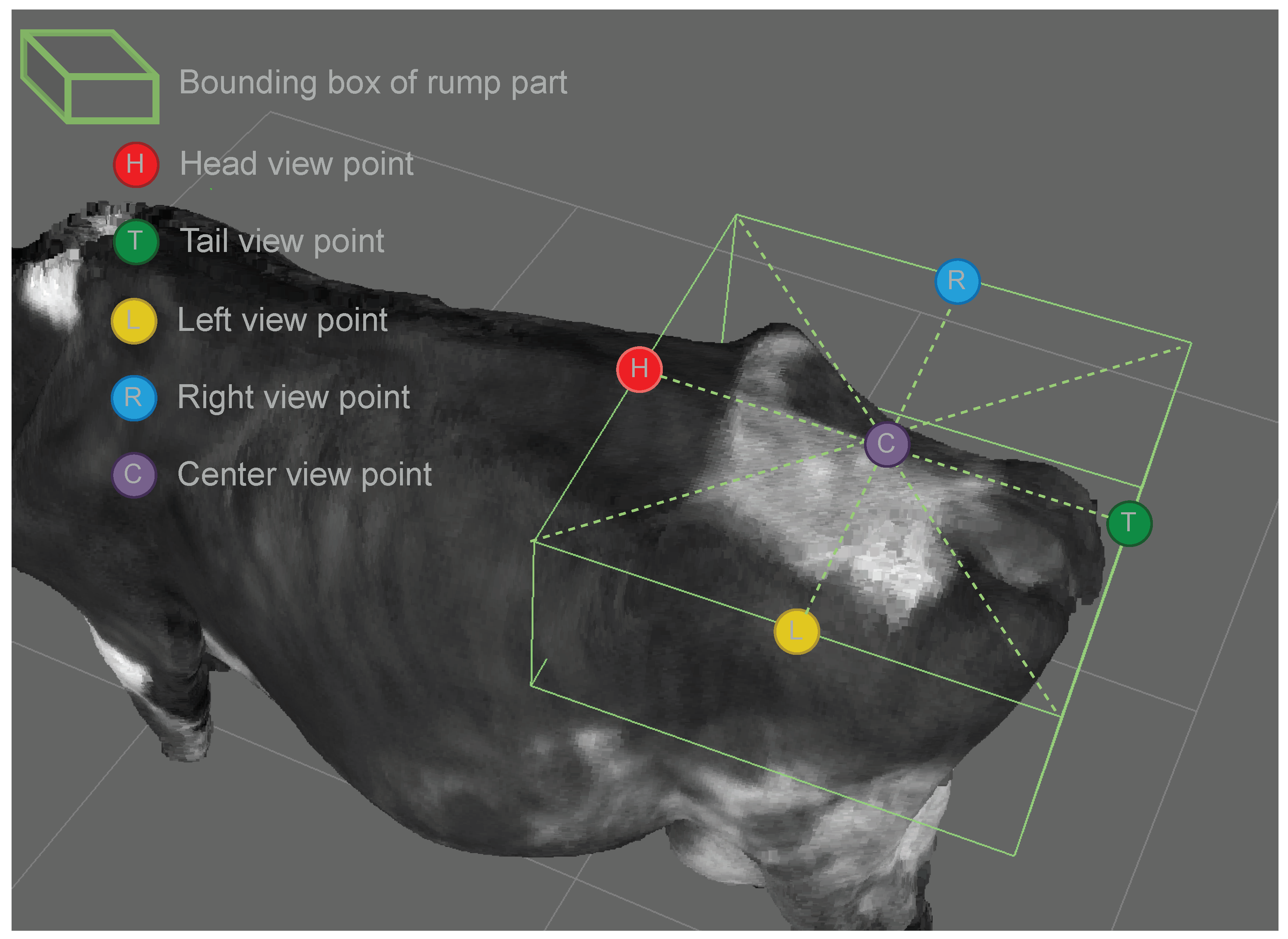
We select 5 different view points: Head (H), Tail (T), Left (L), Right (R), and Center (C) as illustrated in
Figure 4.
After the computation, a point cloud sample
P is transformed into
which can be represented as follows:
The regression problem represented in Equation (
4) can be rewritten as follows:
where
denotes the feature computing function; whiles
and
are the same as in Equation (
8).
3.4. Deep Neural Network Training
The task of estimating BCS of a cow’s body is formulated as a regression problem using point cloud data. To tackle this challenge, we leverage PointNet, a DNN designed for comprehensive 3D data analysis, offering the unique capability to learn both global and local features [
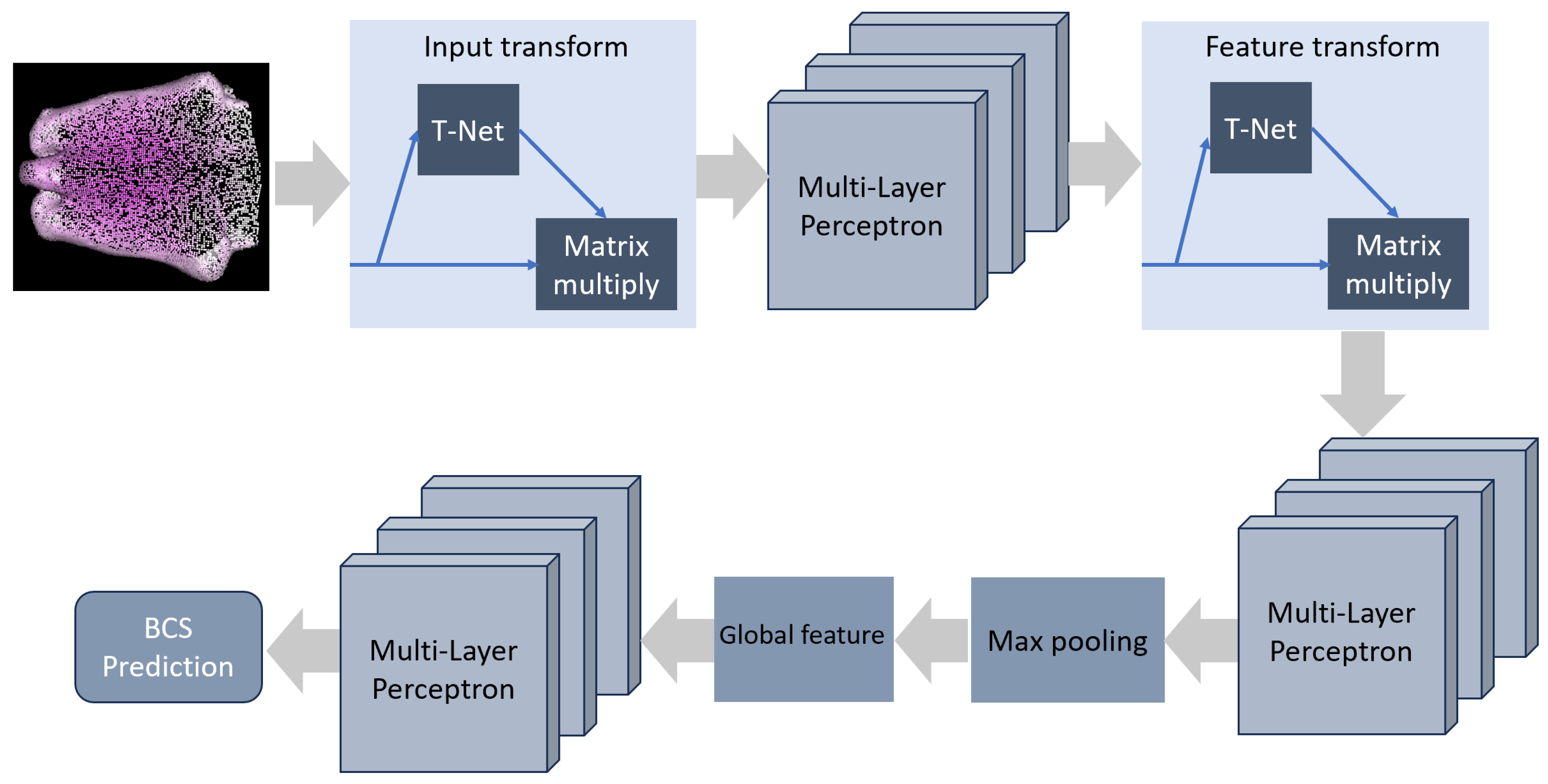
24]. As shown in
Figure 5, the architecture of PointNet is as follows: it incorporates an Input Transform network (T-Net), succeeded by a sequence of Multi-Layer Perceptrons (MLPs) dedicated to local feature extraction. The Input Transform network adeptly captures transformations, ensuring the network’s resilience to variations in input point permutations, rotations, and translations. Following this, a Feature Transform network (T-Net) is employed to augment the network’s ability to handle diverse point orderings. Upon local feature extraction, a global feature vector is derived through max pooling, facilitating the aggregation of information from the entire point cloud. This global feature vector undergoes further processing by a set of MLPs, culminating in the production of the final segmentation mask. This mask assigns class labels to each individual point, effectively completing the task. The synergistic interplay between the Input and Feature Transform networks empowers PointNet to robustly extract features from point cloud data, making it a potent solution for the nuanced task of detecting specific points on a cow’s body.
The initial PointNet models were originally designed for diverse applications, including point cloud part segmentation and point cloud classification. In these applications, the input for PointNet models traditionally consists of three dimensions corresponding to the Cartesian coordinates . However, in the present study, we have adapted and enhanced our DNN-based PointNet model by modifying the input size to encompass 8 dimensions, aligning with the characteristics of the input data. Specifically, the input now incorporates not only the conventional three Cartesian coordinates but also three additional dimensions representing the Gaussian Distance. This modification aims to enrich the model’s capacity to capture and analyze essential features within the point cloud data, facilitating improved performance in tasks such as segmentation and classification.
In the training process, we harnessed a dataset comprising 403 samples of point cloud data from dairy cows, with 15% of the data reserved for rigorous model performance evaluation. The implementation of our PointNet models was conducted using the PyTorch deep learning framework version 1.13, capitalizing on its versatility for seamless model development and optimization. These experiments were orchestrated on a GPU GTX3090, exploiting its parallel processing capabilities to expedite training times. To guide the optimization process, we employed the Mean Squared Error (MSE) loss function for regression tasks. Our training protocol also included a carefully chosen learning rate of 0.001 and the utilization of the Stochastic Gradient Descent (SGD) optimization algorithm. These parameters were fine-tuned to strike a balance between convergence speed and model stability, further enhancing the accuracy and efficiency of our PointNet models for the nuanced analysis of dairy cow point cloud data.
4. Result and Discussion
The outcomes of our experiments, aimed at evaluating the performance of various input data representations and DNN models in the context of BCS, are detailed below. Four distinct input data types were considered: Full Cow Body Point Cloud, Rump Part Point Cloud, POV features, and Depth Map. These were processed by different DNN models, including PointNet [
24], VGG [
25], EfficientNet [
26], and an enhanced PointNet variant incorporating POV features.
Full Cow Body Point Cloud vs. Rump Part Point Cloud
Comparing the results of the PointNet model applied to the full cow body point cloud and the rump part point cloud, it is evident that focusing on the rump part yields superior performance. The Rump Part Point Cloud configuration exhibits a lower Root Mean Squared Error (RMSE) of 0.92, a reduced Mean Absolute Error (MAE) of 0.69, and a slightly lower Mean Absolute Percentage Error (MAPE) of 14.68%, outperforming the Full Cow Body Point Cloud configuration in all metrics.
Depth Map Representations
Introducing Depth Map representations into the mix, the VGG model showcases competitive results with an RMSE of 0.93, MAE of 0.67, and MAPE of 15.59%. In contrast, the EfficientNet model, although proficient, demonstrates slightly higher errors with an RMSE of 1.10, MAE of 0.79, and MAPE of 18.65%. This suggests that the choice of the DNN architecture significantly influences the performance, with VGG exhibiting more favorable outcomes than EfficientNet for this task.
POV Features - Enhanced PointNet
The POV Features - Enhanced PointNet configuration emerges as the most promising, boasting the lowest errors across all metrics. With an RMSE of 0.77, MAE of 0.49, and MAPE of 11.19%, this configuration not only outperforms the other models but also demonstrates the effectiveness of incorporating Point of View features in enhancing BCS prediction accuracy.
Conducting experiments on the same 61 data samples facilitates a robust comparative analysis, the full results are shown in
Table 1. The results underscore the importance of carefully selecting both the input data representation and the DNN model architecture for BCS tasks. While PointNet proves effective, the nuances of the cow’s anatomy, as captured by the rump part, significantly contribute to improved accuracy. Additionally, the incorporation of POV features in the Enhanced PointNet model showcases the potential for leveraging specific anatomical viewpoints to enhance predictive capabilities.
These findings provide valuable insights into the optimization of BCS prediction models, offering guidance for practitioners seeking to deploy efficient and accurate automated BCS assessment systems in dairy farming.
5. Conclusions
This study introduces an innovative automated body condition scoring system for dairy cows, utilizing 3D imaging and deep neural networks. The system proves effective in providing objective and accurate assessments, addressing limitations associated with manual scoring methods. Results from validation indicate reliability and consistency compared to traditional approaches, emphasizing time savings and reduced subjectivity.
The success of this system extends beyond dairy herd management, offering potential applications in precision agriculture. As we navigate the convergence of technology and farming, this research signifies a step towards optimizing animal welfare, improving production outcomes, and promoting sustainability in the dairy industry.
Looking ahead, the study paves the way for future enhancements in automated BCS assessment for dairy cows. Prospective research directions include the integration of multi-modal data for a more comprehensive dataset, exploration of transfer learning techniques, fine-tuning model hyper-parameters, assessing robustness to environmental factors, optimizing for real-time implementation, developing user-friendly interfaces, and collaborating with veterinary experts to refine assessment criteria. These endeavors aim to bolster the accuracy, applicability, and practicality of automated BCS systems, facilitating their seamless integration into dairy farming practices for improved herd management and health monitoring.
Author Contributions
Conceptualization, J.G.L. and S.H.; methodology, S.H.; software, S.H. and N.D.T.; validation, S.S.L., S.H., H.-P.N., H.-S.S., M.A. and S.M.L.; formal analysis, M.A. and C.G.D.; investigation, J.G.L. and M.N.P.; resources, H.-S.S., M.A. and M.N.P.; data curation, S.H., S.S.L., M.A., M.N.P., M.K.B. and H.-P.N.; writing—original draft preparation, S.H. and H.-P.N.; writing—review and editing, S.H. and H.-P.N.; visualization, M.K.B., H.-P.N. and N.D.T.; supervision, J.G.L. and S.S.L.; project administration, J.G.L., M.A. and S.M.L.; funding acquisition, J.G.L., M.N.P., S.S.L., H.-S.S., M.A., S.M.L. and C.G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) and Korea Smart Farm R&D Foundation (KosFarm) through Smart Farm Innovation Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) and Ministry of Science and ICT (MSIT), Rural Development Administration (421011-03).
Institutional Review Board Statement
This animal care and use protocol was reviewed and approved by the IACUC at National Institute of Animal Science (approval number: NIAS 2022-0545).
Data Availability Statement
The datasets generated during this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors, Hoang-Phong Nguyen, Duc Toan Nguyen, Min Ki Baek, and Seungkyu Han were employed by the company ZOOTOS Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI |
Artificial Intelligence |
| BCS |
Body Condition Score |
| CNN |
Convolutional Neural Network |
| DL |
Deep Learning |
| DNN |
Deep Neural Network |
| PCA |
Principle Component Analysis |
| ML |
Machine Learning |
| MLPs |
Multi-Layer Perceptrons |
| MSE |
Mean Squared Error |
| MAE |
Mean Absolute Error |
| MAPE |
Mean Absolute Percentage Error |
| POV |
Point of View |
| RMSE |
Root Mean Squared Error |
| RANSAC |
Random Sample Consensus |
| SGD |
Stochastic Gradient Descent |
References
- Herron, J.; O’Brien, D.; Shalloo, L. Life cycle assessment of pasture-based dairy production systems: Current and future performance. Journal of Dairy Science 2022, 105, 5849–5869. [Google Scholar] [CrossRef] [PubMed]
- Douphrate, D.I.; Hagevoort, G.R.; Nonnenmann, M.W.; Lunner Kolstrup, C.; Reynolds, S.J.; Jakob, M.; Kinsel, M. The dairy industry: a brief description of production practices, trends, and farm characteristics around the world. Journal of agromedicine 2013, 18, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Patton, S. Milk: Its remarkable contribution to human health and well-being; Routledge, 2017. [Google Scholar]
- Truman, C.M.; Campler, M.R.; Costa, J.H.C. Body Condition Score Change throughout Lactation Utilizing an Automated BCS System: A Descriptive Study. Animals 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.; Schutz, M. An interdisciplinary review of body condition scoring for dairy cattle. The professional animal scientist 2008, 24, 507–529. [Google Scholar] [CrossRef]
- Roche, J.R.; Friggens, N.C.; Kay, J.K.; Fisher, M.W.; Stafford, K.J.; Berry, D.P. Invited review: Body condition score and its association with dairy cow productivity, health, and welfare. Journal of dairy science 2009, 92, 5769–5801. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Bhakat, C.; Mondal, S.; Mandal, A. An observational study investigating uniformity of manual body condition scoring in dairy cows. Ind. J. Dairy Sci 2020, 73, 77–80. [Google Scholar] [CrossRef]
- Zieltjens, P. A comparison of an automated body condition scoring system from DeLaval with manual, non-automated, method. Master’s thesis, 2020.
- Bell, M.J.; Maak, M.; Sorley, M.; Proud, R. Comparison of methods for monitoring the body condition of dairy cows. Frontiers in Sustainable Food Systems 2018, 2, 80. [Google Scholar] [CrossRef]
- Wildman, E.; Jones, G.; Wagner, P.; Boman, R.; Troutt Jr, H.; Lesch, T. A dairy cow body condition scoring system and its relationship to selected production characteristics. Journal of dairy science 1982, 65, 495–501. [Google Scholar] [CrossRef]
- Dairy Goat Body Condition Scoring. Available online: https://adga.org/dairy-goat-body-condition-scoring/ (accessed on 11 January 2024).
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press, 2016. [Google Scholar]
- Micle, D.E.; Deiac, F.; Olar, A.; Drența, R.F.; Florean, C.; Coman, I.G.; Arion, F.H. Research on innovative business plan. Smart cattle farming using artificial intelligent robotic process automation. Agriculture 2021, 11, 430. [Google Scholar] [CrossRef]
- Dang, C.G.; Lee, S.S.; Alam, M.; Lee, S.M.; Park, M.N.; Seong, H.S.; Han, S.; Nguyen, H.P.; Baek, M.K.; Lee, J.G.; Pham, V.T. Korean Cattle 3D Reconstruction from Multi-View 3D-Camera System in Real Environment. Sensors 2024, 24. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Dai, B.; Shen, W.; Sun, Y.; Zhao, K.; Zhang, Y. Automatic estimation of dairy cow body condition score based on attention-guided 3D point cloud feature extraction. Computers and Electronics in Agriculture 2023, 206, 107666. [Google Scholar] [CrossRef]
- Yukun, S.; Pengju, H.; Yujie, W.; Ziqi, C.; Yang, L.; Baisheng, D.; Runze, L.; Yonggen, Z. Automatic monitoring system for individual dairy cows based on a deep learning framework that provides identification via body parts and estimation of body condition score. Journal of dairy science 2019, 102, 10140–10151. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J. Implementation of a body condition scoring program in dairy herds. Feeding and managing the transition cow. Proc. Penn. Annu. Conf., Univ. of Pennsylvania, Center for Animal Health and Productivity, 1996.
- Roche, J.; Dillon, P.; Stockdale, C.; Baumgard, L.; VanBaale, M. Relationships among international body condition scoring systems. Journal of dairy science 2004, 87, 3076–3079. [Google Scholar] [CrossRef]
- Dang, C.; Choi, T.; Lee, S.; Lee, S.; Alam, M.; Lee, S.; Han, S.; Hoang, D.T.; Lee, J.; Nguyen, D.T. Case Study: Improving the Quality of Dairy Cow Reconstruction with a Deep Learning-Based Framework. Sensors 2022, 22, 9325. [Google Scholar] [CrossRef]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Communications of the ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Maćkiewicz, A.; Ratajczak, W. Principal components analysis (PCA). Computers & Geosciences 1993, 19, 303–342. [Google Scholar]
- Sager, C.; Zschech, P.; Kuhl, N. LabelCloud: A lightweight labeling tool for domain-agnostic 3D object detection in point clouds. Computer-Aided Design and Applications 2022, 19, 1191–1206. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3d classification and segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 652–660.
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint, 2014; arXiv:1409.1556. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. International conference on machine learning. PMLR, 2019, pp. 6105–6114.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).