1. Introduction
Belowground biodiversity regulates aboveground biodiversity and ecosystem functioning [
1], especially microbial diversity could drive multifunctionality in terrestrial ecosystems, including climate regulation, soil fertility, and food and fiber production [
2,
3], and improve human well-being [
4]. As an important component of terrestrial ecosystems, microbial biodiversity in forests has received much attention as macroorganisms, for example plants [
5,
6]. Also, it has been recommended to apply the existing macro-ecological theory to soil microbial ecology [
7]. Different environmental drivers regulate soil microbial diversity. For instance, temperature and soil carbon regulate soil archaea, while aridity, vegetation attributes, and pH regulate bacteria [
8]. Changes in the soil bacterial community during secondary succession has been found to depend on plant diversity and composition and soil nutrients especially for total organic carbon and total nitrogen [
9], while soil fungal diversity and functionality were found to be driven by plant species in afforestation [
10]. Compared with arable land, forest ecosystems had a more stable and complex microbial network in the karst region [
11].
The karst landscape is mostly distributed in the southwest region of China with a size more than 0.54 million km
2 [
12], which is characterized as being susceptible to disturbances, unstable, and unable to self-adjust [
13,
14,
15] due to slow species turnover and soil poverty [
16]. The karst region has become a hot spot of global greening with substantial increases in vegetation growth and carbon stocks due to ecological engineering [
17]. In addition, natural vegetation restoration has been documented to be superior to managed vegetation restoration for maintenance of multiple ecosystem functions in the karst regions due to the potentially significant role of arbuscular mycorrhizal fungi [
18]. The importance of ecological networks for microbial communities has been the focus of research on natural and agricultural ecosystems [
8,
19]. Karst forests in particular have greater connectivity among bacterial and fungal communities than non-karst forests, which indicated that increased microbial diversity strengthen the complexity of co-occurrence networks [
20]. However, there is a limited knowledge about the magnitude and direction of the response of soil microbial communities to karst forest succession with characterized plant communities and soil properties [
21]. Gaining this knowledge is vital to efforts that increase ecosystem stability and function in the context of international carbon sequestration and carbon neutrality goals.
Soil bacteria and fungi respond differently to plant diversity and plant family composition during the secondary succession of abandoned farmland on the Loess Plateau [
22]. Based on the point and dynamics of woody plant diversity and composition among shrubland, secondary forest, and primary forest in the karst region of Southwest China [
21], we posited that soil microbial (i.e., bacteria and fungi) communities and diversity would respond differently as forest succession. According to the different microbial profiles [
23], we also expected that soil bacterial co-occurrence would have higher connectivity than fungi as forest succession progressed. Here, we collected 11 soil samples in each plot along a restoration gradient in shrub, secondary forest, and primary forest in the karst region of Southwest China. We sequenced amplicons of 16S rRNA gene and ITS gene to obtain information on the community composition and diversity of soil bacteria and fungi. Then, we quantified the responses of the soil microbial community, diversity, and co-occurrence network to karst forest succession. Finally, we investigated the main factors that drove the soil microbial dynamics along karst forest succession.
2. Materials and Methods
2.1. Study Area
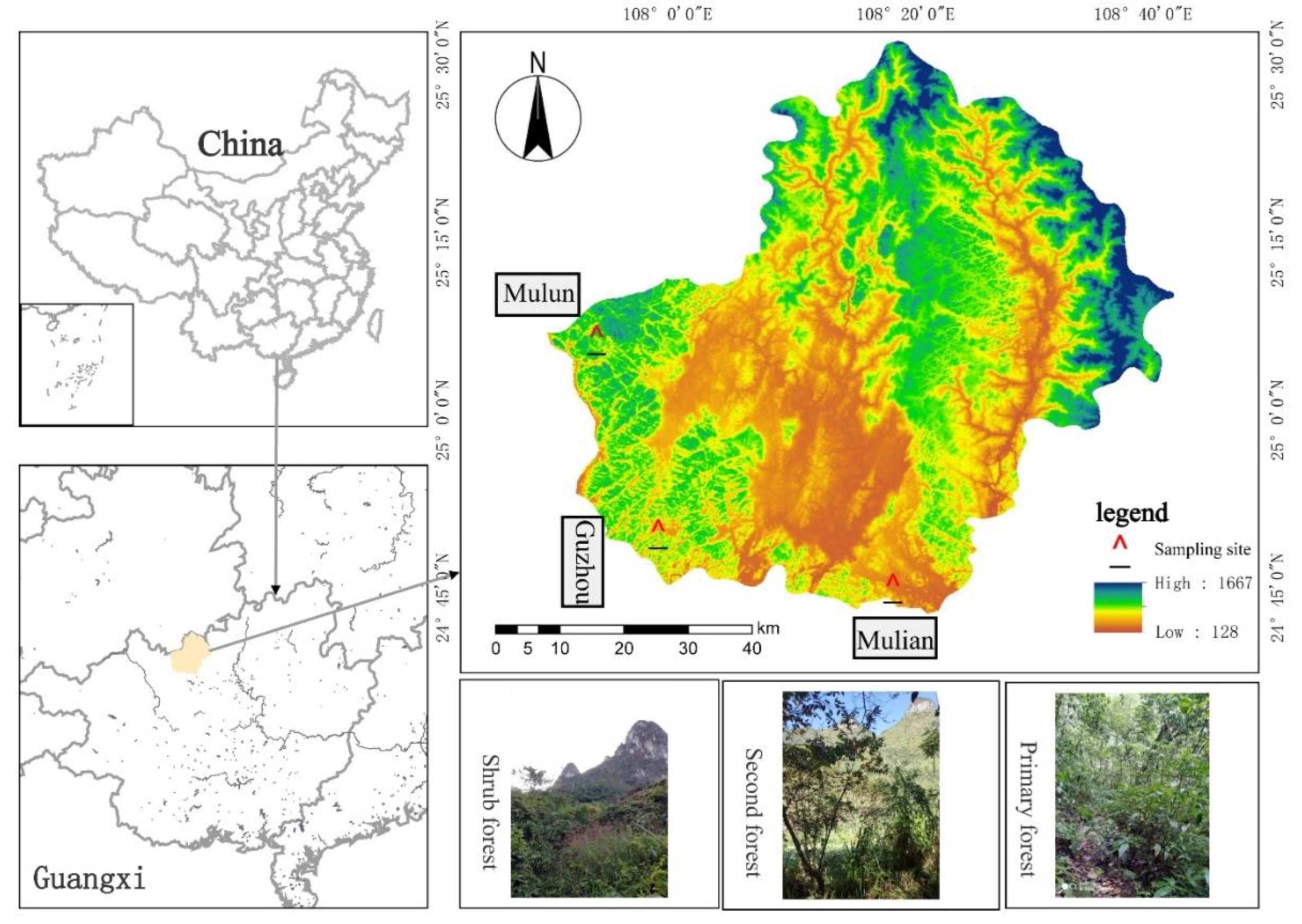
The study area was in Huanjiang Maonan Autonomous County, Guangxi (107°51'- 108° 43' E, 24°44' - 25°33' N), which is in the subtropical monsoon climate zone. The annual average temperature is 19.3 ℃, the annual average sunshine hours are 1451.1 h, and the annual average precipitation is 1529 mm. The shrub, secondary forest, and primary forest are the typical natural forests in the karst region, and the respective plots are built in Mulian Karst Experimental Station (Mulian), Guzhou in Xianan Township (Guzhou), and the Mulun National Nature Reserve (Mulun). The dominant woody vegetation in the shrubland plot in Mulian were Vitex negundo, Alangium chinense, and Ligustrum japonicum (Zhang et al., 2020). Bauhinia brachycarpa, Cipadessa cinerascens, Radermachera sinica, and Toona sinensis were the dominant species in the secondary forest plot in Guzhou. Both the shrubland and secondary forests were typical natural restoration areas after human disturbance. The dominant species in the primary forest in Mulun were Cryptocarya microcarpa, Itoa orientalis, and Brassaiopsis glomerata. Mulun is the best preserved and largest primary karst forest with mixed evergreen and deciduous broadleaf forest [
16]. The soil in the three regions is lime soil, and the site conditions are identical.
2.2. Vegetation Investigation
In 2007, dynamic forest plots of shrubland, secondary forest, and primary forest with an area of 220 m × 40 m were established from the valley to the top of the hills in the Mulian, Guzhou, and Mulun regions, respectively. The plots were divided into 22 quadrants of 20 m × 20 m, which then were divided into 16 sub-quadrants of 5 m × 5 m according to the standard protocol from the Center for Tropical Forest Science (CTFS,
http://www.ctfs.si.edu). All the woody plants with diameter at breast height (DBH) ≥ 1 cm were tagged, identified, measured, and georeferenced in 2007. Then every five years an inventory was conducted, which occurred in 2012, 2017, and 2022. We used woody plant inventory data from 2017 in the middle of the 20 m × 20 m quadrants along the three plots. We used the average DBH, richness, and Shannon-wiener index as vegetation factors in our analyses. DBH was exploited the average of the total woody plants in each plot. Richness index and Shannon index were determined as described in reference [
24].
2.3. Soil Sample Collection and Determination
In October 2019, we took soil samples every 20 m (i.e., the middle sample point) along the middle sample line of the plot from bottom to top, and we measured the soil temperature and volume water content at the sampling point with the soil parameter instrument TDR200. Eight to 10 surface soil samples (0-15 cm) around the sampling points were taken as the soil sample after fully mixing. About 150 g of soil sample from each point was stored in a liquid nitrogen tank and taken to the laboratory for high-throughput sequencing of soil microorganisms. We screened soil samples (about 500 g) through a 10-mesh sieve to remove roots and stones. One part was stored in the freezer (4 ℃) to measure the soil ammonium nitrate and microbial biomass (C, N, P), and the other part was used to determine the soil physio-chemical properties.
Soil pH, organic carbon (SOC), total nitrogen (TN), total phosphorus (TP), total potassium (TK), available nitrogen (AN), available phosphorus (AP), available potassium (AK), exchangeable Ca2+ and Mg2+, NO3-N, and NH4+-N were determined according to reference [
25]. Soil microbial biomass carbon (MBC), nitrogen (MBN), and phosphorus (MBP) were determined using the chloroform fumigation-extraction method [
26].
2.4. DNA Extraction and PCR Amplification
Soil microbial DNA was extracted from each soil sample three times from a 0.5 g fresh soil sample with the Fast soil DNA SPIN Kit (MP Biomedicals, MP, USA). The extracted soil DNA was diluted in 50 μL of sterilized water. Finally, the extracted DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, USA) and was kept at - 80 °C for further analysis.
The hypervariable region V3–V4 of total bacterial 16S rRNA and the internal transcribed spacer (ITS) regions of fungal genes were amplified with the primers 338F/806R [
27] and ITS1F/ITS2R [
28], respectively. The solution for bacterial amplification included 4μL FastPfu Buffer (5 ×), 2 μL dNTPs (2.5 mM), 0.8 μL Forward Primer (5 μM) and Reserve Primer (5 μM), 0.4 μL FastPfu Polymerse (China, Beijing TransGen Biotech Co., Ltd.), 0.2 μL BSA, 10 ng Template DNA, and adding ddH
2O
2 to 20 μL. These samples were denatured at 95 °C for 3 min, amplified by 27 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, followed by extension at 72 °C for 10 min. The qPCR reaction of ITS rRNA were performed in a 20 μL mixture, which contained 2μL Buffer (10 ×), 2 μL dNTPs (2.5 mM), 0.8 μL Forward and Reserve Primers (5 μM), 0.2 μL rTaq Polymerse (China, Shanghai Fusheng Industrial Co., Ltd.), 0.2 μL BSA, 10 ng Template DNA, and ddH2O. These samples were denatured at 95 °C for 3 min, amplified by 35 cycles of 95 °C for 30s, 55 °C for 30 s, and 72 °C for 45s, extended at 72 °C for 10 min, at 10 °C until halted. Each sample was conducted on ABI GeneAmp 9700 system (Applied Biosystems, Waltham, MA, USA) with three duplicates and then the relative amplicons were mixed to a final PCR product. Each mixed gene (i.e., 16S rRNA gene and ITS rRNA gene) sample was undergone by electrophoresis on 2% agarose gel. Bands with DNA fragments of the expected size (301-400 bp for 16S rRNA gene, 201-300 bp for ITS rRNA gene) were assessed with AxyPrep DNA Gel Recovery Kit (Axygen Biosciences (Hangzhou) Co. Ltd., Hangzhou, China). Finally, the amplicon libraries were sequenced on an Illumina’ HiSeq 2000 platform (Illumina, USA) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
2.5. Bioinformatics Processing
The raw gene sequencing reads were demultiplexed, quality-filtered, and then merged using QIIME [
29] according to the three criteria as described in reference [
30]. The operational taxonomic units (OTUs) were clustered at the similarity level of 97% using Uparse v7.0.1 [
31] after chimeric sequences were identified and removed. The taxonomy of the OTUs were identified based on the clustering results using a naïve Bayesian classifier algorithm implemented in mother [
32,
33] against a 16S rRNA database (Silva v138/16s_bacteria) and an ITS rRNA database (unite 7.2/its_fungi) at a 0.7 confidence threshold. Finally, the complete datasets were sent to the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information (NCBI) under the accession numbers of PRJNA 898882 for bacteria and PRJNA 899297 for fungi.
2.6. Statistical Analysis
The α diversity of the soil microbial community was measured using Shannon-Wiener index [
34] and observed richness as the total number of OTUs in each sample normalized to a specific number of reads per sample. The β diversity of the soil microbial communities in different forest types was characterized by non-metric multidimensional scaling (NMDS) based on the Bray-Curtis distance. The inter-group differences were tested using ADONIS analysis, and the stress values were used to assess the goodness of fit [
35]. Specifically, stress >0.2 indicated poor goodness of fit, 0.1<stress<0.2 indicated fair goodness of fit, 0.05<stress<0.1 meant good fitness, and stress <0.05 meant excellent fitness. In our study, stress values (0.0801 for bacteria and 0.153 for fungi) indicated that the NMDS results had good fitness. The Wilcoxon rank sum test was used to analyze the differences in the soil microbial diversity index [
36]. The top 50 genera with relative abundance > 1% were thermally mapped to analyze the soil microbial community structure of each forest. Redundancy analysis based on Bray-Curtis distance (db-RDA) was used to investigate the dominant factors that affected the composition of bacterial and fungal communities [
37]. Variation partition analysis was conducted to quantify the contribution of soil, vegetation, and microbial biomass to the variance variation of soil microbial community [
38]. The above analyses were conducted in R 3.3.4 [
39]. To construct the co-occurrence network of the bacterial and fungal community, we used the relative abundance of the taxa with > 1% relative abundance at genus level in the three forests to calculate the Pearson correlation [
40,
41]. The edges were retained only when their Pearson correlation was >0.7 and adjusted P values was < 0.05. The network was visualized with Gephi 0.9.2.
3. Results
3.1. Plants and Soil Properties during Karst Forest Succession
The average DBH, richness, and Shannon-Wiener index of the woody plants had an upward trend with forest succession from shrubland, to secondary forest, to primary forest. In addition, all the three woody plant properties in the primary forest were significantly higher than those in the shrubland (P<0.05) (
Table 1). The soils had a high soil organic carbon content (>30 g⋅kg-1) in the shrubland and primary forest (
Table 1). The soil properties, except for microbial biomass of carbon (MBC), were significantly different among the three karst forests, and in general soil nutrients in the primary forest were highest (
Table 1).
3.2. Dynamics in Bacterial and Fungal Community Composition and Diversity during Forest Succession
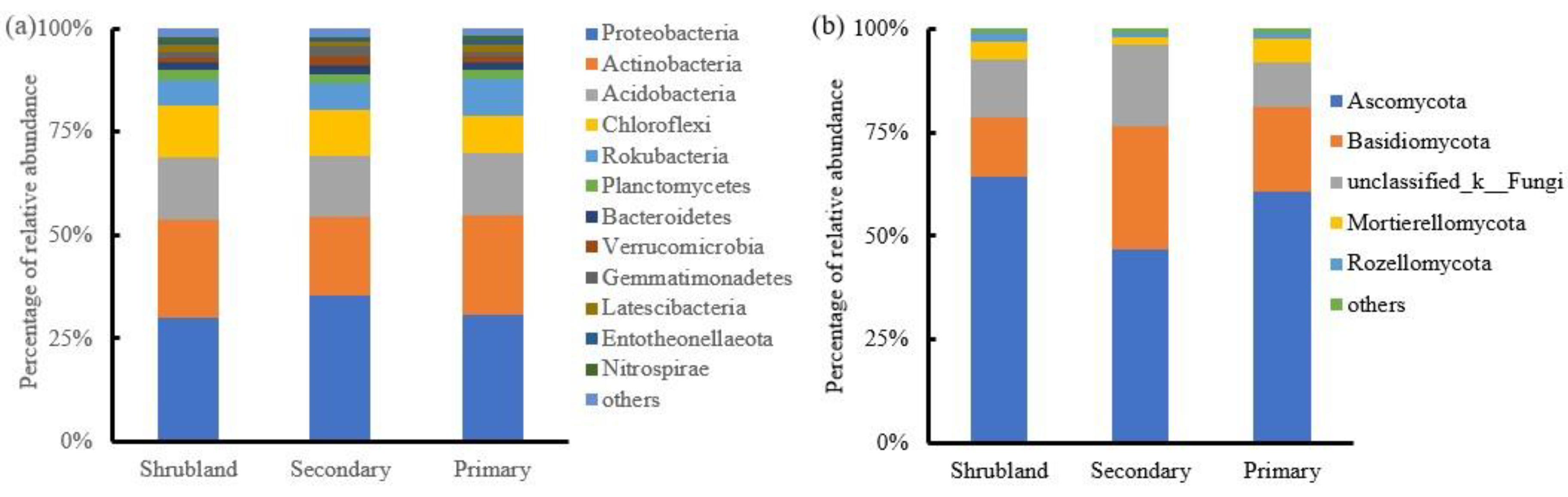
Proteobacteria, Actinobacteria, Acidobacteria, and Chloroflexi were the predominant bacterial phyla in the karst forest soils and accounted for 78.7%-81.3% of the total abundance (
Figure 1a). Ascomycota and Basidiomycota dominated the fungal communities in the karst forests with a relative abundance of 76.4% to 81.2% (
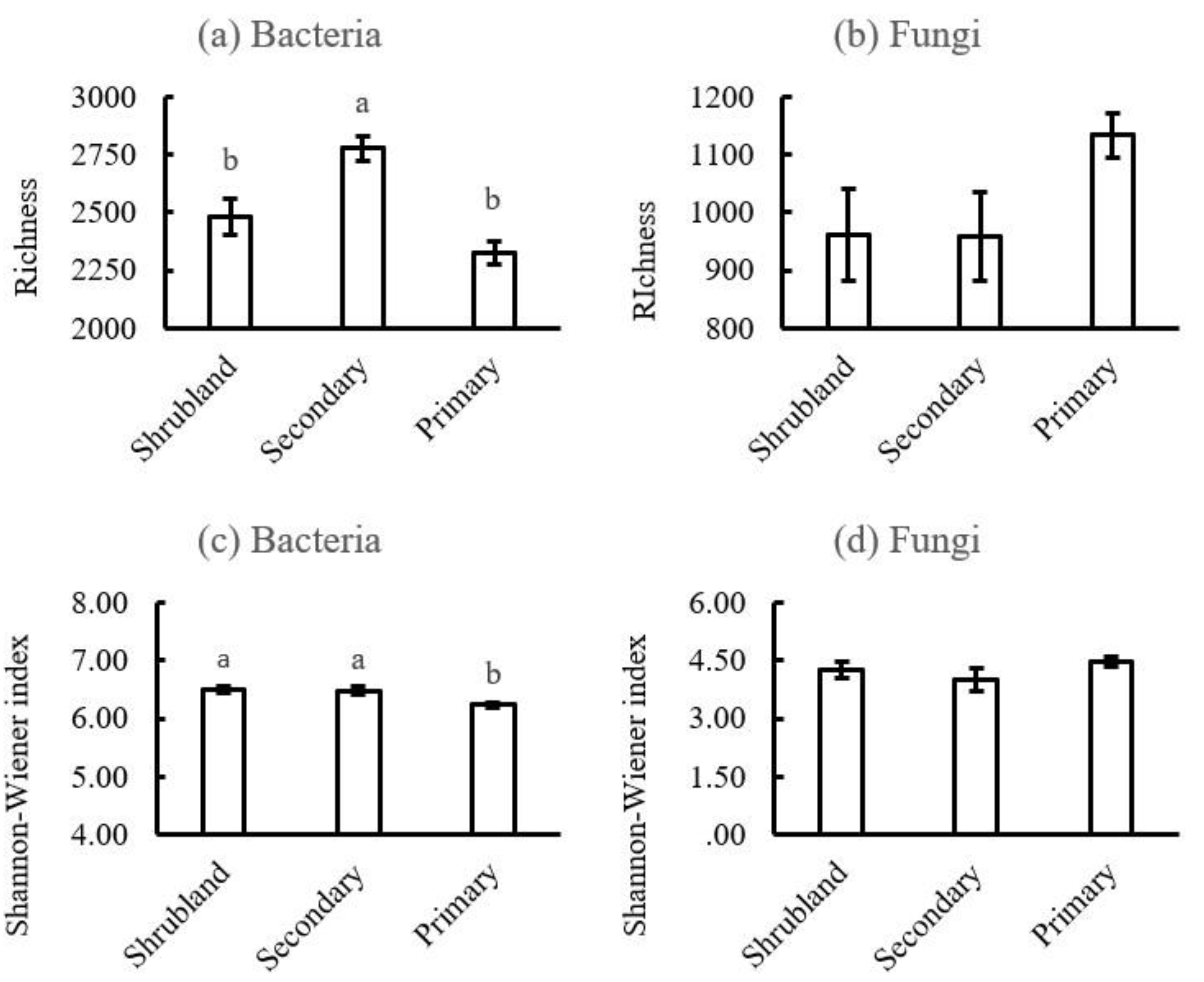
Figure 1b). The α-diversity (based on the richness and Shannon-Wiener index) of fungi had a downward trend at first and then an upward trend during karst forest succession, but there were no significant differences among them (
Figure A2b,d). However, the sobs of soil bacteria at the phylum level were significantly higher in the secondary forest than those in shrubland and primary forest (
Figure A2a), while the Shannon-Wiener index for soil bacteria at the phylum level was significantly lower for the primary forest than for the shrubland and secondary forest (
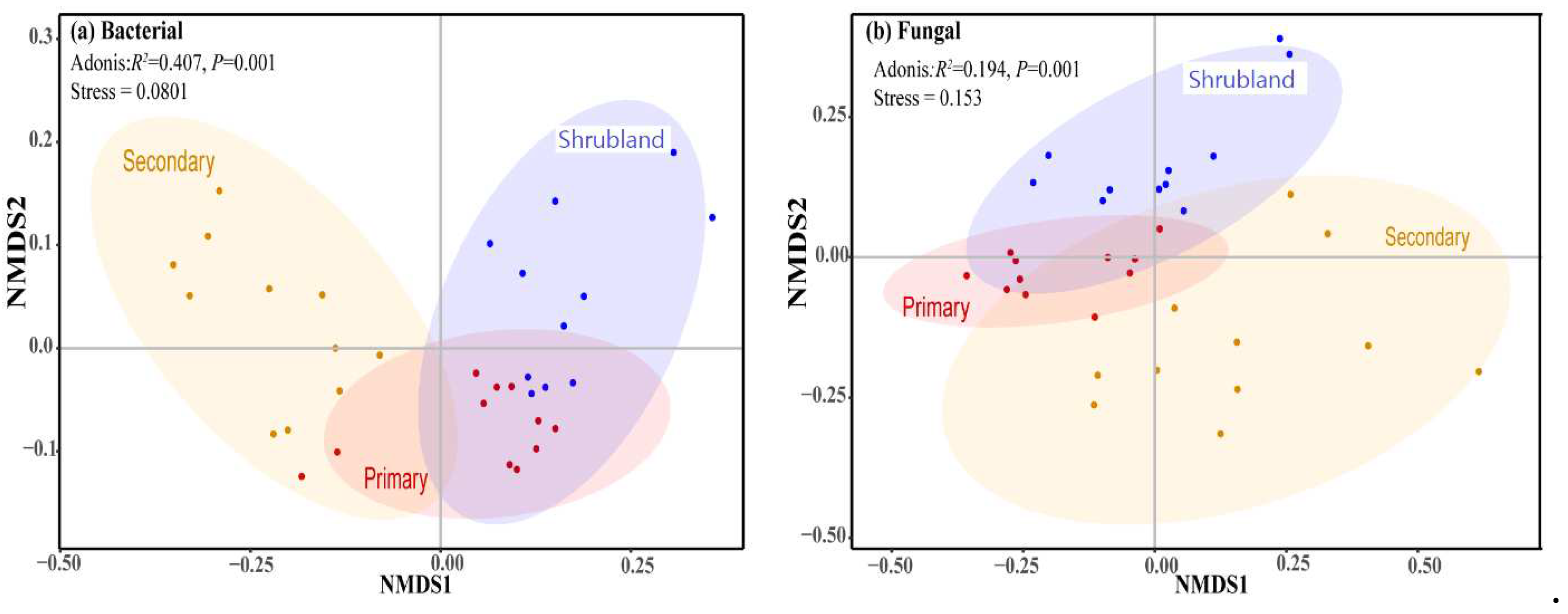
Figure A2c). The NMDS analysis indicated that the soil bacterial community in the primary forest was close to that in the shrubland, with obvious differences from the secondary forest (
Figure 2a, Adonis,
R2=0.407,
P=0.001), and the fungal community could be discriminated among the three forests (
Figure 2b, Adonis,
R2=0.194,
P=0.001).
3.3. Co-Occurrence Networks for Soil Bacteria and Fungi in the Karst Forest Succession
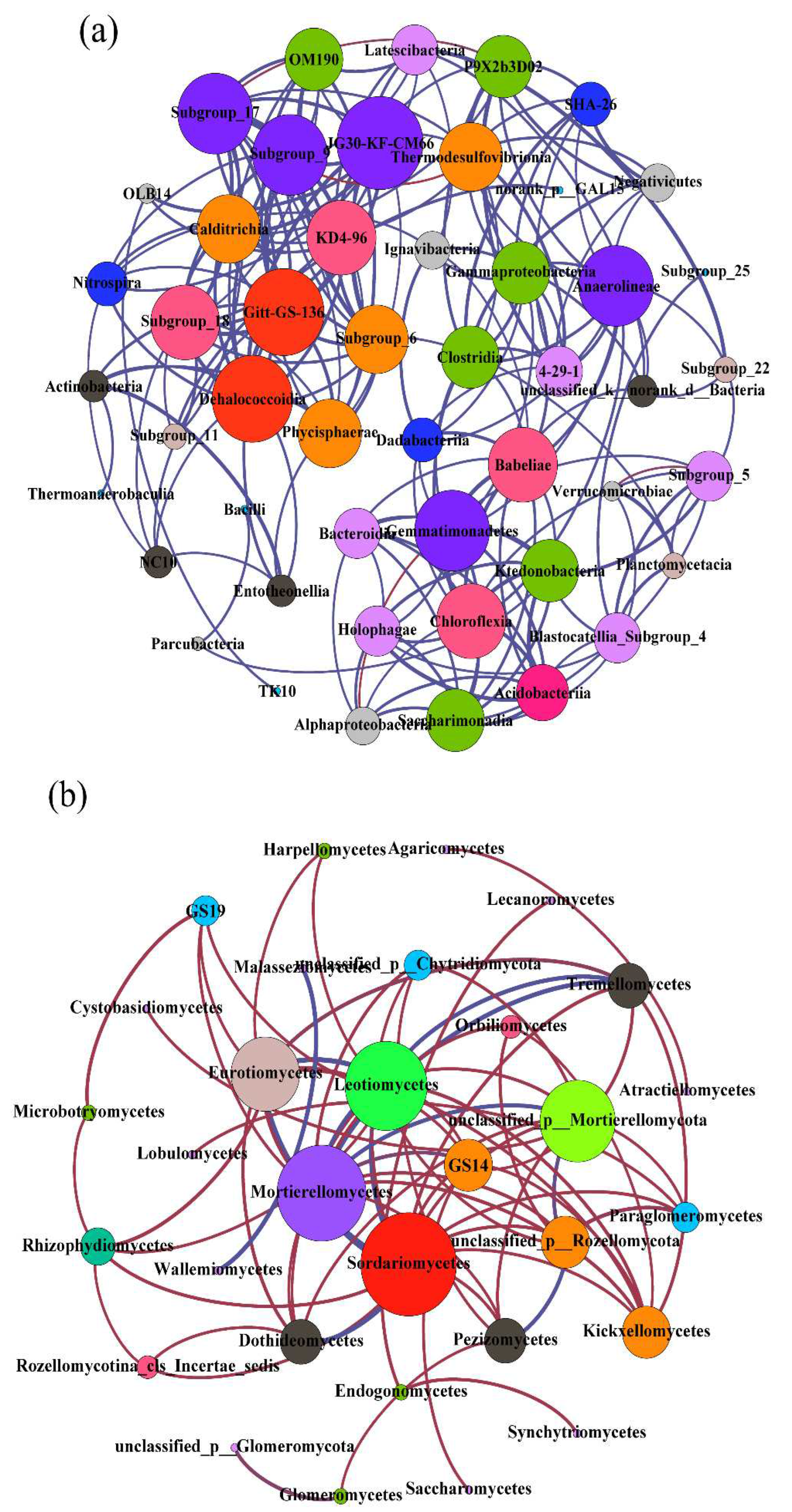
We constructed co-occurrence networks for soil bacteria and fungi for the three karst forests (
Figure 3). The networks were derived from the abundant taxa with a relative abundance >1% at the genus level, which for bacteria and fungi comprised 124 and 24 edges, and 37 and 17 nodes, respectively (
Table A1). JG30-KF-CM66, Subgroup_17, Subgroup_9, Anaerolineae, Dehalococcoidia, Gitt-GS-136, Gemmatimonadetes, and Chloroflexia were the most important nodes in the bacterial co-occurrence network of the three karst forests (
Figure 3a). Sordariomycetes, Mortierllomycetes, Eurotiomycetes, Leotiomycetes, GS-14, and Kickxellomycetes were the most important nodes of fungi in the karst forests (
Figure 3b). The number of nodes, edges, and average path length of bacteria were relatively higher than for fungi in the karst forests (
Figure 3 and
Table A1), which indicated that there was a small but intense correlation among soil fungi in the forest.
3.4. Drivers Regulating Soil Bacterial and Fungal Profiles among Karst Forests
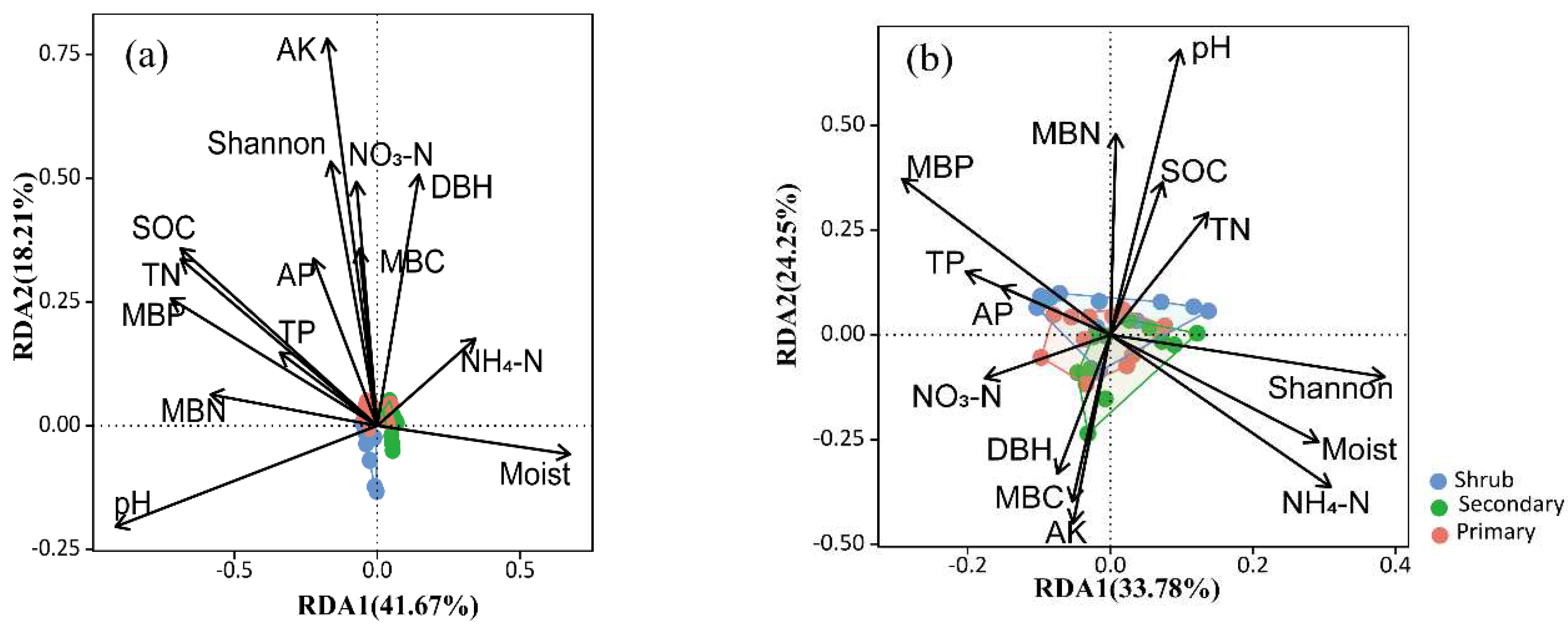
The db-RDA results showed that soil pH, SOC, TN, MBP, moisture, MBN, AK, and the Shannon index of woody plants significantly affected the soil bacterial community in the karst forests (
Figure 4a), and that only soil pH significantly affected the soil fungal community in the karst forests (
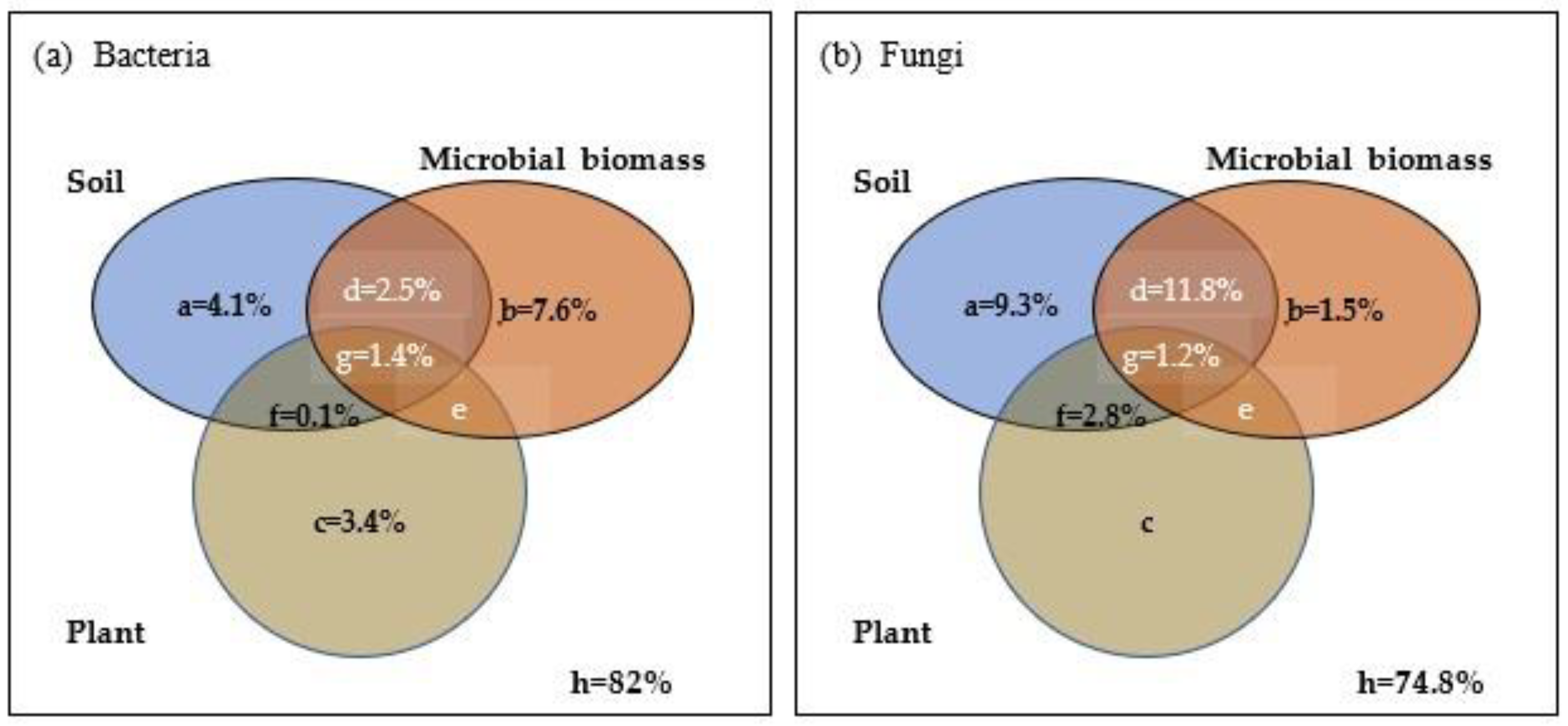
Figure 4b). Further variation partition analysis indicated that soil microbial biomass explained 7.6% of the variation in the soil bacterial community in the karst forests, while soil properties and plant factors explained 4.1% and 3.4% of the variation in the soil bacterial community, respectively (
Figure 5a). Soil properties explained 9.3% of the variation in the soil fungal community in the karst forests (
Figure 5b). Woody plants had little effect on the soil fungal community (
Figure 5b). The unexplained variation was large for both the soil bacterial and fungal communities, which were 82.0% and 74.8%, respectively (
Figure 5).
4. Discussion
4.1. Dynamics in Soil Microbes and Environmental Variables along Karst Forest Succession
Ecological engineering, including natural restoration, has been implemented worldwide [
42]. One such region is in the karst region of southwest China [
14,
16], which has indubitably increased carbon sequestration through accumulation in biomass and soil organic carbon [
27]. Moreover, plant natural succession without human or animal disturbance altered vegetation growth, community composition, and productivity [
21], and in turn changed belowground status and functions [
21,
43]. In our study, Proteobacteria, Actinobacteria, and Acidobactria dominated the bacterial community at the phylum level (
Figure 1a), and Ascomycota and Basidiomycota dominated the fungal communities (
Figure 1b), regardless of the forest succession stage in the karst region. The dominant bacterial and fungal taxa at phylum level were agreed with the previous studies in the karst region [
19,
44], indicating that forest succession had no significant effects on the dominant soil bacterial and fungal community structure. It is worthy to noted that, compared with the shrubland, the Proteobacteria communities were higher in the secondary forest (
Figure 1a), which indicated that soil conditions improved during early plant succession by favoring Proteobacteria in a copiotrophic environment with available labile substrates [
45]. The Protebacteria community increased with secondary succession after abandonment in the Loess Plateau in China [
8] and with vegetation succession along the Franz Josef chronsequence in New Zealand [
46].
We found that the bacterial α-diversity was lowest in the primary forest (Figure A3a,c) despite the high diversity of woody plants (
Table 1), which juxtaposes the view that plant diversity is positively related to soil bacterial diversity [
47,
48]. This contradiction might have been due to the differences in the substrates in the forest ecosystems of our study versus the grasslands studied [
47,
48]. In our study, fungal α-diversity had a downward trend at first and then an upward trend during karst forest succession (Figure A3b,d), which indicated that the different responses of the fungal and bacterial communities to forest succession may be due to their different responses to changing soil properties during forest succession [
49].
4.2. Divergent Patterns of Bacteria and Fungi along Karst Forest Succession
NMDS results showed that the both the soil bacterial and fungal community composition in the primary forest were close to those in the shrubland, compared with the secondary forest (
Figure 2). The phenomenon may be caused by two reasons. On the one hand, it may be the similar soil physical and chemical properties under the shrubland and the primary forest (
Table 1) (i.e., similar soil environment) for microbial survival and growth; on the other hand, it may be different woody plant composition in the primary forest had more proportional evergreen tree species [
20,
50] with higher C:N ratios than deciduous tree species [
51]. Although their litter would be preferred by microorganisms, the amount was relatively less than deciduous tree species, which caused the microbial community composition close to that in the shrubland owning more deciduous trees [
50]. However, in subtropical non-karst regions, the soil bacterial community could be discriminated along forest succession, which may be caused by an increase in the production and accumulation of bacterial residues as forest succession progressed [
52]. The divergence indicated that karst forests might have a more complicated succession process than non-karst forest. Generally, the soil fungal community was discriminated along the forest succession (
Figure 2b), which implied that soil fungal species or taxa varied with successional stages. For example, arbuscular mycorrhizal (AM) fungi are more likely to be fast colonizers of early successional habitats [
53]. Our microbial co-occurrence networks results (
Figure 3 and
Table A1) also indicated less but more intense correlations among soil fungi in the forest than bacteria, which might be caused by more intense and stable interactions between fungal species [
19], such as diazotrophs and arbuscular mycorrhizal fungi [
54]. Our findings also implied that bacteria and fungi might have more diverse trajectories along the forest succession stages in karst regions than in non-karst regions (i.e., the Loess Plateau) [
49].
Researchers have pointed that the abundance of highly connected taxa (e.g., kinless hubs) within soil microbial networks were associated with high functional potential in terrestrial ecosystems [
55]. In our study, the co-occurrence network showed that JG30-KF-CM66, Subgroup_17, Subgroup_9, Anaerolineae, Dehalococcoidia, Gitt-GS-136, Gemmatimonadetes, and Chloroflexia were the most important bacterial nodes, and and Kickxellomycetes were the most important fungal nodes (
Figure 3b). These nodes were mainly connected to other taxa in the networks, which implied that they were highly associated with functional potential. Specifically, JG30-KF-CM66 has been documented to participate in the global cobalamin production through the cobinamide to cobalamin salvage pathway [
56]. Sordariomycetes are comprised of typical saprotrophic fungi which are efficient in decomposing labile C [
57], and Mortierllomycetes respond strongly to easily degradable, N-rich substrates [
58]. Thus, the co-occurrence patterns suggested that species interactions contributed more to soil nutrient processes or functions than microbial diversity [
19].
4.3. Drivers of Soil Microbial Dynamics along Karst Forest Succession
Our db-RDA results and variance partition analysis indicated that soil properties (pH, SOC, TN, moisture, and AK), soil microbial biomass (i.e., MBP, MBN), and plant diversity (Shannon index of woody plants) drove the dynamics of soil bacterial community along karst forest succession (
Figure 4a and
Figure 5a), which supported the view that shifts in bacterial community structure along plant secondary succession is most driven by changes in soil nutrients and plant diversity and composition [
8,
21]. Our findings also showed the phenomenon that woody plant diversity and biomass (DBH is highly positively related biomass [
59]) could trigger negative responses in soil bacterial diversity during forest succession (
Table A2). However, in unaffected grasslands [
47] and during plant secondary succession after farmland was abandoned [
8,
48], plant diversity was positively related with soil bacterial diversity. This finding indicated that the relationship between plants diversity and bacterial diversity would change along the succession stages. A possible reason for this change is that in the unaffected grasslands or early succession stages, plant diversity provided niches for bacteria. In the late succession stage (i.e., forest), woody plant biomass (DBH as the most important variables [
59]) was also negatively significantly correlated with soil bacterial diversity (
Table A2), which implied that higher woody plant diversity and biomass might be prone to provide stronger plant-soil feedbacks for the stability rather than diversity of soil bacteria.
As forest progressed, plant regenerated and greatly affected soil properties such as pH, organic inputs, and available nutrients (
Table 1). SOC, TN, AN, NO
3--N, and MBN significantly correlated with soil bacterial alpha diversity (
Table A2), indicating that soil carbon and nitrogen was important to the bacterial diversity [
60]. It is interesting that NH
4+-N did not significantly correlated with the relative abundances of the dominant bacterial phyla, while the reverse was true for NO
3-N (
Figure 4a,
Table A2), which agreed with findings that this variation in the bacterial communities might be caused by N fractions during forest succession [
61].
Plant diversity has been documented to affect soil fungal communities at the global scale [
62], which was supported by research on plant secondary succession on the Loess Plateau in China [
21]. In general, plant diversity and composition could affect fungal composition and diversity by providing diverse food resources (i.e., root exudates and litter) [
63,
64]. For example, Ascomycota and Basidiomycota participate in the decomposition and rhizodeposition of organic substrates [
63,
65]. However, it was observed that soil properties especially pH mostly explained the response of the soil fungal community to forest succession (
Figure 4b and
Figure 5b). Also, Ascomycota and Basidiomycota were significantly correlated with most of the soil properties, but not woody plant diversity (
Table A2). This finding indicated that fungal community compositions (i.e., dominant phyla, Ascomycota and Basidiomycota) and diversity responded significantly to forest succession that depended on soil pH, C, and N dynamics. This phenomenon might be explained by soil property dynamics during forest succession under the subtropical climate and unique karst habitat.
5. Conclusions
Forest succession had different effects on soil bacterial and fungal diversity, community composition, and co-occurrence patterns. Soil bacterial diversity in the secondary forest significantly differed from the shrubland and primary forest, while fungal diversity was clearly discriminated among the three stages of karst forest succession. Co-occurrence patterns indicated that fungi had less but more intense relationships than bacteria among the karst forests, which indicating that bacteria and fungi exhibited diverse strategies to forest succession. Moreover, soil properties (i.e., pH, SOC, TN, AK, MBP, MBN) and woody plant diversity (Shannon index of woody plants) collectively mediated the bacterial community, but soil properties especially pH controlled the fungal community. Therefore, changes in woody plant-induced soil nutrient status would predict the dynamics of soil bacterial and fungal community composition and diversity during forest succession.
Author Contributions
Conceptualization, Wanxia Peng, Tongqing Song, and Hu Du; methodology, Huijun Chen, and WX Peng; investigation, F Wang and HJ Chen; writing—original draft preparation, Q Peng, and M Song; writing—review and editing, Q Peng, M Song, and WX Peng; supervision, TQ Song, and Fuping Zeng. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by National Natural Science Foundation of China, grant numbers 31971487 and 42277245, and Bagui Scholarship Program of Guangxi Zhuang Autonomous Region.
Acknowledgments
The authors especially thank the help and assistance from Management Center for Guangxi Mulun National Nature Reserve.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
Figure A1.
Location of the study area.
Figure A1.
Location of the study area.
Figure A2.
Richness and Shannon-Wiener index of soil bacteria (a and c) and fungi (b and d) in the three karst forests. Different letters indicated significant differences between the two forest soils at P<0.05 level. Shrubland, shrubland; Secondary, secondary forest; Primary, primary forest.
Figure A2.
Richness and Shannon-Wiener index of soil bacteria (a and c) and fungi (b and d) in the three karst forests. Different letters indicated significant differences between the two forest soils at P<0.05 level. Shrubland, shrubland; Secondary, secondary forest; Primary, primary forest.
Table A1.
Topology parameters of soil bacterial and fungal networks in the three karst forests.
Table A1.
Topology parameters of soil bacterial and fungal networks in the three karst forests.
| Parameters |
Bacteria network |
Fungal network |
| Number of nodes |
37 |
17 |
| Number of edges |
124 |
24 |
| Average density |
0.186 |
0.176 |
| Transitivity |
0.581 |
0.476 |
| Network diameter |
7 |
4 |
| Average path length |
2.752 |
2.088 |
| Connectivity |
1 |
0 |
Table A2.
Pearson correlation between microbial diversity, major bacterial and fungal phyla, and environmental factors in the karst forests.
Table A2.
Pearson correlation between microbial diversity, major bacterial and fungal phyla, and environmental factors in the karst forests.
| |
BShannon |
FShannon |
Proteobacteria |
Actinobacteria |
Acidobacteria |
Chloroflexi |
Rokubacteria |
Ascomycota |
Basidiomycota |
Mortierellomycota |
| pH |
-.224 |
.136 |
-.457** |
.553** |
.078 |
-.039 |
.240 |
.574** |
-.452** |
.012 |
| SOC |
-.650** |
.200 |
-.197 |
.589** |
-.110 |
-.464** |
.424* |
.524** |
-.295 |
.118 |
| TN |
-.618** |
.117 |
-.225 |
.609** |
-.102 |
-.398* |
.411* |
.478** |
-.261 |
.000 |
| TP |
-.220 |
-.049 |
-.146 |
.280 |
-.055 |
-.151 |
.204 |
.031 |
-.045 |
.402* |
| TK |
.394 |
-.252 |
.381* |
-.466** |
-.020 |
.137 |
-.411* |
-.532** |
.326 |
-.229 |
| AN |
-.748** |
.275 |
-.065 |
.492** |
-.149 |
-.577** |
.447** |
.437*
|
-.202 |
.054 |
| AP |
-.323 |
.328 |
-.069 |
.192 |
.006 |
-.354 |
.215 |
.285 |
-.163 |
.021 |
| AK |
-.486** |
.038 |
-.157 |
.443 |
.093 |
-.555** |
.331 |
-.031 |
.218 |
-.367 |
| Ca |
-.503** |
.043 |
-.264 |
.568** |
-.086 |
-.290 |
.394* |
.542** |
-.355* |
-.076 |
| Mg |
-.297 |
.148 |
-.462** |
.578** |
.058 |
-.108 |
.341 |
.530** |
-.294 |
.123 |
| Moist |
.104 |
-.094 |
.528** |
-.622** |
-.229 |
.011 |
.075 |
-.257 |
.018 |
.107 |
| Tem |
-.253 |
.367* |
-.217 |
.308 |
.039 |
-.077 |
-.067 |
.521** |
-.361* |
.113 |
| NH4-N |
.036 |
-.293 |
.172 |
-.028 |
-.044 |
-.025 |
-.215 |
-.201 |
.209 |
-.429* |
| NO3-N |
-.565** |
.251 |
.111 |
.032 |
-.093 |
-.504** |
.515** |
-.002 |
.008 |
.398* |
| MBC |
-.042 |
-.296 |
-.270 |
.181 |
.244 |
-.017 |
.078 |
.115 |
.160 |
-.530** |
| MBN |
-.578** |
.061 |
-.197 |
.510 |
-.018 |
-.481** |
.436* |
.393 |
-.140 |
-.023 |
| MBP |
-.238 |
.165 |
-.429* |
.482** |
.074 |
-.088 |
.225 |
.463** |
-.290 |
.088 |
| DBH |
-.357* |
.001 |
.112 |
.069 |
-.120 |
-.328 |
.321 |
-.031 |
.214 |
-.031 |
| S |
-.666** |
.095 |
-.018 |
.456** |
-.053 |
-.620** |
.446** |
.283 |
-.150 |
-.180 |
| Shannon |
-.535** |
-.028 |
.057 |
.386* |
-.092 |
-.452** |
.253 |
.212 |
-.111 |
-.350* |
| |
Proteobacteria |
Actinobacteria |
Acidobacteria |
Chloroflexi |
Rokubacteria |
Ascomycota |
Basidiomycota |
Mortierellomycota |
| Actinobacteria |
-.247 |
|
|
|
|
|
|
|
| Acidobacteria |
-.793** |
-.252 |
|
|
|
|
|
|
| Chloroflexi |
-.269 |
-.491** |
.347* |
|
|
|
|
|
| Rokubacteria |
-.103 |
.181 |
-.066 |
-.513** |
|
|
|
|
| Ascomycota |
-.254 |
.217 |
.153 |
-.172 |
.222 |
|
|
|
| Basidiomycota |
.014 |
-.059 |
.032 |
.048 |
-.020 |
-.783** |
|
|
| Mortierellomycota |
.145 |
-.045 |
-.183 |
-.236 |
.065 |
.095 |
-.154 |
|
| pH |
-.457** |
.553** |
.078 |
-.039 |
.240 |
.574** |
-.452** |
.012 |
| SOC |
-.197 |
.589** |
-.110 |
-.464** |
.424* |
.524** |
-.295 |
.118 |
| TN |
-.225 |
.609** |
-.102 |
-.398* |
.411* |
.478** |
-.261 |
.000 |
| TP |
-.146 |
.280 |
-.055 |
-.151 |
.204 |
.031 |
-.045 |
.402* |
| AP |
-.069 |
.192 |
.006 |
-.354* |
.215 |
.285 |
-.163 |
.021 |
| AK |
-.157 |
.443** |
.093 |
-.555** |
.331 |
-.031 |
.218 |
-.367* |
| Moist |
.528** |
-.622** |
-.229 |
.011 |
.075 |
-.257 |
.018 |
.107 |
| NH4-N |
.172 |
-.028 |
-.044 |
-.025 |
-.215 |
-.201 |
.209 |
-.429* |
| NO3-N |
.111 |
.032 |
-.093 |
-.504** |
.515**
|
-.002 |
.008 |
.398* |
| MBC |
-.270 |
.181 |
.244 |
-.017 |
.078 |
.115 |
.160 |
-.530** |
| MBN |
-.197 |
.510** |
-.018 |
-.481** |
.436* |
.393* |
-.140 |
-.023 |
| MBP |
-.429* |
.482** |
.074 |
-.088 |
.225 |
.463** |
-.290 |
.088 |
| DBH |
.112 |
.069 |
-.120 |
-.328 |
.321 |
-.031 |
.214 |
-.031 |
| Shannon |
.057 |
.386* |
-.092 |
-.452** |
.253 |
.212 |
-.111 |
-.350* |
References
- Bardgett, R.D.; van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, BK. Microbial diversity drivers multifuncionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef]
- Yang, G.W.; Wagg, C.; Veresoglou, S.D.; Hempel, S.; Rillig, M.C. How soil biota drive ecosystem stability. Trends Plant Sci. 2018, 23, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Wall, D.H.; Nielsen, U.N.; Six, J. Soil biodiversity and human health. Nature 2015, 528, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Condit, R.; Pitman, N.; Leigh Jr., E.G.; Chave J.; Terborgh, J.; Foster, R.B.; Núñez, V.P.; Aguilar, S.; Valencia, R.; Villa, G.; Muller-Landau, H.C.; Losos, E.; Hubbell, S.P. Beta-diversity in tropical forest trees. Science. 2002; 295, 666–669. [CrossRef]
- Wright, J.S. Plant diversity in tropical forests: a review of mechanisms of species coexistence. Oecologia 2002, 130, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.H.; Wang, C.K.; Jiang, L.F.; Luo, Y.Q. Trends in soil microbial communities during secondary succession. Soil Biol Biochem. 2017, 115, 92–99. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Reith, F.; Dennis, P.G.; Hamonts, K.; Powell, J.R.; Young, A.; Singh, B.K.; Bissett, A. Ecological drivers of soil microbial diversity and soil biological networks in the Southern Hemisphere. Ecology 2018, 99, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Z.; Bai, L.; Wang, J.Y.; Deng, J.; Ren, C.J.; Han, X.H.; Yang, G.H.; Wang, J. Change in soil bacterial community during secondary succession depend on plant and soil characteristics. Catena 2019, 173, 246–252. [Google Scholar] [CrossRef]
- Gil-Martínez, M.; López-García, Á.; Domínguez, M.T.; Kjøller, R.; Navarro-Fernández, C.M.; Rosendahl, S.; Marañón, T. Soil fungal diversity and functionality are driven by plant species used in phytoremediation. Soil Biol. Biochem. 2021, 153, 108102. [Google Scholar] [CrossRef]
- Cheng, X.Y.; Yun, Y.; Wang, H.M.; Ma, L.Y.; Tian, W.; Man, B.Y.; Liu, C.Y. Contrasting bacterial communities and their assembly processes in karst soils under different land use. Sci. Total Environ. 2021, 751, 142263. [Google Scholar] [CrossRef]
- Jiang, Z.; Lian, Y.; Qin, X. Rocky desertification in Southwest China: impacts, causes, and restoration. Earth-Sci. Rev. 2014, 132, 1–12. [Google Scholar] [CrossRef]
- Liu, S.J.; Zhang, W.; Wang, K.L.; Pan, F.J.; Yang, S.; Shu, S.Y. Factors controlling accumulation of soil organic carbon along vegetation succession in a typical karst region in Southwest China. Sci. Total Environ. 2015, 521, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; Wang, K.L.; Song, T.Q.; Zeng, F.P.; Wang, J.R. Controlling and restoration models of complex degradation of vulnerable karst ecosystem. Acta Ecologica Sinica 2008, 28(2), 811–820, (in Chinese with English abstract). [Google Scholar]
- Wang, K.L.; Zhang, C.H.; Chen, H.S.; Yue, Y.M.; Zhang, W.; Zhang, M.Y.; Qi, X.K.; Fu, Z.Y. Karst landscapes of China: patterns, ecosystem processes and services. Landscape Ecol. 2019, 34, 2743–2763. [Google Scholar] [CrossRef]
- Song, T.Q.; Wang, K.L.; Zeng, F.P.; Peng, W.X.; Du, H. Plants and Environment in Karst Areas of Southwest China; China Science Press: Beijing, China, 2015; pp. 115–123. (in Chinese) [Google Scholar]
- Tong, X.W.; Brandt, M.; Yue, Y.M.; Horion, S.; Wang, K.L.; Keermaecker, W.D.; Tian, F.; Schuegers, G.; Xiao, X.M.; Luo, Y.Q.; Chen, C.; Myneni, R.; Shi, Z.; Chen, H.S.; Fensholt, R. Increased vegetation growth and carbon stock in China karst via ecological engineering. Nat. Sustain. 2018, 1, 44–50. [Google Scholar] [CrossRef]
- Hu, P.L.; Xiao, J.; Zhang, W.; Xiao, L.M.; Yang, R.; Xiao, D.; Zhao, J.; Wang, K.L. Response of soil microbial communities to natural and managed vegetation restoration in a subtropical karst region. Catena 2020, 195, 104849. [Google Scholar] [CrossRef]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4814. [Google Scholar] [CrossRef]
- Xiao, D.; He, X.Y.; Zhang, W.; Hu, P.L.; Sun, M.M.; Wang, K.L. Comparision of bacterial and fungal diversity and network connectivity in karst and non-karst forests in southwest China. Sci. Total Environ. 2022, 822, 153179. [Google Scholar] [CrossRef]
- Zhang, F.; Du, H.; Zeng, F.P.; Peng, W.X.; Song, T.Q. Changes of woody community structure and diversity in karst peak-cluster depressions in southwest china. Acta Ecologica Sinica 2020, 40(12), 4094–4104, (in Chinese with English abstract). [Google Scholar]
- Zhong, Z.K.; Zhang, X.Y.; Wang, X.; Fu, S.Y.; Wu, S.J.; Lu, X.Q.; Ren, C.J.; Han, X.H.; Yang, G.H. Soil bacteria and fungi respond differently to plant diversity and plant family composition during the secondary succession of abandoned farmland on the Loess Plateau, China. Plant Soil 2020, 448, 183–200. [Google Scholar] [CrossRef]
- Powell, J.R.; Karunaratne, S.; Campbell, C.D.; Yao, H.; Robinson, L.; Singh, B.K. Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat. Commun. 2015, 6, 8444. [Google Scholar] [CrossRef] [PubMed]
- Green, J.J.; Dawson, L.A.; Proctor, J.; Duff, E.I.; Elston, D.A. Fine root dynamics in a tropical rain forest is influenced by rainfall. Plant Soil 2005, 276, 23–32. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis. 3rd edn. China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Wu, J.S.; Lin, Q.M.; Huang, Q.Y.; He, J.Z.; Xiao, H.A. Measurement Method and Application of Soil Microbial Biomass. Beijing: China Meteorological Press: Beijing, China, 2006; pp. 54–84. [Google Scholar]
- Xu, N.; Tan, G.C.; Wang, H.Y.; Gai, X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Bastin, J.F.; Finegold, Y.; Garcia, C.; Mollicone, D.; Rezende, M.; Routh, D.; Zohner, C.M.; Crowther, T.W. The global tree restoration potential. Science 2019, 365, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.I.; Miletto, M.; Taylor, J.W.; Bruns, T.D. , Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J 2013, 7(7), 1262–1273. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; Gormley, N.; Gilbert, J.A.; Smith, G.; Knight, R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Gołębiewski, M.; Deja-Sikora, E.; Cichosz, M.; Tretyn, A.; Wróbel, B. 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb. Ecol. 2014, 67, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10(10), 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; Sahl, J.W.; Stres, B.; Thallinger, G.G.; VanHorn, D.J.; Weber, C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, Illinois, 1949; pp. 1–117. [Google Scholar]
- Minchin, P. R. Simulation of multidimensional community patterns: towards a comprehensive model. Vegetatio 1987, 1(3), 145–156. [Google Scholar] [CrossRef]
- Hogg, R.V.; Tanis, E.A. Probability and Statistical Inference, 7th Ed. Prentice Hall, 2006. [Google Scholar]
- Clarke, K.; Gorley, R.; Somerfield, P.; Warwick, R. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. PRIMER-E: Plymouth, UK, 2014. [Google Scholar]
- Hoffman, G.E.; Schadt, E.E. Variance partition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics. 2016, 17(1), 483. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34(17), 884–890. [Google Scholar] [CrossRef] [PubMed]
-
R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computin: Vienna, Austria, 2016.
- Clemmensen, K.E.; Bahr, A.; Ovaskainen, O.; Dahlberg, A.; Ekblad, A.; Wallander, H.; Stenlid, J.; Finlay, R.D.; Wardle, D.A.; Lindahl, B.D. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 2013, 339, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLOS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.; Jumpponen, A.; Schlatter, D.C.; Paulitz, T.C.; Gardener, B.B.M.; Kinkel, L.L.; Garrett, K. Microbiome networks: A systems framework for identifying candidate microbial assemblages for disease management. Phytopathology 2016, 106, 1083–1096. [Google Scholar] [CrossRef]
- Lu, F.; Hu, H.F.; Sun, W.J.; Zhu, J.J.; Liu, G.B.; Zhou, W.M.; Zhang, Q.F.; Shi, P.L.; Liu, X. P.; Wu, X.; Zhang, L.; Wei, X.H.; Dai, L.M.; Zhang, K.R.; Sun, Y.R.; Xue, S.; Zhang, W. J.; Xiong, D.P.; Deng, L.; Liu, B.J.; Zhou, L.; Zhang, C.; Zheng, X.; Cao, J.S.; Huang, Y.; He, N.P.; Zhou, G.Y.; Bai, Y.F.; Xie, Z.Q.; Tang, Z.Y.; Wu, B.F.; Fang, J.Y.; Liu, G.H.; Yu, G.R. Effects of national ecological restoration projects on carbon sequestration in China from 2001 to 2010. Proc. Natl. Acad. Sci. U.S.A 2018, 115, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.U.; Bignell, D.E.; Brown, V.K.; Brussard, L.; Dangerfield, J.M.; Wall, D.H.; Wardle, D.A.; Coleman, D.C.; Giller, K.E.; Lavelle, P.; Van der Putten, W.H.; De Ruiter, P.C.; Rusek, J.; Silver, W.L.; Tiedje, J.M.; Wolters, V. Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: patterns, mechanisms and feedbacks. BioScience 2000, 50, 1049–1061. [Google Scholar] [CrossRef]
- Peng, W.X.; Zhu, Y.F.; Song, M.; Du, H.; Song, T.Q.; Zeng, F.P.; Zhang, F.; Wang, K.L.; Luo, Y.Q.; Zhang, J.Y. The spatial distribution and drivers of soil microbial richness and diversity in a karst broadleaf forest. For. Ecol. Manage. 2019, 449, 117241. [Google Scholar] [CrossRef]
- Li, H.; Ye, D.D.; Wang, X.G.; Settles, M.L.; Wang, J.; Hao, Z.Q.; Zhou, L.S.; Dong, P.; Jiang, Y.; Ma, Z.S. Soil bacterial communities of different natural forest types in Northeast China. Plant Soil 2014, 383, 203–216. [Google Scholar] [CrossRef]
- Jangid, K.; Whitman, W.B.; Condron, L.M.; Turner, B.L.; Williams, N.A. Soil bacterial community succession during long-term ecosystem development. Mol. Ecol. 2013, 22, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Liu, X.; Song, L.; Lin, X.; Zhang, H.; Shen, C.; Chu, H. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 2016, 92, 41–49. [Google Scholar] [CrossRef]
- Ren, C.J.; Chen, J.; Deng, J.; Zhao, F.Z.; Han, X.H.; Yang, G.H.; Tong, X.G.; Feng, Y.Z.; Shelton, S.; Ren, G.X. Response of microbial diversity to C:N:P stoichiometry in fine root and microbial biomass following afforestation. Biol. Fertil. Soils 2017, 53(4), 457–468. [Google Scholar] [CrossRef]
- Ren, C.J.; Liu, W.C.; Zhao, F.Z.; Zhong, Z.K.; Deng, J.; Han, X.H.; Yang, G.H.; Feng, Y.Z.; Ren, G.X. Soil bacterial and fungal diversity and composition respond differently to forest development. Catena 2019, 181, 104071. [Google Scholar] [CrossRef]
- Song, T.Q.; Peng, W.X.; Zeng, F.P.; Wang, K.L.; Cao, H.L.; Li, X.K.; Qin, W.G.; Tan, W.N.; Liu, L. Community composition and biodiversity characteristics of forests in Karst cluster-peak-depression region. Biodiversity Science 2010, 18(4), 355–364, (in Chinese with English abstract). [Google Scholar]
- Cools, N.; Vesterdal, L.; De Vos, B.; Vanguelova, E.; Hansen, K. Tree species is the major factor explaining C:N ratios in European forest soils. For. Ecol. Manage. 2014, 311, 3–16. [Google Scholar] [CrossRef]
- Shao, S.; Zhao, Y.; Zhang, W.; Hu, G.Q.; Xie, H.T.; Yan, J.H.; Han, S.J.; He, H.B.; Zhang, X.D. Linkage of microbial residue dynamics with soil organic carbon accumulation during subtropical forest succession. Soil Biol. Biochem. 2017, 114, 114–120. [Google Scholar] [CrossRef]
- García de León, D.; Moora, M.; Öpik, M.; Jairus, T.; Neuenkamp, L.; Vasar, M.; Bueno, C.G.; Gerz, M.; Davison, J.; Zobel, M. Dispersal of arbuscular mycorrhizal fungi and plants during succession. Acta Oecol. 2016, 77, 128–135. [Google Scholar] [CrossRef]
- Xiao, D.; He, X.Y.; Zhang, W.; Cheng, M.; Hu, P.; Wang, K.L. Diazotroph and arbuscular mycorrhizal fungal diversity and community composition responses to karst and non-karst soils. Appl. Soil Ecol. 2022, 170, 104227. [Google Scholar] [CrossRef]
- Shi, Y.; Delgado-Baquerizo, M.; Li, Y.T.; Yang, Y.F.; Zhu, Y.G.; Peñuelas, J.; Chu, H.Y. Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ. Int. 2020, 142, 105869. [Google Scholar] [CrossRef]
- Mehrshad, M.; Salcher, M.M.; Okazaki, Y.; Nakano, S.; Šimek, K.; Andrei, A.; Ghai, R. Hidden in plain sight-highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome 2018, 6, 176. [Google Scholar] [CrossRef]
- Bödeker, I.T.M.; Lindahl, B.D.; Olson, A.; Clemmensen, K.E. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 2016, 30, 1967–1978. [Google Scholar] [CrossRef]
- Liu, X.; Hannula, S.E.; Li, X.; Hundscheid, M.P.J.; Klein Gunnewiek, P.J.A.; Clocchiatti, A.; Ding, W.; de Boer, W. Decomposing cover crops modify root- associated microbiome composition and disease tolerance of cash crop seedlings. Soil Biol. Biochem. 2021, 160, 108343. [Google Scholar] [CrossRef]
- Fang, J.Y.; Guo, Z.D.; Hu, H.F.; Kato, T.; Muraoka, H.; Son, Y. Forest biomass carbon sinks in East Asia, with special reference to the relative contributions of forest expansion and forest growth. Glob. Chang Biol. 2014, 20, 2019–2030. [Google Scholar] [CrossRef] [PubMed]
- Jeanbille, M.; Buée, M.; Bach, C.; Cébron, A.; Frey-Klett, P.; Turpault, M.P.; Uroz, S. Soil parameters drive the structure, diversity and metabolic potentials of the bacterial communities across temperate beech forest soil sequences. Microb. Ecol. 2016, 71(2), 482–493. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Xue, S.; Wang, G. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Prober, S.M.; Leff, J.W.; Bates, S.T.; Borer, E.T.; Firn, J.; Harpole, W.S.; Lind, E.M.; Seabloom, E.W.; Adler, P.B.; Bakker, J.D. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 2015, 18, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Cline, L.C.; Zak, D.R. Soil microbial communities are shaped by plant-driven changes in resource availability during secondary succession. Ecology 2015, 96, 3374–3385. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Peng, C.; Huang, C.; Wang, K.; Liu, Q.; Liu, Y.; Hai, X.; Shangguan, Z. Drivers of soil microbial metabolic limitation changes along a vegetation restoration gradient on the Loess Plateau, China. Geoderma 2019, 353, 188–200. [Google Scholar] [CrossRef]
- Clemmensen, K.E.; Bahr, A.; Ovaskainen, O.; Dahlberg, A.; Ekblad, A.; Wallander, H.; Stenlid, J.; Finlay, R.D.; Wardle, D.A.; Lindahl, B.D. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 2013, 339, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).