1. Introduction
This brief computational study focuses on assessing the binding energies of various natural substances against four highly pathogenic bacterial strains—Mycobacterium tuberculosis, Helicobacter pylori, Haemophilus influenzae, and Pseudomonas aeruginosa. Molecular docking techniques [
1,
2,
3,
4] were employed to investigate the potential interactions between these natural compounds and crucial targets within the selected bacterial strains.
Molecular docking techniques was to investigate the binding energies of various natural substances against specific targets crucial for the viability of four dangerous bacterial strains—Mycobacterium tuberculosis, Helicobacter pylori, Haemophilus influenzae, and Pseudomonas aeruginosa. The studied targets include Aminoglycoside 2’-N-acetyltransferase, Thioredoxin Reductase, N-acetylneuraminate Lyase, and Glucose-1-phosphate Thymidylyltransferase, representing key proteins essential for the survival of these bacteria. The outcomes aim to contribute insights into potential natural substance candidates for the development of antimicrobial strategies against these pathogenic strains.
The control of Mycobacterium tuberculosis necessitates a comprehensive approach, including early diagnosis, appropriate treatment with antimicrobial drugs, vaccination, and public health measures to prevent transmission. The emergence of drug-resistant strains underscores the need for ongoing research and the development of new therapeutic approaches [
5,
6]. Helicobacter pylori is a bacterium that infects the stomach and upper small intestine, causing conditions such as gastritis and peptic ulcers [
7]. Haemophilus influenzae is a Gram-negative bacterium that can cause a range of infections in humans [
8].
Pseudomonas aeruginosa is a versatile and opportunistic Gram-negative bacterium that poses a significant threat to human health, particularly in hospital settings and among immunocompromised individuals [
9].
2. Material and Methods
-Glucose-1-phosphate thymidylyltransferas (PDB Code: 1fxo, Pseudomonas aeruginosa); Grid box Coordinates of binding Center X ( 61,4158), Y( 15,76), Z(-23,3845).
-N-acetylneuraminate lyase (PDB Code: 1f73, Haemophilus influenzae); Grid box Coordinates of binding Center X ( 22,3873), Y( 33,8121), Z(23,5284).
-Thioredoxin reductase (PDB Code: 2q0l, Helicobacter pylori); Grid box Coordinates of binding Center X ( 15,7801), Y( 13,535), Z(-23,1844).
-Aminoglycoside 2’-N-acetyltransferase (PDB Code: 1m4d, Mycobacterium tuberculosis); Grid box Coordinates of binding Center X ( 20,692), Y( 29,2649), Z(13,4987).
3. Results and Discussion
For the first time is explored the antimicrobial potential of Amentoflavone against four clinically significant bacterial strains—Mycobacterium tuberculosis, Helicobacter pylori, Haemophilus influenzae, and Pseudomonas aeruginosa—through molecular docking studies [
1,
2].
Mycobacterium tuberculosis (M. tuberculosis) is a bacterial species responsible for tuberculosis (TB) in humans. TB is an infectious disease that is both contagious and potentially severe, predominantly impacting the lungs but also capable of affecting various other organs. M. tuberculosis is characterized as an aerobic, slow-growing bacterium, with transmission occurring through the air when an infected individual coughs or sneezes, releasing respiratory droplets containing the bacteria [
5,
6].
Helicobacter pylori is associated with several stomach diseases, including gastritis and peptic ulcers. This bacterium is known for its ability to survive in the acidic environment of the stomach, causing inflammation and damage to the gastric mucosa. The presence of Helicobacter pylori is often confirmed through diagnostic tests such as gastroscopy or breath tests. Treatment of Helicobacter pylori infections commonly involves the use of antibiotics and antacid drugs [
7].
Haemophilus influenzae is a gram-negative bacterium that can cause various infections, including ear infections, pneumonia and meningitis. There are different types of Haemophilus influenzae, identified based on the presence or absence of an antigen called capsule. Some strains of this bacterium, known as Hib (Haemophilus influenzae type b), are associated with more serious illnesses, particularly in children. Transmission occurs through respiratory droplets and the bacterium can colonize different areas of the body, particularly the upper respiratory tract [
8].
Pseudomonas aeruginosa is an opportunistic and pathogenic Gram-negative bacterium that can cause a wide range of infections in humans. It is known for its resistance to many antibiotics and can infect people with compromised immune systems, wounds, compromised lungs, or implanted medical devices. This bacterium can cause lung infections, urinary tract infections, skin and soft tissue infections, and other serious conditions, especially in hospital settings. Diagnosis often involves analysis of clinical specimens and antibiotic sensitivity testing. Treatment can be challenging due to bacterial resistance, requiring thoughtful use of available antibiotics [
9].
Conducting molecular docking studies, we have evaluated the binding interactions between Amentoflavone and key protein targets critical for the viability of these bacteria. The results indicate promising binding affinities, particularly within the active sites of Aminoglycoside 2’-N-acetyltransferase, Thioredoxin Reductase, N-acetylneuraminate Lyase, and Glucose-1-phosphate Thymidylyltransferase in the respective bacterial strains. While these in silico findings offer preliminary insights, further in vitro and in vivo experiments are imperative to confirm the biological relevance and effectiveness of Amentoflavone as a potential antimicrobial agent against these bacterial pathogens.
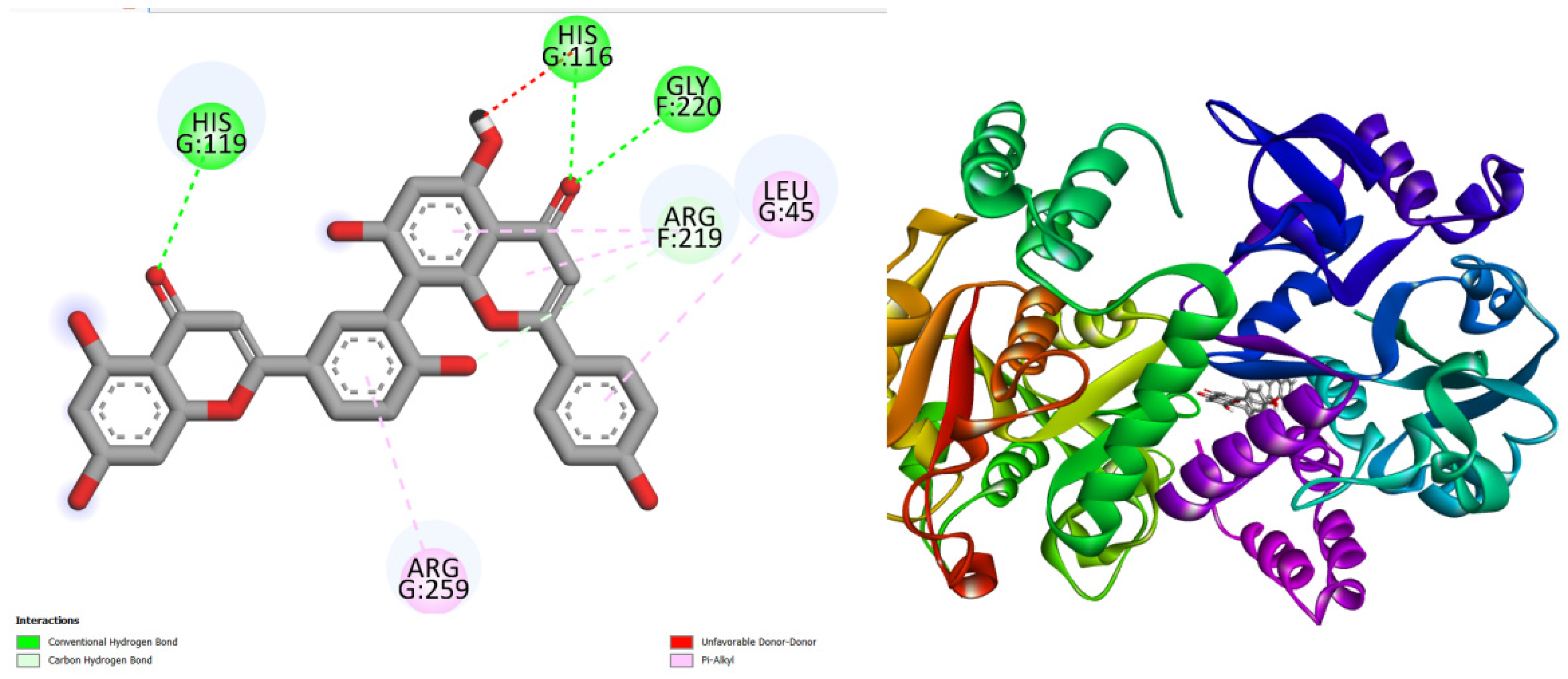
Figure 1.
displays the docking outcomes of Glucose-1-phosphate thymidylyltransferas in conjunction with Amentoflavone -11 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
Figure 1.
displays the docking outcomes of Glucose-1-phosphate thymidylyltransferas in conjunction with Amentoflavone -11 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
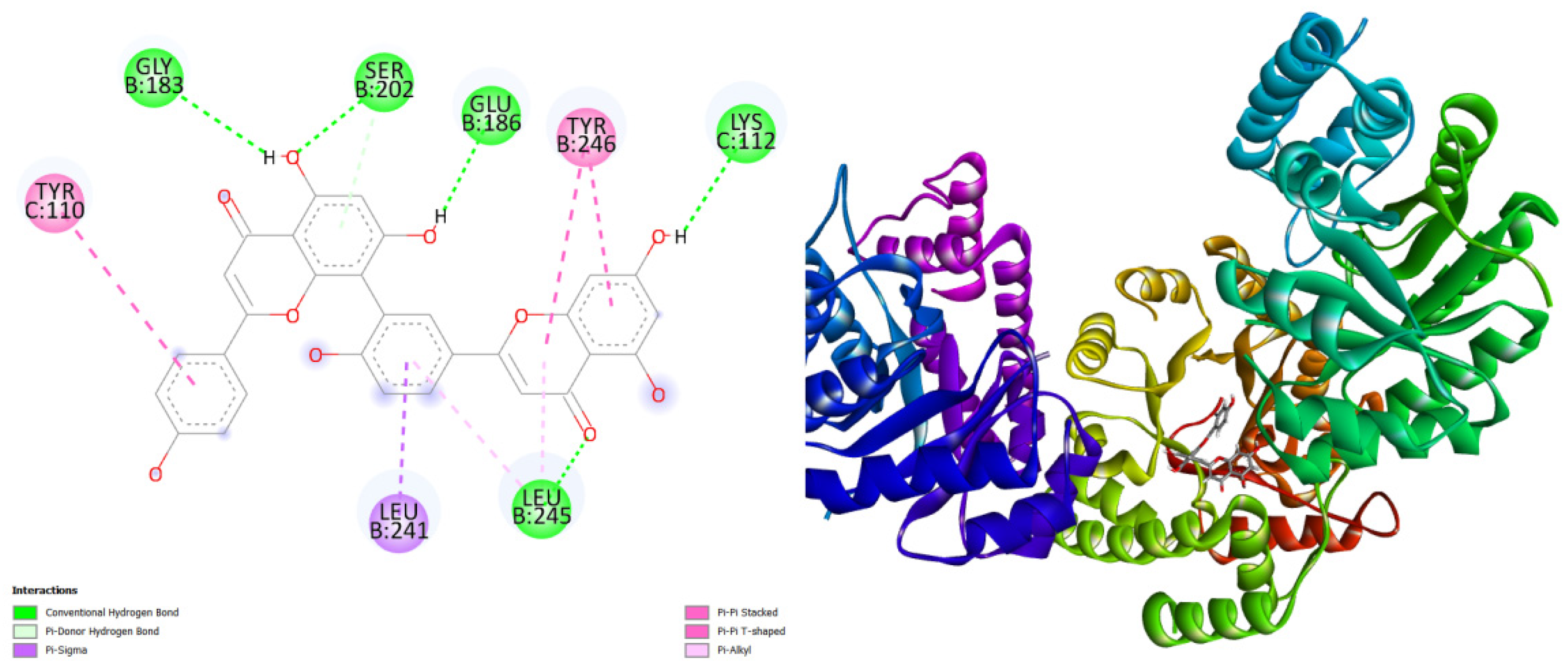
Figure 2.
displays the docking outcomes of N-acetylneuraminate lyase in conjunction with Amentoflavone -10 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
Figure 2.
displays the docking outcomes of N-acetylneuraminate lyase in conjunction with Amentoflavone -10 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
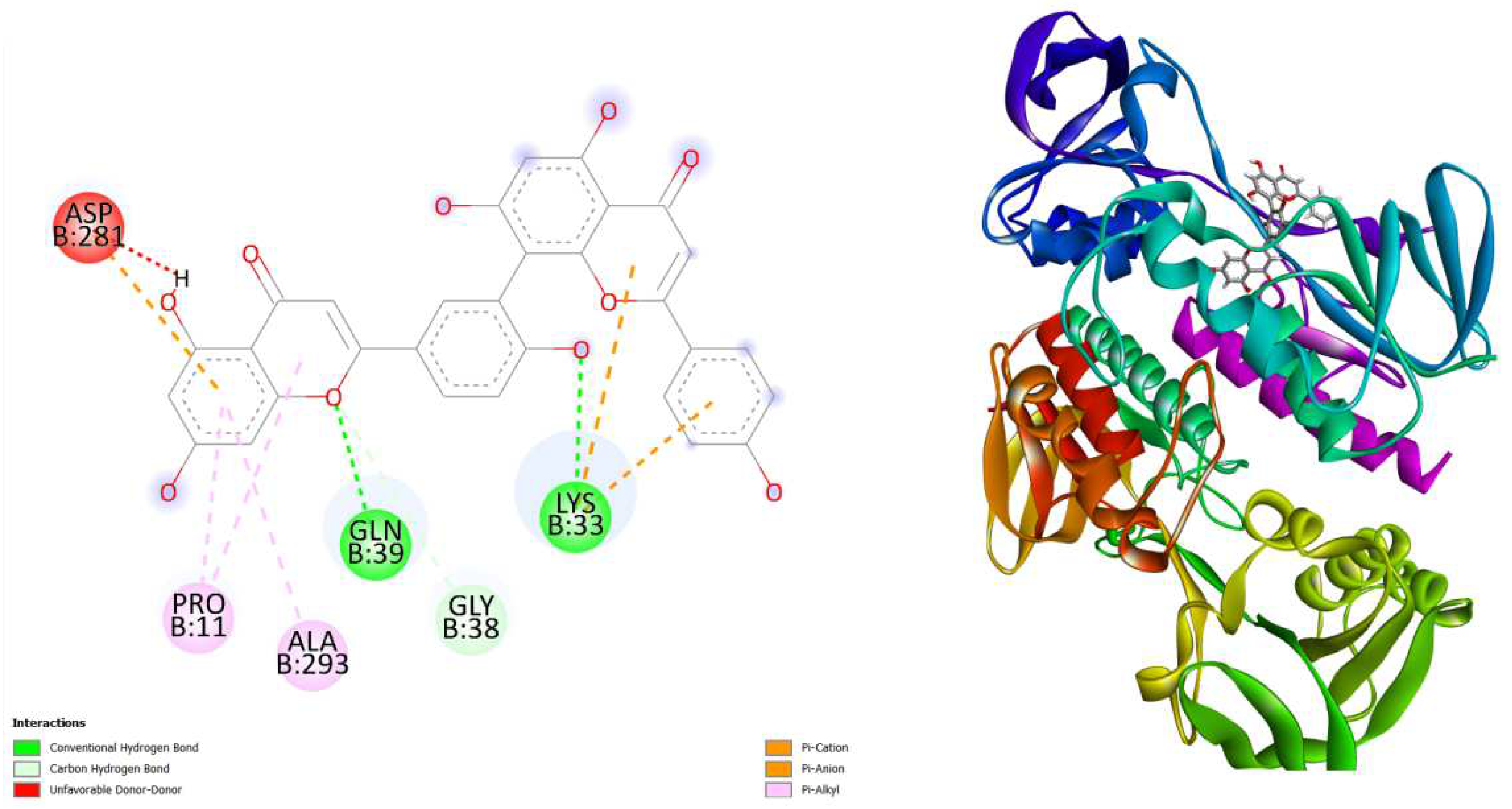
Figure 3.
displays the docking outcomes of Thioredoxin reductase in conjunction with Amentoflavone -10.5 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
Figure 3.
displays the docking outcomes of Thioredoxin reductase in conjunction with Amentoflavone -10.5 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
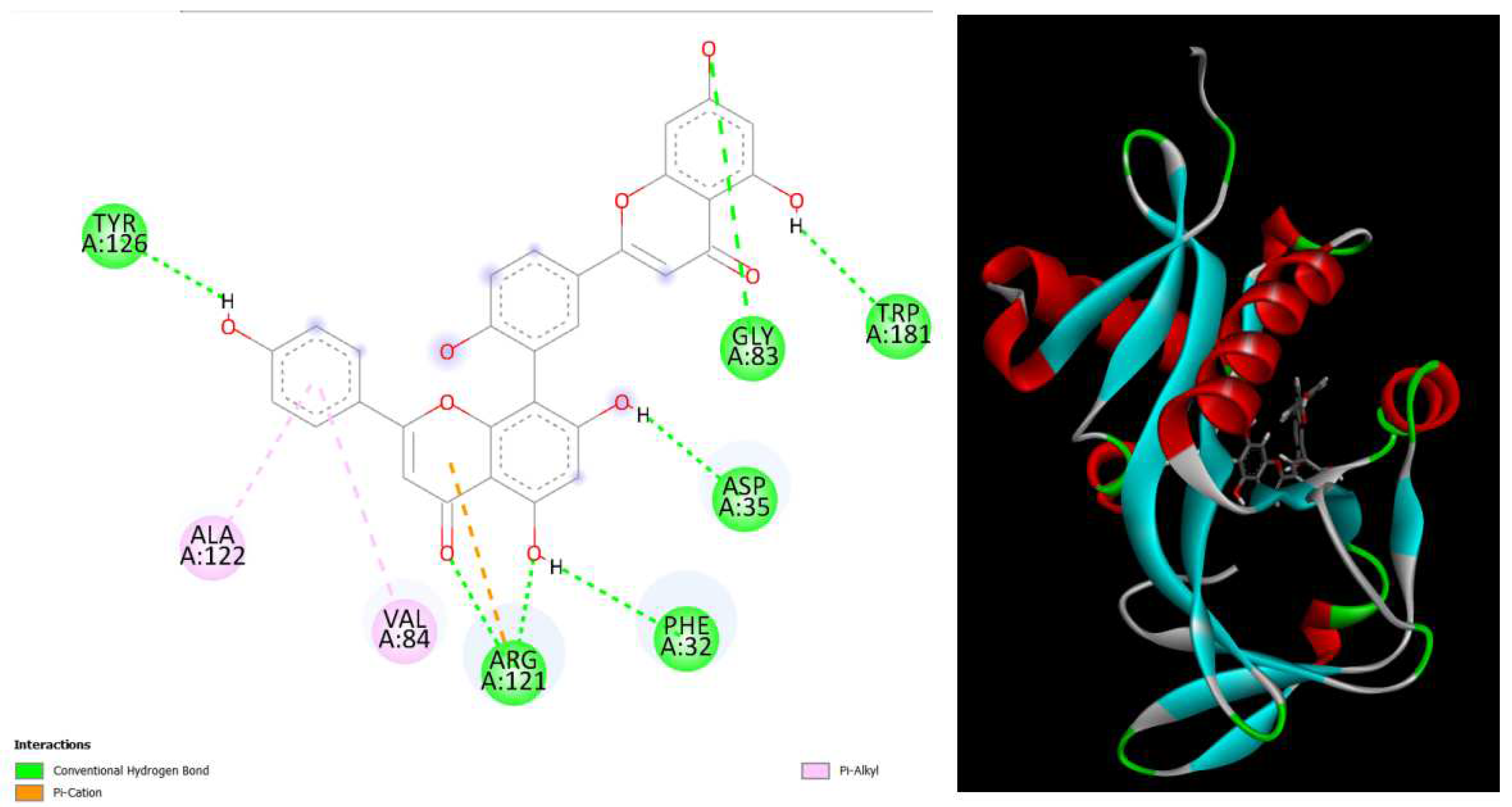
Figure 4.
displays the docking outcomes of Aminoglycoside 2’-N-acetyltransferase in conjunction with Amentoflavone -10.4 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
Figure 4.
displays the docking outcomes of Aminoglycoside 2’-N-acetyltransferase in conjunction with Amentoflavone -10.4 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
4. Conclusion
In conclusion, this molecular docking study sheds light on the potential antimicrobial efficacy of Amentoflavone against four clinically significant bacterial strains—Mycobacterium tuberculosis, Helicobacter pylori, Haemophilus influenzae, and Pseudomonas aeruginosa. The favorable binding interactions observed between Amentoflavone and key protein targets associated with bacterial survival indicate its promising role as a potential antimicrobial agent. However, it is crucial to underscore that these computational findings serve as a preliminary step, and further in vitro and in vivo experiments are imperative to validate the actual antimicrobial activity and safety of Amentoflavone
References
- Fan, Jiyu, Ailing Fu, and Le Zhang. “Progress in molecular docking.” Quantitative Biology 7 (2019): 83-89. [CrossRef]
- Agarwal, S., & Mehrotra, R. J. J. C. (2016). An overview of molecular docking. JSM chem, 4(2), 1024-1028.
- Stanzione, F., Giangreco, I., & Cole, J. C. (2021). Use of molecular docking computational tools in drug discovery. Progress in Medicinal Chemistry, 60, 273-343. [CrossRef]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 31(2), 455-461. [CrossRef]
- Ranjitha, J., Rajan, A., & Shankar, V. (2020). Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. Journal of infection and public health, 13(9), 1255-1264. [CrossRef]
- Dulberger, C. L., Rubin, E. J., & Boutte, C. C. (2020). The mycobacterial cell envelope—a moving target. Nature Reviews Microbiology, 18(1), 47-59. [CrossRef]
- Mezmale, L., Coelho, L. G., Bordin, D., & Leja, M. (2020). Epidemiology of Helicobacter pylori. Helicobacter, 25, e12734. [CrossRef]
- Brueggemann, A. B., van Rensburg, M. J. J., Shaw, D., McCarthy, N. D., Jolley, K. A., Maiden, M. C., ... & Zhou, F. (2021). Changes in the incidence of invasive disease due to Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis during the COVID-19 pandemic in 26 countries and territories in the Invasive Respiratory Infection Surveillance Initiative: a prospective analysis of surveillance data. The Lancet Digital Health, 3(6), e360-e370. [CrossRef]
- Reynolds, D., & Kollef, M. (2021). The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: an update. Drugs, 81(18), 2117-2131. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).