Submitted:
22 January 2024
Posted:
23 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. HIV life cycle and correlated target for drug discovery
2.1. Overview of the life cycle of HIV-1
2.2. Viral entry inhibitors
2.3. Reverse transcriptase inhibitors
2.4. Protease inhibitors
2.5. Integrase inhibitors
3. CRISPR/Cas9 based gene editing in HIV treatment
3.1. Gene editing targets host cell
3.2. Gene editing targets HIV genome
3.3. Clinical trials of gene editing applied in HIV treatment
4. Conclusions and future perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef]
- Gallo, R.C.; Salahuddin, S.Z.; Popovic, M.; Shearer, G.M.; Kaplan, M.; Haynes, B.F.; Palker, T.J.; Redfield, R.; Oleske, J.; Safai, B.; et al. Frequent Detection and Isolation of Cytopathic Retroviruses (HTLV-III) from Patients with AIDS and at Risk for AIDS. Science 1984, 224, 500–503. [Google Scholar] [CrossRef]
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int J Biol Macromol 2021, 172, 524–541. [Google Scholar] [CrossRef]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2012, 2, a007161. [Google Scholar] [CrossRef]
- Menendez-Arias, L. Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res 2013, 98, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, R.; Klecker, R.W.; Weinhold, K.J.; Markham, P.D.; Lyerly, H.K.; Durack, D.T.; Gelmann, E.; Lehrman, S.N.; Blum, R.M.; Barry, D.W.; et al. Administration of 3'-azido-3'-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. Lancet 1986, 1, 575–580. [Google Scholar] [CrossRef] [PubMed]
- McLeod, G.X.; Hammer, S.M. Zidovudine: five years later. Ann Intern Med 1992, 117, 487–501. [Google Scholar] [CrossRef]
- Zhao, A.V.; Crutchley, R.D.; Guduru, R.C.; Ton, K.; Lam, T.; Min, A.C. A clinical review of HIV integrase strand transfer inhibitors (INSTIs) for the prevention and treatment of HIV-1 infection. Retrovirology 2022, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Mbhele, N.; Chimukangara, B.; Gordon, M. HIV-1 integrase strand transfer inhibitors: a review of current drugs, recent advances and drug resistance. Int J Antimicrob Agents 2021, 57, 106343. [Google Scholar] [CrossRef] [PubMed]
- Lecher, S.L.; Fonjungo, P.; Ellenberger, D.; Toure, C.A.; Alemnji, G.; Bowen, N.; Basiye, F.; Beukes, A.; Carmona, S.; de Klerk, M.; et al. HIV Viral Load Monitoring Among Patients Receiving Antiretroviral Therapy - Eight Sub-Saharan Africa Countries, 2013-2018. MMWR Morb Mortal Wkly Rep 2021, 70, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Han, W.M.; Law, M.G.; Egger, M.; Wools-Kaloustian, K.; Moore, R.; McGowan, C.; Kumarasamy, N.; Desmonde, S.; Edmonds, A.; Davies, M.A.; et al. Global estimates of viral suppression in children and adolescents and adults on antiretroviral therapy adjusted for missing viral load measurements: a multiregional, retrospective cohort study in 31 countries. Lancet HIV 2021, 8, e766–e775. [Google Scholar] [CrossRef]
- Eisinger, R.W.; Dieffenbach, C.W.; Fauci, A.S. HIV Viral Load and Transmissibility of HIV Infection: Undetectable Equals Untransmittable. JAMA 2019, 321, 451–452. [Google Scholar] [CrossRef]
- LeMessurier, J.; Traversy, G.; Varsaneux, O.; Weekes, M.; Avey, M.T.; Niragira, O.; Gervais, R.; Guyatt, G.; Rodin, R. Risk of sexual transmission of human immunodeficiency virus with antiretroviral therapy, suppressed viral load and condom use: a systematic review. CMAJ 2018, 190, E1350–E1360. [Google Scholar] [CrossRef]
- Cohen, M.S.; Chen, Y.Q.; McCauley, M.; Gamble, T.; Hosseinipour, M.C.; Kumarasamy, N.; Hakim, J.G.; Kumwenda, J.; Grinsztejn, B.; Pilotto, J.H.; et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 2011, 365, 493–505. [Google Scholar] [CrossRef]
- Cohen, M.S.; Chen, Y.Q.; McCauley, M.; Gamble, T.; Hosseinipour, M.C.; Kumarasamy, N.; Hakim, J.G.; Kumwenda, J.; Grinsztejn, B.; Pilotto, J.H. , et al. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N Engl J Med 2016, 375, 830–839. [Google Scholar] [CrossRef]
- Mujugira, A.; Celum, C.; Coombs, R.W.; Campbell, J.D.; Ndase, P.; Ronald, A.; Were, E.; Bukusi, E.A.; Mugo, N.; Kiarie, J. , et al. HIV Transmission Risk Persists During the First 6 Months of Antiretroviral Therapy. J Acquir Immune Defic Syndr 2016, 72, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, L.; Hou, Y.; Zhao, Y.; Dou, Z.; Ma, Y.; Zhang, D.; Wu, Y.; Zhao, D.; Liu, Z. , et al. Immune restoration in HIV-1-infected patients after 12 years of antiretroviral therapy: a real-world observational study. Emerg Microbes Infect 2020, 9, 2550–2561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.X.; Jiao, Y.M.; Zhang, C.; Song, J.W.; Fan, X.; Xu, R.N.; Huang, H.H.; Zhang, J.Y.; Wang, L.F.; Zhou, C.B. , et al. HIV Reservoir Decay and CD4 Recovery Associated With High CD8 Counts in Immune Restored Patients on Long-Term ART. Front Immunol 2020, 11, 1541. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.K.; Cole, S.R.; Breger, T.L.; Rudolph, J.E.; Filiatreau, L.M.; Buchacz, K.; Humes, E.; Rebeiro, P.F.; D'Souza, G.; Gill, M.J. , et al. Mortality Among Persons Entering HIV Care Compared With the General U.S. Population : An Observational Study. Ann Intern Med 2021, 174, 1197–1206. [Google Scholar] [CrossRef]
- Fontela, C.; Aguinaga, A.; Moreno-Iribas, C.; Reparaz, J.; Rivero, M.; Gracia, M.; Floristan, Y.; Fresan, U.; Miguel, R.S.; Ezpeleta, C. , et al. Trends and causes of mortality in a population-based cohort of HIV-infected adults in Spain: comparison with the general population. Sci Rep 2020, 10, 8922. [Google Scholar] [CrossRef] [PubMed]
- Reis, K.G.; Desderius, B.; Kingery, J.; Kirabo, A.; Makubi, A.; Myalla, C.; Lee, M.H.; Kapiga, S.; Peck, R.N. Blood pressure, T cells, and mortality in people with HIV in Tanzania during the first 2 years of antiretroviral therapy. J Clin Hypertens (Greenwich) 2020, 22, 1554–1562. [Google Scholar] [CrossRef]
- Pannus, P.; Rutsaert, S.; De Wit, S.; Allard, S.D.; Vanham, G.; Cole, B.; Nescoi, C.; Aerts, J.; De Spiegelaere, W.; Tsoumanis, A. , et al. Rapid viral rebound after analytical treatment interruption in patients with very small HIV reservoir and minimal on-going viral transcription. J Int AIDS Soc 2020, 23, e25453. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Aga, E.; Bosch, R.J.; Pilkinton, M.; Kroon, E.; MacLaren, L.; Keefer, M.; Fox, L.; Barr, L.; Acosta, E. , et al. Time to Viral Rebound After Interruption of Modern Antiretroviral Therapies. Clin Infect Dis 2022, 74, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Kolakowska, A.; Maresca, A.F.; Collins, I.J.; Cailhol, J. Update on Adverse Effects of HIV Integrase Inhibitors. Curr Treat Options Infect Dis 2019, 11, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Liu, K.; Liu, H.; Hu, Y.; Zhang, Z.; Qin, J.; Xu, Q.; Peng, K.; Jin, X.; Wang, J.H. , et al. Corrigendum to "Trend of HIV-1 drug resistance in China: A systematic review and meta-analysis of data accumulated over 17 years (2001-2017)" [EClinicalMedicine 18 (2020) 100238]. EClinicalMedicine 2021, 33, 100696. [Google Scholar] [CrossRef] [PubMed]
- Chimukangara, B.; Lessells, R.J.; Rhee, S.Y.; Giandhari, J.; Kharsany, A.B.M.; Naidoo, K.; Lewis, L.; Cawood, C.; Khanyile, D.; Ayalew, K.A. , et al. Trends in Pretreatment HIV-1 Drug Resistance in Antiretroviral Therapy-naive Adults in South Africa, 2000-2016: A Pooled Sequence Analysis. EClinicalMedicine 2019, 9, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, N.; von Siebenthal, C.; Vongrad, V.; Turk, T.; Neumann, K.; Beerenwinkel, N.; Bogojeska, J.; Fellay, J.; Roth, V.; Kok, Y.L. , et al. Determinants of HIV-1 reservoir size and long-term dynamics during suppressive ART. Nat Commun 2019, 10, 3193. [Google Scholar] [CrossRef] [PubMed]
- Bandera, A.; Gori, A.; Clerici, M.; Sironi, M. Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance. Curr Opin Pharmacol 2019, 48, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Bour, S.; Geleziunas, R.; Wainberg, M.A. The human immunodeficiency virus type 1 (HIV-1) CD4 receptor and its central role in promotion of HIV-1 infection. Microbiol Rev 1995, 59, 63–93. [Google Scholar] [CrossRef]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef]
- Berger, E.A.; Murphy, P.M.; Farber, J.M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 1999, 17, 657–700. [Google Scholar] [CrossRef]
- Chan, D.C.; Fass, D.; Berger, J.M.; Kim, P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997, 89, 263–273. [Google Scholar] [CrossRef]
- 33. Telesnitsky, A. and S.P. Goff, Reverse Transcriptase and the Generation of Retroviral DNA, in Retroviruses, J.M. Coffin, S.H. Hughes, and H.E. Varmus, Editors. 1997: Cold Spring Harbor (NY).
- Lusic, M.; Siliciano, R.F. Nuclear landscape of HIV-1 infection and integration. Nat Rev Microbiol 2017, 15, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Saad, J.S. The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation. Viruses 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Rein, A. RNA Packaging in HIV. Trends Microbiol 2019, 27, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 2011, 410, 582–608. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K. Maturation of retroviruses. Curr Opin Virol 2019, 36, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Cai, Y.; Chen, B. HIV-1 Entry and Membrane Fusion Inhibitors. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.V. Enfuvirtide, a new drug for HIV infection. Lancet 2003, 361, 1577–1578. [Google Scholar] [CrossRef] [PubMed]
- He, Y. Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion. Curr Pharm Des 2013, 19, 1800–1809. [Google Scholar] [CrossRef]
- Patel, I.H.; Zhang, X.; Nieforth, K.; Salgo, M.; Buss, N. Pharmacokinetics, pharmacodynamics and drug interaction potential of enfuvirtide. Clin Pharmacokinet 2005, 44, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Lalezari, J.P.; Henry, K.; O'Hearn, M.; Montaner, J.S.; Piliero, P.J.; Trottier, B.; Walmsley, S.; Cohen, C.; Kuritzkes, D.R.; Eron, J.J., Jr. , et al. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N Engl J Med 2003, 348, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Dorr, P.; Westby, M.; Dobbs, S.; Griffin, P.; Irvine, B.; Macartney, M.; Mori, J.; Rickett, G.; Smith-Burchnell, C.; Napier, C. , et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 2005, 49, 4721–4732. [Google Scholar] [CrossRef]
- Tan, Q.; Zhu, Y.; Li, J.; Chen, Z.; Han, G.W.; Kufareva, I.; Li, T.; Ma, L.; Fenalti, G.; Li, J. , et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 2013, 341, 1387–1390. [Google Scholar] [CrossRef]
- Tiraboschi, J.M.; Niubo, J.; Curto, J.; Podzamczer, D. Maraviroc concentrations in cerebrospinal fluid in HIV-infected patients. J Acquir Immune Defic Syndr 2010, 55, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Tiraboschi, J.M.; Niubo, J.; Curto, J.; Podzamczer, D. Maraviroc concentrations in seminal plasma in HIV-infected patients. J Acquir Immune Defic Syndr 2010, 55, e35–36. [Google Scholar] [CrossRef]
- Dumond, J.B.; Patterson, K.B.; Pecha, A.L.; Werner, R.E.; Andrews, E.; Damle, B.; Tressler, R.; Worsley, J.; Kashuba, A.D. Maraviroc concentrates in the cervicovaginal fluid and vaginal tissue of HIV-negative women. J Acquir Immune Defic Syndr 2009, 51, 546–553. [Google Scholar] [CrossRef]
- Fatkenheuer, G.; Nelson, M.; Lazzarin, A.; Konourina, I.; Hoepelman, A.I.; Lampiris, H.; Hirschel, B.; Tebas, P.; Raffi, F.; Trottier, B. , et al. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med 2008, 359, 1442–1455. [Google Scholar] [CrossRef]
- Beccari, M.V.; Mogle, B.T.; Sidman, E.F.; Mastro, K.A.; Asiago-Reddy, E.; Kufel, W.D. Ibalizumab, a Novel Monoclonal Antibody for the Management of Multidrug-Resistant HIV-1 Infection. Antimicrob Agents Chemother 2019, 63. [Google Scholar] [CrossRef]
- Markham, A. Ibalizumab: First Global Approval. Drugs 2018, 78, 781–785. [Google Scholar] [CrossRef]
- Pace, C.S.; Fordyce, M.W.; Franco, D.; Kao, C.Y.; Seaman, M.S.; Ho, D.D. Anti-CD4 monoclonal antibody ibalizumab exhibits breadth and potency against HIV-1, with natural resistance mediated by the loss of a V5 glycan in envelope. J Acquir Immune Defic Syndr 2013, 62, 1–9. [Google Scholar] [CrossRef]
- Sluis-Cremer, N.; Arion, D.; Parniak, M.A. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell Mol Life Sci 2000, 57, 1408–1422. [Google Scholar] [CrossRef]
- Saag, M.S.; Gandhi, R.T.; Hoy, J.F.; Landovitz, R.J.; Thompson, M.A.; Sax, P.E.; Smith, D.M.; Benson, C.A.; Buchbinder, S.P.; Del Rio, C. , et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2020 Recommendations of the International Antiviral Society-USA Panel. JAMA 2020, 324, 1651–1669. [Google Scholar] [CrossRef]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 1992, 256, 1783–1790. [Google Scholar] [CrossRef]
- Tronchet, J.M.; Seman, M. Nonnucleoside inhibitors of HIV-1 reverse transcriptase: from the biology of reverse transcription to molecular design. Curr Top Med Chem 2003, 3, 1496–1511. [Google Scholar] [CrossRef]
- De Clercq, E. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antiviral Res 1998, 38, 153–179. [Google Scholar] [CrossRef] [PubMed]
- Kohl, N.E.; Emini, E.A.; Schleif, W.A.; Davis, L.J.; Heimbach, J.C.; Dixon, R.A.; Scolnick, E.M.; Sigal, I.S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A 1988, 85, 4686–4690. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.; Faulds, D. Saquinavir. A review of its pharmacology and clinical potential in the management of HIV infection. Drugs 1996, 52, 93–112. [Google Scholar] [CrossRef]
- Bozzette, S.A.; Ake, C.F.; Tam, H.K.; Chang, S.W.; Louis, T.A. Cardiovascular and cerebrovascular events in patients treated for human immunodeficiency virus infection. N Engl J Med 2003, 348, 702–710. [Google Scholar] [CrossRef]
- Holmberg, S.D.; Moorman, A.C.; Williamson, J.M.; Tong, T.C.; Ward, D.J.; Wood, K.C.; Greenberg, A.E.; Janssen, R.S.; investigators, H.I.V.O.S. Protease inhibitors and cardiovascular outcomes in patients with HIV-1. Lancet 2002, 360, 1747–1748. [Google Scholar] [CrossRef] [PubMed]
- Alvi, R.M.; Neilan, A.M.; Tariq, N.; Awadalla, M.; Afshar, M.; Banerji, D.; Rokicki, A.; Mulligan, C.; Triant, V.A.; Zanni, M.V. , et al. Protease Inhibitors and Cardiovascular Outcomes in Patients With HIV and Heart Failure. J Am Coll Cardiol 2018, 72, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Paton, N.I.; Kityo, C.; Hoppe, A.; Reid, A.; Kambugu, A.; Lugemwa, A.; van Oosterhout, J.J.; Kiconco, M.; Siika, A.; Mwebaze, R. , et al. Assessment of second-line antiretroviral regimens for HIV therapy in Africa. N Engl J Med 2014, 371, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Sham, H.L.; Zhao, C.; Li, L.; Betebenner, D.A.; Saldivar, A.; Vasavanonda, S.; Kempf, D.J.; Plattner, J.J.; Norbeck, D.W. Novel lopinavir analogues incorporating non-Aromatic P-1 side chains--synthesis and structure--activity relationships. Bioorg Med Chem Lett 2002, 12, 3101–3103. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, C.; Stray, K.; Tsai, L.; Williams, M.; Yang, Z.Y.; Cannizzaro, C.; Leavitt, S.A.; Liu, X.; Wang, K.; Murray, B.P. , et al. In vitro characterization of GS-8374, a novel phosphonate-containing inhibitor of HIV-1 protease with a favorable resistance profile. Antimicrob Agents Chemother 2011, 55, 1366–1376. [Google Scholar] [CrossRef] [PubMed]
- Hruz, P.W.; Yan, Q.; Tsai, L.; Koster, J.; Xu, L.; Cihlar, T.; Callebaut, C. GS-8374, a novel HIV protease inhibitor, does not alter glucose homeostasis in cultured adipocytes or in a healthy-rodent model system. Antimicrob Agents Chemother 2011, 55, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- LaFemina, R.L.; Schneider, C.L.; Robbins, H.L.; Callahan, P.L.; LeGrow, K.; Roth, E.; Schleif, W.A.; Emini, E.A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol 1992, 66, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Felock, P.J.; Hastings, J.C.; Blau, C.U.; Hazuda, D.J. The role of manganese in promoting multimerization and assembly of human immunodeficiency virus type 1 integrase as a catalytically active complex on immobilized long terminal repeat substrates. J Virol 1996, 70, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Mizuuchi, K.; Craigie, R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 1991, 67, 1211–1221. [Google Scholar] [CrossRef]
- Grobler, J.A.; Stillmock, K.; Hu, B.; Witmer, M.; Felock, P.; Espeseth, A.S.; Wolfe, A.; Egbertson, M.; Bourgeois, M.; Melamed, J. , et al. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci U S A 2002, 99, 6661–6666. [Google Scholar] [CrossRef]
- Scarsi, K.K.; Havens, J.P.; Podany, A.T.; Avedissian, S.N.; Fletcher, C.V. HIV-1 Integrase Inhibitors: A Comparative Review of Efficacy and Safety. Drugs 2020, 80, 1649–1676. [Google Scholar] [CrossRef]
- Payra, S.; Manjhi, P.K.; Singh, S.; Kumar, R.; Singh, S.K.; Kumar, A.; Maharshi, V. HIV cure: Are we going to make history? HIV Med 2023. [Google Scholar] [CrossRef]
- Allers, K.; Schneider, T. CCR5Delta32 mutation and HIV infection: basis for curative HIV therapy. Curr Opin Virol 2015, 14, 24–29. [Google Scholar] [CrossRef] [PubMed]
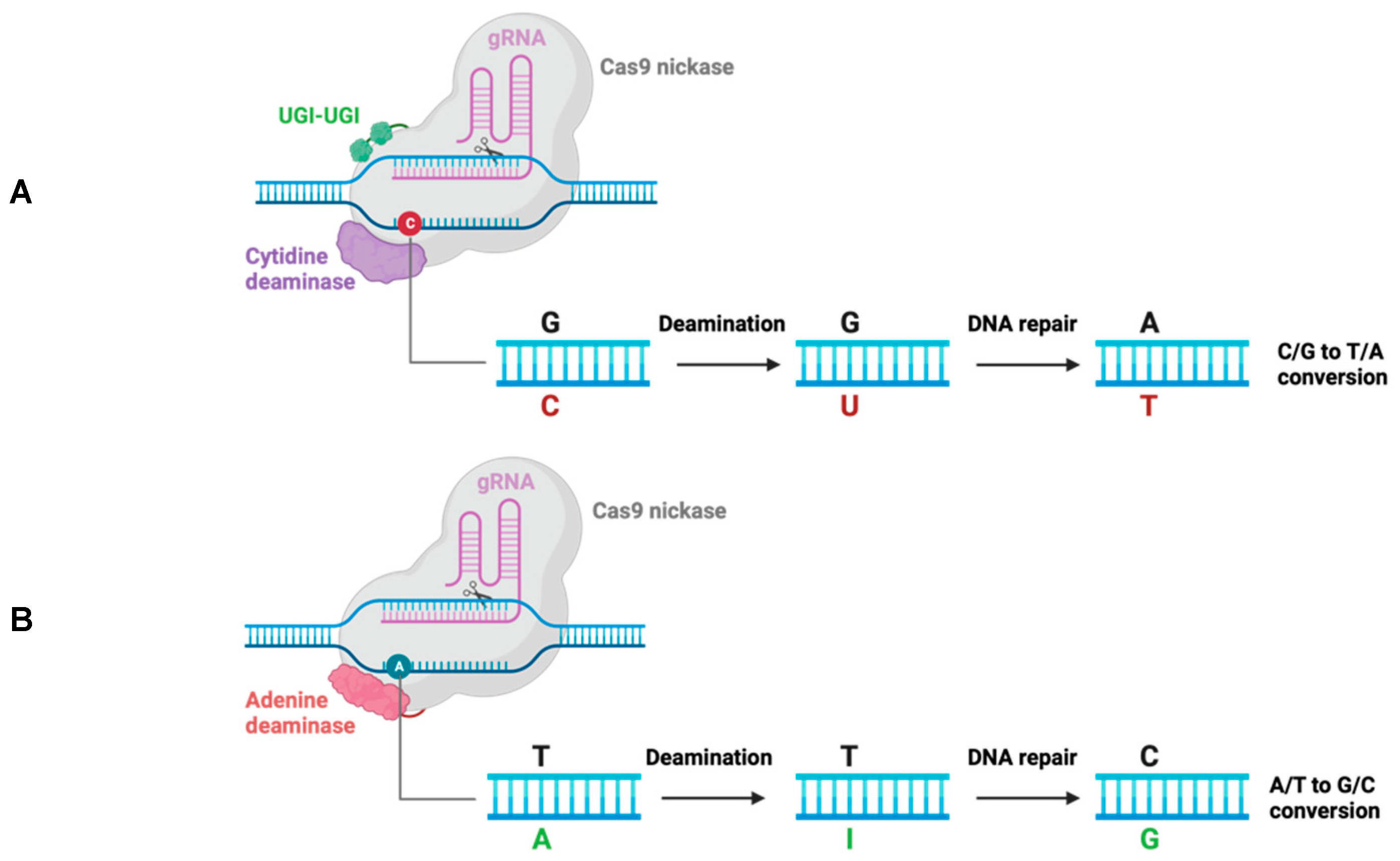
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, H.; Gao, Y.; Chen, Z.; Xie, L.; Liu, Y.; Liu, Y.; Wang, X.; Li, H.; Lai, W. , et al. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol Ther 2017, 25, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S. , et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4(+) T cells from HIV-1 infection. Cell Biosci 2017, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Knipping, F.; Newby, G.A.; Eide, C.R.; McElroy, A.N.; Nielsen, S.C.; Smith, K.; Fang, Y.; Cornu, T.I.; Costa, C.; Gutierrez-Guerrero, A. , et al. Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol Ther 2022, 30, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Chinnapaiyan, S.; Santiago, M.J.; Panda, K.; Rahman, M.S.; Alluin, J.; Rossi, J.; Unwalla, H.J. A conditional RNA Pol II mono-promoter drives HIV-inducible, CRISPR-mediated cyclin T1 suppression and HIV inhibition. Mol Ther Nucleic Acids 2023, 32, 553–565. [Google Scholar] [CrossRef]
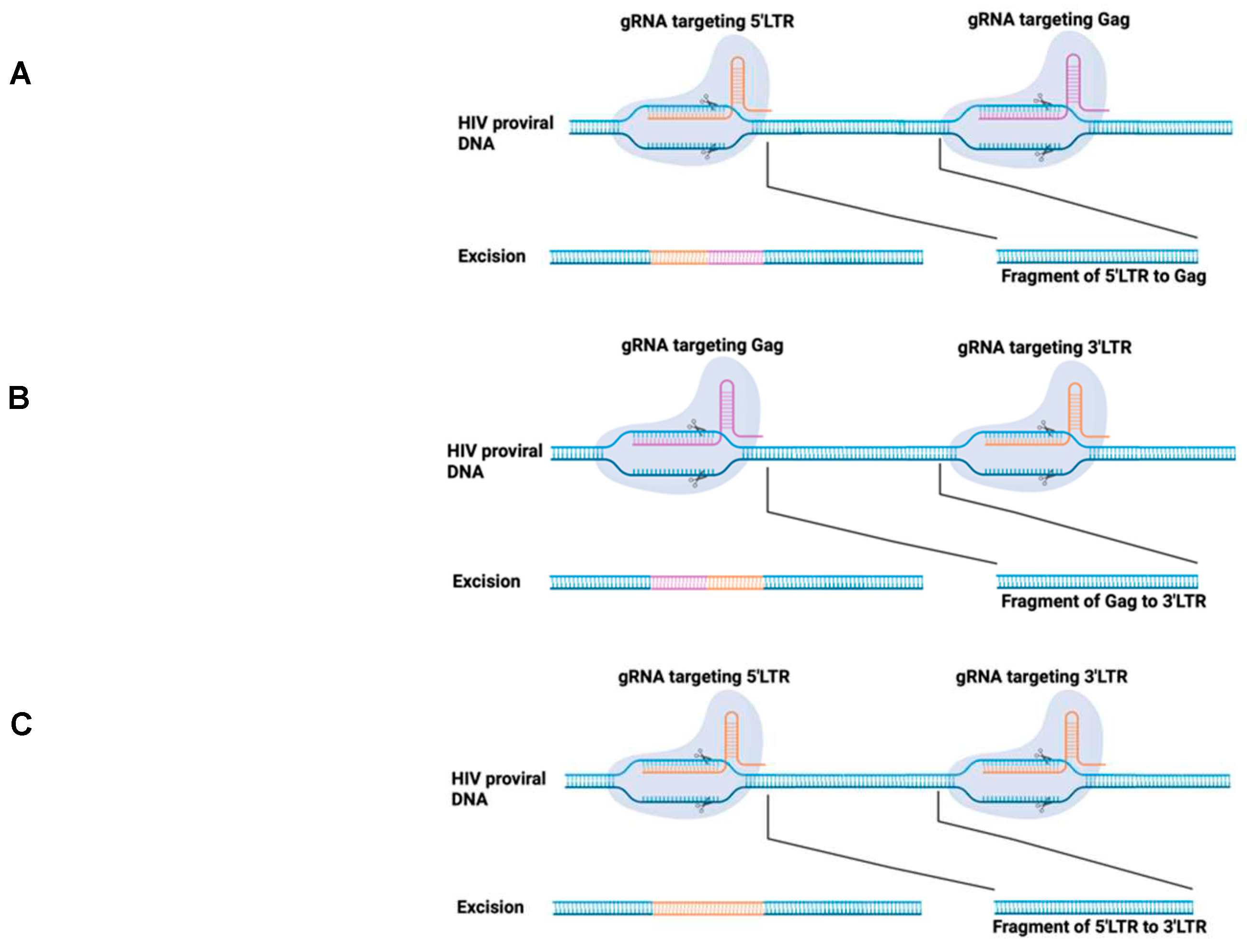
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 2013, 3, 2510. [Google Scholar] [CrossRef]
- Kaminski, R.; Chen, Y.; Fischer, T.; Tedaldi, E.; Napoli, A.; Zhang, Y.; Karn, J.; Hu, W.; Khalili, K. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci Rep 2016, 6, 22555. [Google Scholar] [CrossRef]
- Liao, H.K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.J. , et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 2015, 6, 6413. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lei, R.; Le Duff, Y.; Li, J.; Guo, F.; Wainberg, M.A.; Liang, C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 2015, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Chen, C.; Kaminski, R.; Su, H.; Mancuso, P.; Sillman, B.; Zhang, C.; Liao, S.; Sravanam, S.; Liu, H. , et al. CRISPR editing of CCR5 and HIV-1 facilitates viral elimination in antiretroviral drug-suppressed virus-infected humanized mice. Proc Natl Acad Sci U S A 2023, 120, e2217887120. [Google Scholar] [CrossRef] [PubMed]
- https://www.fda.gov/news-events/press-announcements/fda-approves-first-gene-therapies-treat-patients-sickle-cell-disease.
- Burdo, T.H.; Chen, C.; Kaminski, R.; Sariyer, I.K.; Mancuso, P.; Donadoni, M.; Smith, M.D.; Sariyer, R.; Caocci, M.; Liao, S., et al.; et al. Preclinical safety and biodistribution of CRISPR targeting SIV in non-human primates. Gene Ther 2023. [Google Scholar] [CrossRef]
- Hamann, M.V.; Beschorner, N.; Vu, X.K.; Hauber, I.; Lange, U.C.; Traenkle, B.; Kaiser, P.D.; Foth, D.; Schneider, C.; Buning, H. , et al. Improved targeting of human CD4+ T cells by nanobody-modified AAV2 gene therapy vectors. PLoS One 2021, 16, e0261269. [Google Scholar] [CrossRef]
- Levy, J.M.; Yeh, W.H.; Pendse, N.; Davis, J.R.; Hennessey, E.; Butcher, R.; Koblan, L.W.; Comander, J.; Liu, Q.; Liu, D.R. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng 2020, 4, 97–110. [Google Scholar] [CrossRef]
| Year | Trade name | Generic name | Molecule type | Target | Manufacturer | Class |
|---|---|---|---|---|---|---|
| 2003 | Fuzeon | Enfuvirtide (T20) | Peptide | GP41 | Trimeris/Roche | FI |
| 2007 | Selzentry | Maraviroc (MVC) | Small molecule | CCR5 | Pfizer | CRI |
| 2018 | Trogarzo | Ibalizumab-uiyk (IBA) | monoclonal antibody | CD4 | TaiMed Biologics | RI |
| 1987 | Retrovir | Zidovudine (AZT) | Small molecule | RT | GlaxoSmithKline | NRTI |
| 1991 | Videx | Didanosine (ddI) | Small molecule | RT | Bristol-Myers Squibb | NRTI |
| 1992 | Hivid | Zalcitabine (ddC) | Small molecule | RT | Roche | NRTI |
| 1994 | Zerit | Stavudine (d4T) | Small molecule | RT | Bristol-Myers Squibb | NRTI |
| 1995 | Epivir | Lamivudine (3TC) | Small molecule | RT | GlaxoSmithKline | NRTI |
| 2003 | Fuzeon | Enfuvirtide (T20) | Peptide | GP41 | Trimeris/Roche | FI |
| 2007 | Selzentry | Maraviroc (MVC) | Small molecule | CCR5 | Pfizer | CRI |
| 2018 | Trogarzo | Ibalizumab-uiyk (IBA) | monoclonal antibody | CD4 | TaiMed Biologics | RI |
| 1996 | Viramune | Nevirapine (NVP) | Small molecule | RT | Boehringer Ingelheim | NNRTI |
| 1997 | Rescriptor | Delavirdine (DLV) | Small molecule | RT | ViiV Healthcare | NNRTI |
| 1998 | Sustiva | Efavirenz (EFV) | Small molecule | RT | DuPont Pharmaceuticals | NNRTI |
| 1998 | Ziagen | Abacavir (ABC) | Small molecule | RT | ViiV Healthcare | NRTI |
| 2001 | Viread | Tenofovir disoproxil (TDF) | Small molecule | RT | Gilead | NRTI |
| 2003 | Emtriva | Emtricitabine (FTC) | Small molecule | RT | Gilead | NRTI |
| 2008 | Intelence | Etravirine (ETR) | Small molecule | RT | Johnson & Johnson | NNRTI |
| 2011 | Edurant | Rilpivirine (RPV) | Small molecule | RT | Tibotec | NNRTI |
| 2018 | Pifeltro | Doravirine (DOR) | Small molecule | RT | Merck | NNRTI |
| 1995 | Invirase | Saquinavir (SQV) | Small molecule | PR | Roche | PI |
| 1996 | Crixivan | Indinavir sulfate (IDV) | Small molecule | PR | Merck | PI |
| 1996 | Norvir | Ritonavir (RTV) | Small molecule | PR | Abbott | PI |
| 1997 | Viracept | Nelfinavir (NFV) | Small molecule | PR | Agouron Pharmaceuticals | PI |
| 1999 | Agenerase | Amprenavir (APV) | Small molecule | PR | GlaxoSmithKline | PI |
| 2000 | Kaletra | Lopinavir (LPV) | Small molecule | PR | Abbott | PI |
| 2003 | Lexiva | Fosamprenavir (FPV) | Small molecule | PR | GlaxoSmithKline | PI |
| 2003 | Reyataz | Atazanavir (ATV) | Small molecule | PR | Bristol-Myers Squibb | PI |
| 2005 | Aptivus | Tipranavir (TPV) | Small molecule | PR | Boehringer Ingelheim | PI |
| 2006 | Prezista | Darunavir (DRV) | Small molecule | PR | Tibotec | PI |
| 2007 | Isentress | Raltegravir (RAL) | Small molecule | IN | Merck | InSTI |
| 2013 | Tivicay | Dolutegravir (DTG) | Small molecule | IN | ViiV Healthcare | InSTI |
| 2014 | Vitekta | Elvitegravir (EVG) | Small molecule | IN | Gilead | InSTI |
| 2018 | Biktarvy | Bictegravir (BIC) | Small molecule | IN | Gilead | InSTI |
| 2021 | Vocabria | Cabotegravir (CAB) | Small molecule | IN | ViiV Healthcare | InSTI |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




