Submitted:
25 January 2024
Posted:
26 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
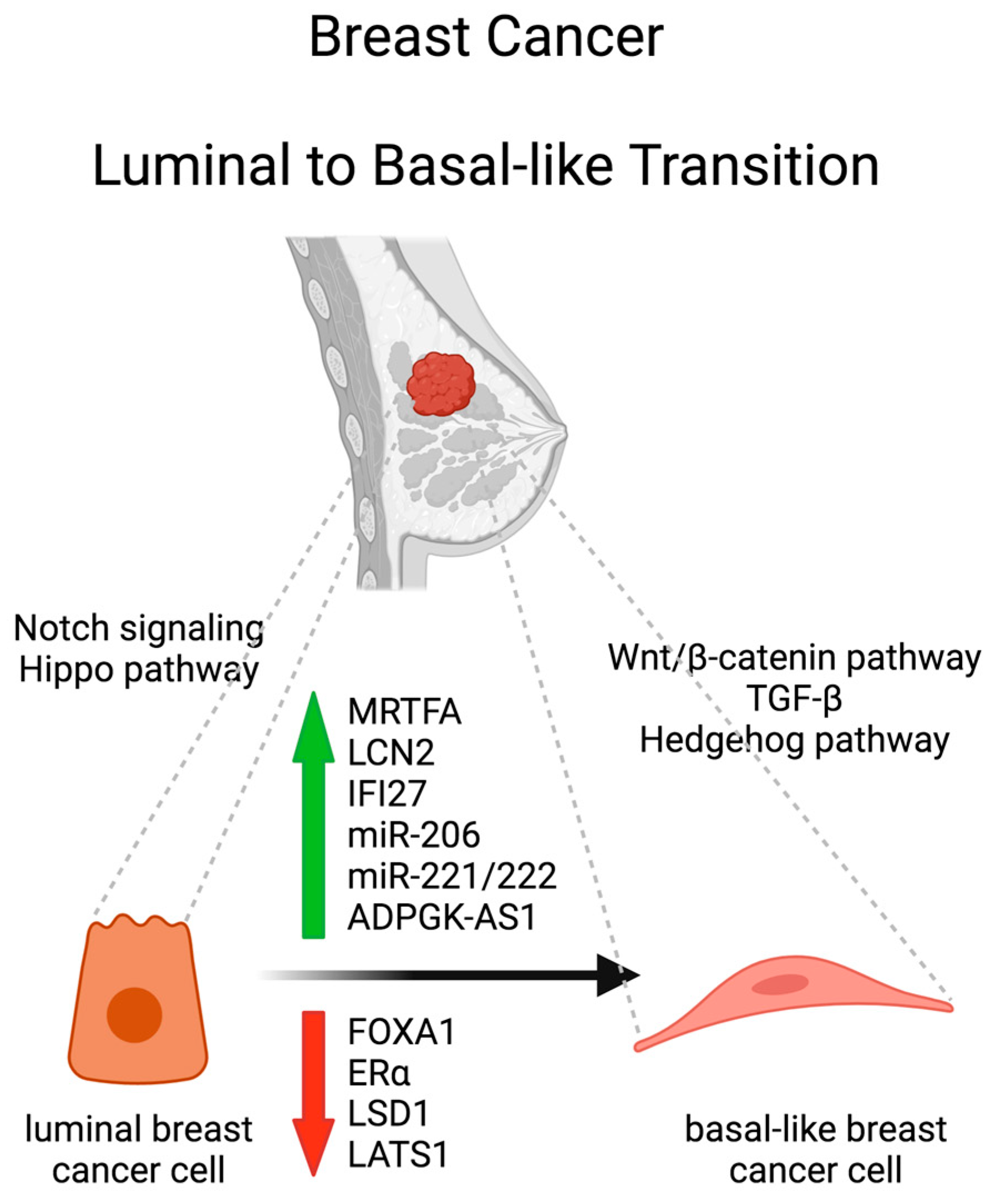
2. Luminal to Basal-Like Transition (LBT) in Breast Cancer
2.1. Transcriptional Regulation
2.2. Epigenetic Modifications, Chromatin Remodeling and Non-Coding RNAs
2.3. Signaling Pathways and Microenvironmental Factors
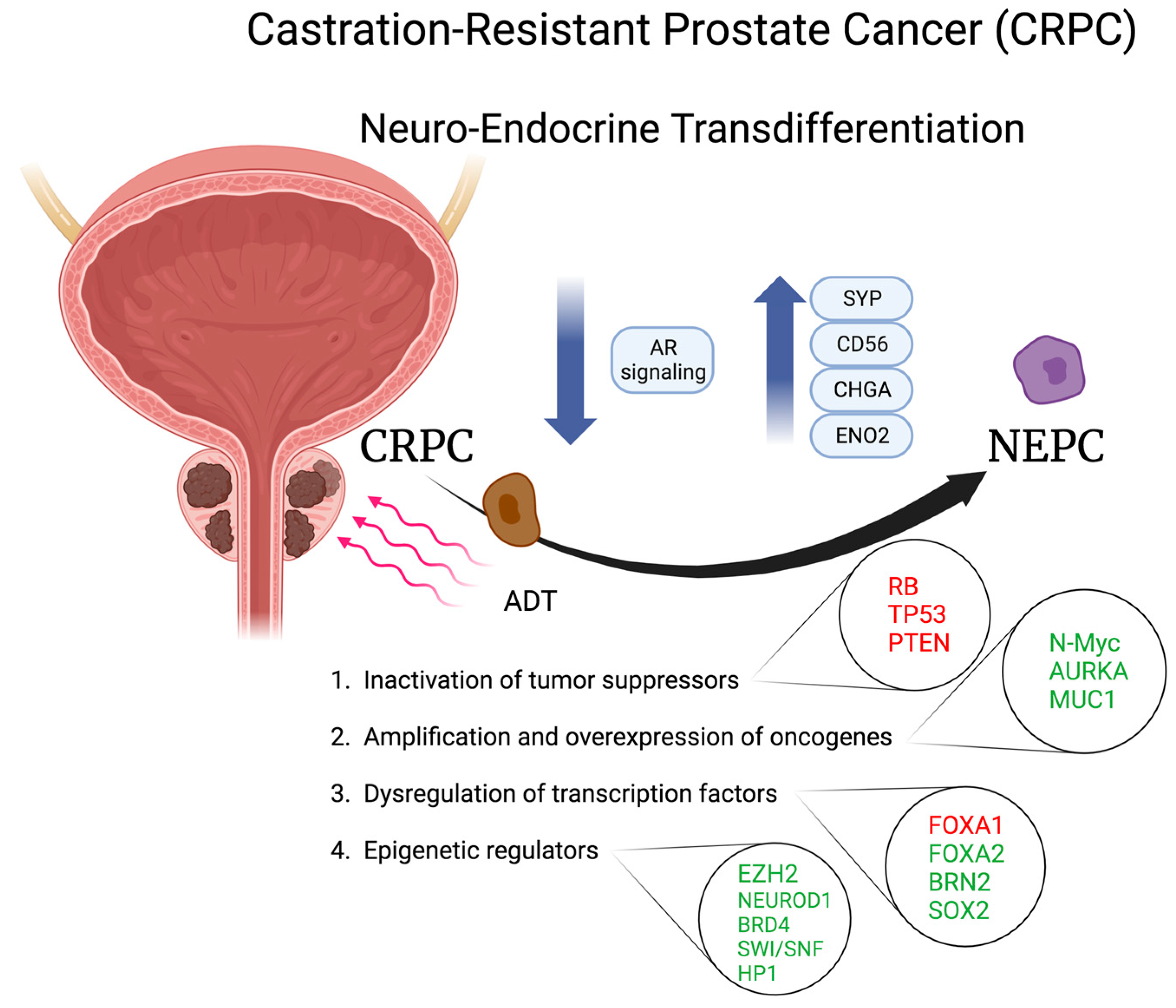
3. Neuro-Endocrine Transdifferentiation (NET) of Prostate Cancer
3.1. Inactivation of Tumor Suppressors
3.2. Amplification and Overexpression of Oncogenes
3.3. Dysregulation of Transcriptional Factors
3.4. Epigenetic Regulators and Chromatin Remodelling
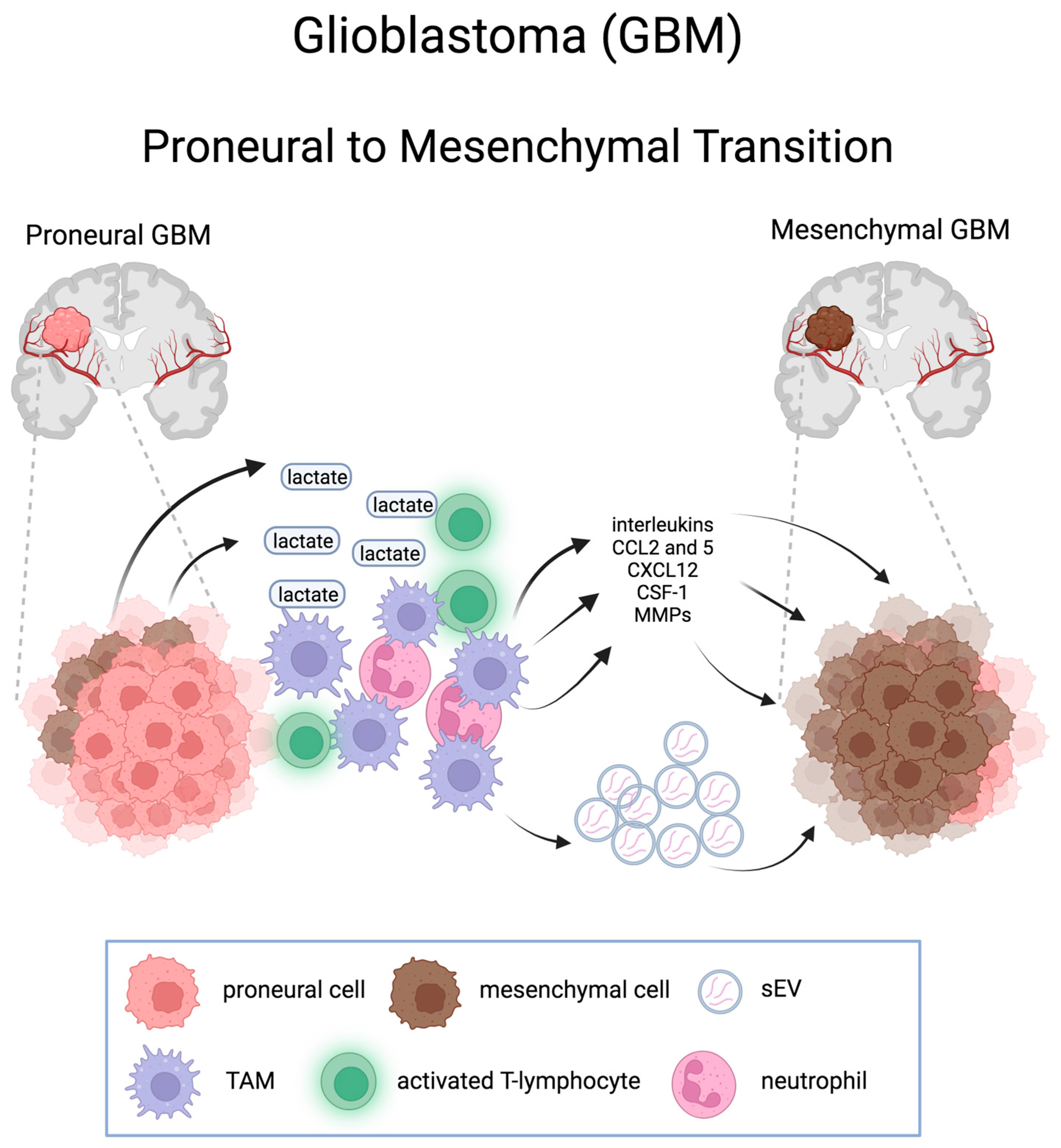
4. Proneural-to-Mesenchymal Transition in Glioblastoma
4.1. Role of Stem Cells in PMT
4.2. Epigenetic and Transcription Factors
4.3. Post-Transcriptional Mechanisms (Non-Coding RNAs and Proteostatic Mechanisms)
4.4. Metabolic Alterations in PMT
4.4. GBM Microenvironment: Immune Cells, Chemokines, and Extracellular Vesicles
4.5. Reversal of PMT
5. Acinar-to-Ductal Transdifferentiation in Pancreatic Carcinomas
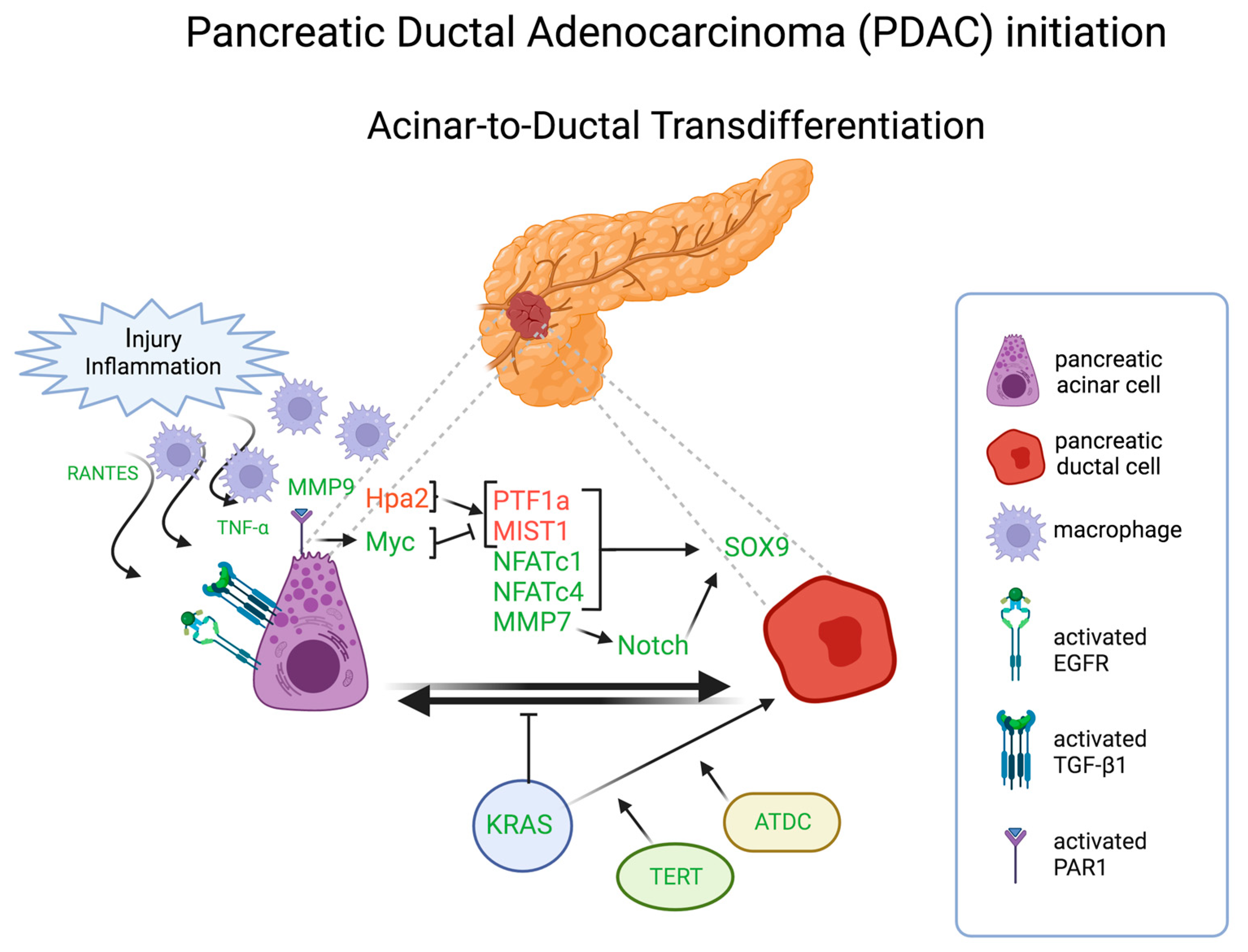
5.1. Trancriptional factors
5.2. Oncogenic signaling and tumor suppressor genes
5.3. Metabolic pathways
5.4. Microenvironment crosstalk
6. Clinical perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Buttitta, L.A.; Edgar, B.A. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol 2007, 19, 697–704. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness. Cancers (Basel) 2020, 12, 3716. [Google Scholar] [CrossRef]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol. 2020, 21, 268–283. [Google Scholar] [CrossRef]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J Clin Oncol 2014, 5, 412–424. [Google Scholar] [CrossRef]
- Rakha, E.A.; Reis-Filho, J.S.; Ellis, I.O. Basal-Like Breast Cancer: A Critical Review. J Clin Oncol 2008, 26, 2568–2581. [Google Scholar] [CrossRef]
- Badowska-Kozakiewicz, A.M.; Budzik, M.P. Immunohistochemical characteristics of basal-like breast cancer. Contemp Oncol (Pozn). 2016, 20, 436–443. [Google Scholar] [CrossRef]
- Gupta, P.B.; Fillmore, C.M.; Jiang, G.; Shapira, S.D.; Tao, K.; Kuperwasser, C.; Lander, E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011, 146, 633–644. [Google Scholar] [CrossRef]
- Bernardo, G.M.; Bebek, G.; Ginther, C.L.; Sizemore, S.T.; Lozada, K.L.; Miedler, J.D.; Anderson, L.A.; Godwin, A.K.; Abdul-Karim, F.W.; Slamon, D.J.; et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 2013, 32, 554–63. [Google Scholar] [CrossRef] [PubMed]
- Anzai, E.; Hirata, K.; Shibazaki, M.; Yamada, C.; Morii, M.; Honda, T.; Yamaguchi, N.; Yamaguchi, N. FOXA1 Induces E-Cadherin Expression at the Protein Level via Suppression of Slug in Epithelial Breast Cancer Cells. Biol Pharm Bull 2017, 40, 1483–148. [Google Scholar] [CrossRef] [PubMed]
- Jehanno, C.; Fernandez-Calero, T.; Habauzit, D.; Avner, S.; Percevault, F.; Jullion, E.; Le Goff, P.; Coissieux, M.M.; Muenst, S.; Marin, M.; et al. Nuclear accumulation of MKL1 in luminal breast cancer cells impairs genomic activity of ERα and is associated with endocrine resistance. Biochim Biophys Acta Gene Regul Mech 2020, 1863, 194507. [Google Scholar] [CrossRef]
- Jehanno, C.; Percevault, F.; Boujrad, N.; Le Goff, P.; Fontaine, C.; Arnal, J.F.; Primig, M.; Pakdel, F.; Michel, D.; Métivier, R.; et al. Nuclear translocation of MRTFA in MCF7 breast cancer cells shifts ERα nuclear/genomic to extra-nuclear/non genomic actions. Mol Cell Endocrinol 2021, 530, 111282. [Google Scholar] [CrossRef]
- Ariazi, E.A.; Taylor, J.C.; Black, M.A.; Nicolas, E.; Slifker, M.J.; Azzam, D.J.; Boyd, J. A New Role for ERα: Silencing via DNA Methylation of Basal, Stem Cell, and EMT Genes. Mol Cancer Res 2017, 15, 152–164. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, T.; Zhao, B.; Liu, Z.; Qian, R.; Zhang, J.; Shi, Y.; Wan, Y.; Li, Z.; Hu, X. E239K mutation abolishes the suppressive effects of lysine-specific demethylase 1 on migration and invasion of MCF7 cells. Cancer Sci 2022, 113, 489–499. [Google Scholar] [CrossRef]
- Pegoraro, S.; Ros, G.; Piazza, S.; Sommaggio, R.; Ciani, Y.; Rosato, A.; Sgarra, R.; Del Sal, G.; Manfioletti, G. HMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemness. Oncotarget 2013, 4, 1293–1308. [Google Scholar] [CrossRef]
- Yang, E.; Cisowski, J.; Nguyen, N.; O'Callaghan, K.; Xu, J.; Agarwal, A.; Kuliopulos, A.; Covic, L. Dysregulated protease activated receptor 1 (PAR1) promotes metastatic phenotype in breast cancer through HMGA2. Oncogene 2016, 35, 1529–1540. [Google Scholar] [CrossRef]
- Aylon, Y.; Furth, N.; Mallel, G.; Friedlander, G.; Nataraj, N.B.; Dong, M.; Hassin, O.; Zoabi, R.; Cohen, B.; Drendel, V.; et al. Breast cancer plasticity is restricted by a LATS1-NCOR1 repressive axis. Nat Commun 2022, 13, 7199. [Google Scholar] [CrossRef]
- Guttilla, I.K.; Adams, B.D.; White, B.A. ERα, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol Metab 2012, 23, 73–82. [Google Scholar] [CrossRef]
- Ghaforui-Fard, S.; Vafaee, R.; Taheri, M. Taurine-upregulated gene 1: A functional long noncoding RNA in tumorigenesis. J Cell Physiol 2019, 234, 17100–17112. [Google Scholar] [CrossRef]
- Yang, J.; Wu, W.; Wu, M.; Ding, J. Long noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and EMT process through regulating miR-3196/OTX1 axis in breast cancer. In Vitro Cell Dev Biol Anim 2019, 55, 522–532. [Google Scholar] [CrossRef]
- Bouras, T.; Pal, B.; Vaillant, F.; Harburg, G.; Asselin-Labat, M.L.; Oakes, S.R.; Lindeman, G.J.; Visvader, J.E. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 2008, 3, 429–441. [Google Scholar] [CrossRef]
- DiMeo, T.A.; Anderson, K.; Phadke, P.; Fan, C.; Perou, C.M.; Naber, S.; Kuperwasser, C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res 2009, 69, 5364–5373. [Google Scholar] [CrossRef]
- Mani, S.A.; Yang, J.; Brooks, M.; Schwaninger, G.; Zhou, A.; Miura, N.; Kutok, J.L.; Hartwell, K.; Richardson, A.L.; Weinberg, R.A. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A 2007, 104, 10069–10074. [Google Scholar] [CrossRef]
- Fedele, M.; Cerchia, L.; Chiappetta, G. The Epithelial-to-Mesenchymal Transition in Breast Cancer: Focus on Basal-Like Carcinomas. Cancers (Basel) 2017, 9, 134. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015, 12, 445–64. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, C.H.; Tan, L.D.; Wang, Q.S.; Li, X.Q.; Feng, Y.M. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br J Cancer 2014, 110, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Clauser, K.R.; Tam, W.L.; Fröse, J.; Ye, X.; Eaton, E.N.; Reinhardt, F.; Donnenberg, V.S.; Bhargava, R.; Carr, S.A.; et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol 2014, 16, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Valeta-Magara, A.; Gadi, A.; Volta, V.; Walters, B.; Arju, R.; Giashuddin, S.; Zhong, H.; Schneider, R.J. Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network. Cancer Res 2019, 79, 3360–3371. [Google Scholar] [CrossRef]
- Liu, S.; Alabi, B.R.; Yin, Q.; Stoyanova, T. Molecular mechanisms underlying the development of neuroendocrine prostate cancer. Semin Cancer Biol 2022, 86, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Spandonaro, F.; D'Angela, D.; Polistena, B.; Bruzzi, P.; Iacovelli, R.; Luccarini, I.; Stagni, P.; Brigido, A. Prevalence of Prostate Cancer at Different Clinical Stages in Italy: Estimated Burden of Disease Based on a Modelling Study. Biology (Basel) 2021, 10, 210. [Google Scholar] [CrossRef]
- Zou, M.; Toivanen, R.; Mitrofanova, A.; Floch, N.; Hayati, S.; Sun, Y.; Le Magnen, C.; Chester, D.; Mostaghel, E.A.; Califano, A.; et al. Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-Resistant Prostate Cancer. Cancer Discov 2017, 7, 736–749. [Google Scholar] [CrossRef]
- Lee, J.K.; Phillips, J.W.; Smith, B.A.; Park, J.W.; Stoyanova, T.; McCaffrey, E.F.; Baertsch, R.; Sokolov, A.; Meyerowitz, J.G.; Mathis, C.; et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell 2016, 29, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Liu, Y.; Liao, L.; Li, W.; Danielpour, D.; Xu, J. Neuroendocrine prostate carcinoma cells originate from the p63-expressing basal cells but not the pre-existing adenocarcinoma cells in mice. Cell Res 2019, 29, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Dong, B. , Miao J., Wang Y., Luo W., Ji Z., Lai H., Zhang M., Cheng X., Wang J., Fang Y., et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun Biol 2020, 3, 778. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Lee, J.K.; Sheu, K.M.; Wang, L.; Balanis, N.G.; Nguyen, K.; Smith, B.A.; Cheng, C.; Tsai, B.L.; Cheng, D.; et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 2018, 362, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Abdulfatah, E.; Reichert, Z.R.; Davenport, M.S.; Chinnaiyan, A.M.; Dadhania, V.; Wang, X.; Mannan, R.; Kunju, L.P.; Hollenbeck, B.K.; Montgomery, J.S.; et al. De novo neuroendocrine transdifferentiation in primary prostate cancer-a phenotype associated with advanced clinico-pathologic features and aggressive outcome. Med Oncol 2021, 38, 26. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.S.K.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.Y.; Rosario, S. , Wang Y., Mu P., Seshadri M., Goodrich Z.W., Goodrich M.M., Labbé D.P., Gomez E.C., Wang J., et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef]
- Mu, P.; Zhang, Z.; Benelli, M.; Karthaus, W.R.; Hoover, E.; Chen, C.C.; Wongvipat, J.; Ku, S.Y.; Gao, D.; Cao, Z.; et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017, 355, 84–88. [Google Scholar] [CrossRef]
- Beltran, H.; Rickman, D.S.; Park, K.; Chae, S.S.; Sboner, A.; MacDonald, T.Y.; Wang, Y.; Sheikh, K.L.; Terry, S.; Tagawa, S.T.; et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 2011, 1, 487–495. [Google Scholar] [CrossRef]
- Dardenne, E.; Beltran, H.; Benelli, M.; Gayvert, K.; Berger, A.; Puca, L.; Cyrta, J.; Sboner, A.; Noorzad, Z.; MacDonald, T.; et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 2016, 30, 563–577. [Google Scholar] [CrossRef]
- Zhang, X.W.; Li, J.Y.; Li, L.; Hu, W.Q.; Tao, Y.; Gao, W.Y.; Ye, Z.N.; Jia, H.Y.; Wang, J.N.; Miao, X.K.; et al. Neurokinin-1 receptor drives PKCɑ-AURKA/N-Myc signaling to facilitate the neuroendocrine progression of prostate cancer. Cell Death Dis 2023, 14, 384. [Google Scholar] [CrossRef]
- Yasumizu, Y.; Rajabi, H.; Jin, C.; Hata, T.; Pitroda, S.; Long, M.D.; Hagiwara, M.; Li, W.; Hu, Q.; Liu, S.; et al. MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat Commun 2020, 11, 338. [Google Scholar] [CrossRef]
- Hagiwara, M.; Yasumizu, Y.; Yamashita, N.; Rajabi, H.; Fushimi, A.; Long, M.D.; Li, W.; Bhattacharya, A.; Ahmad, R.; Oya, M.; et al. MUC1-C activates the BAF (mSWI/SNF) complex in prostate cancer stem cells. Cancer Res 2021, 81, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, W.; Zhang, Y.; Yuan, X.; Xu, K.; Yu, J.; Chen, Z.; Beroukhim, R.; Wang, H.; Lupien, M.; et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 2009, 138, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jin, H.; Zhao, J.C.; Yang, Y.A.; Li, Y.; Yang, X.; Dong, X.; Yu, J. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene 2017, 36, 4072–4080. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Nakayama, K.; Cardiff, R.D.; Borowsky, A.D.; Kaul, K.; Williams, R.; Krajewski, S.; Mercola, D.; Carpenter, P.M.; Bowtell, D.; et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 2010, 18, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Pang, J.; Wang, L.A.; Huang, Z.; Xu, J.; Yang, X.; Xie, Q.; Huang, Y.; Tang, T.; Tong, D.; et al. Histone demethylase PHF8 drives neuroendocrine prostate cancer progression by epigenetically upregulating FOXA2. J Pathol 2021, 253, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.L.; Thaper, D.; Vahid, S.; Davies, A.; Ketola, K.; Kuruma, H.; Jama, R.; Nip, K.M.; Angeles, A.; Johnson, F.; et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov 2017, 7, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, D.; Yang, T.L.; Tabatabai, Z.L.; Majid, S.; Dahiya, R.; Tanaka, Y.; Saini, S. BRN4 is a novel driver of neuroendocrine differentiation in castration-resistant prostate cancer and is selectively released in extracellular vesicles with BRN2. Clin Cancer Res 2019, 25, 6532–6545. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Zhang, P.; Liang, C.; Yu, Y.; Wang, X. ASCL1-mediated ferroptosis resistance enhances the progress of castration-resistant prostate cancer to neurosecretory prostate cancer. Free Radic Biol Med 2023, 205, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Baca, S.C.; Takeda, D.Y.; Seo, J.H.; Hwang, J.; Ku, S.Y.; Arafeh, R.; Arnoff, T.; Agarwal, S.; Bell, C.; O’Connor, E.; et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat Commun 2021, 12, 1979. [Google Scholar] [CrossRef] [PubMed]
- Brady, N.J.; Bagadion, A.M; Singh, R.; Conteduca, V.; Van Emmenis, L.; Arceci, E.; Pakula, H.; Carelli, R.; Khani, F.; Bakht, M.; et al. Temporal evolution of cellular heterogeneity during the progression to advanced AR-negative prostate cancer. Nat Commun 2021, 12, 3372. [Google Scholar] [CrossRef] [PubMed]
- Rubin, M.A.; Bristow, R.G.; Thienger, P.D.; Dive, C.; Imielinski, M. Impact of Lineage Plasticity to and from a Neuroendocrine Phenotype on Progression and Response in Prostate and Lung Cancers. Mol Cell 2020, 80, 562–577. [Google Scholar] [CrossRef]
- Song, Z.; Cao, Q.; Guo, B.; Zhao, Y.; Li, X.; Lou, N.; Zhu, C.; Luo, G.; Peng, S.; Li, G.; et al. Overexpression of RACGAP1 by E2F1 Promotes Neuroendocrine Differentiation of Prostate Cancer by Stabilizing EZH2 Expression. Aging Dis 2023. [Google Scholar] [CrossRef]
- Sreekumar, A.; Saini, S. Role of transcription factors and chromatin modifiers in driving lineage reprogramming in treatment-induced neuroendocrine prostate cancer. Front Cell Dev Biol 2023, 11, 1075707. [Google Scholar] [CrossRef] [PubMed]
- Nouruzi, S.; Ganguli, D.; Tabrizian, N.; Kobelev, M.; Sivak, O.; Namekawa, T.; Thaper, D.; Baca, S.C.; Freedman, M.L.; Aguda, A.; et al. ASCL1 activates neuronal stem cell-like lineage programming through remodeling of the chromatin landscape in prostate cancer. Nat Commun 2022, 13, 2282. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; Barthel, F.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e6. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Bergmann, N.; Delbridge, C.; Gempt, J.; Feuchtinger, A.; Walch, A.; Schirmer, L.; Bunk, W.; Aschenbrenner, T.; Liesche-Starnecker, F.; Schlegel, J. The Intratumoral Heterogeneity Reflects the Intertumoral Subtypes of Glioblastoma Multiforme: A Regional Immunohistochemistry Analysis. Front Oncol 2020, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Segerman, A.; Niklasson, M.; Haglund, C.; Bergström, T.; Jarvius, M.; Xie, Y.; Westermark, A.; Sönmez, D.; Hermansson, A.; Kastemar, M.; et al. Clonal Variation in Drug and Radiation Response among Glioma-Initiating Cells Is Linked to Proneural-Mesenchymal Transition. Cell Rep 2016, 17, 2994–3009. [Google Scholar] [CrossRef] [PubMed]
- Phillips, HS.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Company, C.; Dramaretska, Y.; Barozzi, I.; Göhrig, A.; Kertalli, S.; Großmann, M.; Naumann, H.; Sanchez-Bailon, M.P.; Hulsman, D.; et al. Phenotypic Mapping of Pathologic Cross-Talk between Glioblastoma and Innate Immune Cells by Synthetic Genetic Tracing. Cancer Discov 2021, 11, 754–777. [Google Scholar] [CrossRef]
- Aldape, K.; Zadeh, G.; Mansouri, S.; Reifenberger, G.; von Deimling, A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 2015, 129, 829–848. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Kosty, J.; Lu, F.; Kupp, R.; Mehta, S.; Lu, Q.R. Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas. Cell Cycle 2017, 16, 1654–1660. [Google Scholar] [CrossRef]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef]
- Fedele, M.; Cerchia, L.; Pegoraro, S.; Sgarra, R.; Manfioletti, G. Proneural-Mesenchymal Transition: Phenotypic Plasticity to Acquire Multitherapy Resistance in Glioblastoma. Int J Mol Sci 2019, 20, 274. [Google Scholar] [CrossRef]
- Osborn, A.G.; Louis, D.N.; Poussaint, T.Y.; Linscott, L.L.; Salzman, K.L. The 2021 World Health Organization Classification of Tumors of the Central Nervous System: What Neuroradiologists Need to Know. AJNR Am J Neuroradiol 2022, 43, 928–937. [Google Scholar] [CrossRef]
- Dénes, A.; Bontell, T.O.; Barchéus, H.; Vega, S.F.; Carén, H.; Lindskog, C.; Jakola, A.S.; Smits, A. The clinical value of proneural, classical and mesenchymal protein signatures in WHO 2021 adult-type diffuse lower-grade gliomas. PLoS One 2023, 18, e0285732. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- Loras, A.; Gonzalez-Bonet, L.G.; Gutierrez-Arroyo, J.L.; Martinez-Cadenas, C.; Marques-Torrejon, M.A. Neural Stem Cells as Potential Glioblastoma Cells of Origin. Life (Basel) 2023, 13, 905. [Google Scholar] [CrossRef] [PubMed]
- Suvà, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, T. N.; Kukekov, V.G.; Laywell, E.D.; Suslov, O.N.; Vrionis, F.D.; Steindler, D.A. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002, 39, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Kappadakunnel, M.; Eskin, A.; Dong, J.; Nelson, S.F.; Mischel, P.S.; Liau, L.M.; Ngheimphu, P.; Lai, A.; Cloughesy, T.F.; Goldin, J.; et al. Stem cell associated gene expression in glioblastoma multiforme: relationship to survival and the subventricular zone. J Neurooncol 2020, 96, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Kim, L.J.Y.; Wu, Q.; Wallace, L.C.; Prager, B.C.; Sanvoranart, T.; Gimple, R.C.; Wang, X.; Mack, S.C.; Miller, T.E.; et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med 2017, 23, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Joshi, K.; Li, J.; Kim, S.H.; Li, P.; Santana-Santos, L.; Luthra, S.; Chandran, U.R.; Benos, P.V.; Smith, L.; et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A 2013, 110, 8644–8649. [Google Scholar] [CrossRef]
- Pine, A.R.; Cirigliano, S.M.; Nicholson, J.G.; Hu, Y.; Linkous, A.; Miyaguchi, K.; Edwards, L.; Singhania, R.; Schwartz, T.H.; Ramakrishna, R.; et al. Tumor Microenvironment Is Critical for the Maintenance of Cellular States Found in Primary Glioblastomas. Cancer Discov 2020, 10, 964–979. [Google Scholar] [CrossRef]
- Kalebic, N.; Namba, T. Inheritance and flexibility of cell polarity: a clue for understanding human brain development and evolution. Development 2021, 148, dev199417. [Google Scholar] [CrossRef]
- Keller, M.; Blom, M.; Conze, L.L.; Guo, M.; Hägerstrand, D.; Aspenström, P. Altered cytoskeletal status in the transition from proneural to mesenchymal glioblastoma subtypes. Sci Rep 2022, 12, 9838. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Salazar, K.L.; Balasubramaniyan, V.; Wani, K.; Heathcock, L.; Hollingsworth, F.; James, J.D.; Gumin, J.; Diefes, K.L.; Kim, S.H.; et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev 2011, 25, 2594–2609. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Cui, X.; Wang, Q.; Fang, C.; Tan, Y.; Wang, Y.; Yi, K.; Yang, C.; You, H.; Shang, R.; et al. RUNX1 contributes to the mesenchymal subtype of glioblastoma in a TGFbeta pathway-dependent manner. Cell Death Dis 2019, 10, 877. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Wang, J.; Waghmare, I.; Sartini, S.; Coviello, V.; Zhang, Z.; Kim, S.H.; Mohyeldin, A.; Pavlyukov, M.S.; Minata, M.; et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res 2016, 76, 7219–7230. [Google Scholar] [CrossRef] [PubMed]
- Guardia, G.D.A.; Correa, B.R.; Araujo, P.R.; Qiao, M.; Burns, S.; Penalva, L.O.F.; Galante, P.A.F. Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles. NPJ Genom Med 2020, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Xu, J.; Fan, Y.; Qi, Y.; Wang, S.; Zhao, S.; Guo, X.; Xue, H.; Deng, L.; Zhao, R.; et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPβ degradation to facilitate proneural-to-mesenchymal transition. J Exp Clin Cancer Res 2022, 41, 223. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, Z.; Xu, J.; Wang, H.; Guo, Q.; Li, B.; Li, M.; Xu, H.; Qi, Y.; Zhao, S.; et al. SPI1-mediated MIR222HG transcription promotes proneural-to-mesenchymal transition of glioma stem cells and immunosuppressive polarization of macrophages. Theranostics 2023, 13, 3310–3329. [Google Scholar] [CrossRef]
- Qiu, W.; Xiao, Z.; Yang, Y.; Jiang, L.; Song, S.; Qi, X.; Chen, Y.; Yang, H.; Liu, J.; Chu, L. USP10 deubiquitinates RUNX1 and promotes proneural-to-mesenchymal transition in glioblastoma. Cell Death Dis 2023, 14, 207. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, S.; Li, H.L.; Luo, H.; Wu, X.; Lu, J.; Wang, H.W.; Chen, Y.; Chen, D.; Wu, W.T.; et al. FOSL1 promotes proneural-to-mesenchymal transition of glioblastoma stem cells via UBC9/CYLD/NF-κB axis. Mol Ther 2022, 30, 2568–2583. [Google Scholar] [CrossRef]
- Fedele, M.; Sgarra, R.; Battista, S.; Cerchia, L.; Manfioletti, G. The Epithelial-Mesenchymal Transition at the Crossroads between Metabolism and Tumor Progression. Int J Mol Sci 2022, 23, 800. [Google Scholar] [CrossRef]
- Li, L.; Yang, M.; Pu, X.; Tang, Y.; Fei, F.; Li, Z.; Hou, H.; Chen, Q.; Wang, Q.; Wu, Y.; et al. ALKBH5-PYCR2 Positive Feedback Loop Promotes Proneural-Mesenchymal Transition Via Proline Synthesis In GBM. J Cancer 2023, 14, 1579–1591. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr Biol 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Chen, M.; Ren, R.; Lin, W.; Xiang, L.; Zhao, Z.; Shao, B. Exploring the oncostatin M (OSM) feed-forward signaling of glioblastoma via STAT3 in pan-cancer analysis. Cancer Cell Int 2021, 21, 565. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.B.; Lee, S.; Harmanci, A.S.; Patel, R.; Latha, K.; Yang, Y.; Marisetty, A.; Lee, H.K.; Heimberger, A.B.; Fuller, G.N.; et al. CXCR4 expression is associated with proneural-to-mesenchymal transition in glioblastoma. Int J Cancer 2023, 152, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Soni, N.; Pinero, G.; Giotti, B.; Eddins, D.J.; Lindblad, K.E.; Ross, J.L.; Puigdelloses Vallcorba, M.; Joshi, T.; Angione, A.; et al. Monocyte depletion enhances neutrophil influx and proneural to mesenchymal transition in glioblastoma. Nat Commun 2023, 14, 1839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, J.; Chen, Z.; Wang, H.; Xue, H.; Yang, C.; Guo, Q.; Qi, Y.; Guo, X.; Qian, M.; et al. Transfer of MicroRNA via Macrophage-Derived Extracellular Vesicles Promotes Proneural-to-Mesenchymal Transition in Glioma Stem Cells. Cancer Immunol Res 2020, 8, 966–981. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Varn, F.S.; Park, S.H.; Yoon, B.W.; Park, H.R.; Lee, C.; Verhaak, R.G.W.; Paek, S.H. Perspective of mesenchymal transformation in glioblastoma. Acta Neuropathol Commun 2021, 9, 50. [Google Scholar] [CrossRef]
- Pavlyukov, M.S.; Yu, H.; Bastola, S.; Minata, M.; Shender, V.O.; Lee, Y.; Zhang, S.; Wang, J.; Komarova, S.; Wang, J.; et al. Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma Via Intercellular Transfer of Splicing Factors. Cancer Cell 2018, 34, 119–135.e10. [Google Scholar] [CrossRef] [PubMed]
- Park, N.I.; Guilhamon, P.; Desai, K.; McAdam, R.F.; Langille, E.; O’Connor, M.; Lan, X.; Whetstone, H.; Coutinho, F.J.; Vanner, R.J.; et al. ASCL1 reorganizes chromatin to direct neuronal fate and suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell 2017, 21, 209–224.e7. [Google Scholar] [CrossRef]
- Narayanan, A.; Gagliardi, F.; Gallotti, A.L.; Mazzoleni, S.; Cominelli, M.; Fagnocchi, L.; Pala, M.; Piras, I.S.; Zordan, P.; Moretta, N.; et al. The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Cell Death Differ 2019, 26, 1813–1831. [Google Scholar] [CrossRef]
- Haltom, A.R.; Hassen, W.E.; Hensel, J.; Kim, J.; Sugimoto, H.; Li, B.; McAndrews, K.M.; Conner, M.R.; Kirtley, M.L.; Luo, X.; et al. Engineered exosomes targeting MYC reverse the proneural-mesenchymal transition and extend survival of glioblastoma. Extracell Vesicle 2022, 1, 100014. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J Clin 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Storz, P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2017, 14, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Burclaff, J.; Mills, J.C. Plasticity of differentiated cells in wound repair and tumorigenesis, part I: stomach and pancreas. Dis Model Mech 2018, 11, dmm033373. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.C.; Sansom, O.J. Reserve stem cells: Differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci Signal 2015, 8, re8. [Google Scholar] [CrossRef]
- Jiang, M.; Azevedo-Pouly, A.C.; Deering, T.G.; Hoang, C.Q.; DiRenzo, D.; Hess, D.A.; Konieczny, S.F.; Swift, G.H.; MacDonald, R.J. MIST1 and PTF1 collaborate in feed-forward regulatory loops that maintain the pancreatic acinar phenotype in adult mice. Mol Cell Biol 2016, 36, 2945–2955. [Google Scholar] [CrossRef]
- Krah, N.M.; Narayanan, S.M.; Yugawa, D.E.; Straley, J.A.; Wright, C.V.E.; MacDonald, R.J.; Murtaugh, L.C. Prevention and reversion of pancreatic tumorigenesis through a differentiation-based mechanism. Dev Cell 2019, 50, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Krah, N.M.; De La O, J.-P.; Swift, G.H.; Hoang, C.Q.; Willet, S.G.; Chen Pan, F.; Cash, G.M.; Bronner, M.P.; Wright, C.V.; MacDonald, R.J.; et al. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. eLife 2015, 4, e07125. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; DiRenzo, D.; Qu, C.; Barney, D.; Miley, D.; Konieczny, S.F. Maintenance of acinar cell organization is critical to preventing Krasinduced acinar-ductal metaplasia. Oncogene 2013, 32, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Kopp, J.L.; von Figura, G.; Mayes, E.; Liu, F.-F.; Dubois, C.L.; Morris, J.P.; Pan, F.C.; Akiyama, H.; Wright, C.V.; Jensen, K.; et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012, 22, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Julian, L.M.; McDonald, A.C.; Stanford, W.L. Direct reprogramming with SOX factors: masters of cell fate. Curr Opin Genet Dev 2017, 46, 24–36. [Google Scholar] [CrossRef]
- Grimm, D.; Bauer, J.; Wise, P.; Krüger, M.; Simonsen, U.; Wehland, M.; Infanger, M.; Corydon, T.J. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol 2020, 67, 122–53. [Google Scholar] [CrossRef]
- Chen, N.M.; Singh, G.; Koenig, A.; Liou, G.Y.; Storz, P.; Zhang, J.S.; Regul, L.; Nagarajan, S.; Kühnemuth, B.; A Johnsen, S.A.; et al. NFATc1 Links EGFR Signaling to Induction of Sox9 Transcription and Acinar-Ductal Transdifferentiation in the Pancreas. Gastroenterology 2015, 148, 1024–1034.e9. [Google Scholar] [CrossRef]
- Chen, N.M.; Neesse, A.; Dyck, M.L.; Steuber, B.; Koenig, A.O.; Lubeseder-Martellato, C.; Winter, T.; Forster, T.; Bohnenberger, H.; Kitz, J.; et al. Context-Dependent Epigenetic Regulation of Nuclear Factor of Activated T Cells 1 in Pancreatic Plasticity. Gastroenterology 2017, 152, 1507–1520.e15. [Google Scholar] [CrossRef]
- Hessmann, E.; Zhang, J.S.; Chen, N.M.; Hasselluhn, M.; Geou-Yarh Liou, G.Y.; Storz, P.; Ellenrieder, V.; Billadeau, D.D.; Koenig, A. NFATc4 Regulates Sox9 Gene Expression in Acinar Cell Plasticity and Pancreatic Cancer Initiation. Stem Cells Int, 2016, 2016, 5272498. [Google Scholar] [CrossRef]
- Li, S.; Xie, K. Ductal metaplasia in pancreas. Biochim Biophys Acta Rev Cancer 2022, 1877, 188698. [Google Scholar] [CrossRef] [PubMed]
- Means, A.L.; Meszoely, I.M.; Suzuki, K.; Miyamoto, Y.; Rustgi, A.K.; Coffey Jr, R.J.; Wright, C.V.E.; Stoffers, D.A.; Leach, S.D. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development 2005, 132, 3767–3776. [Google Scholar] [CrossRef]
- Sawey, E.T.; Johnson, J.A.; Crawford, H.C. Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc Natl Acad Sci U S A 2007, 104, 19327–19332. [Google Scholar] [CrossRef]
- Liu, J.; Akanuma, N.; Liu, C.; Naji, A.; Halff, G.A.; Washburn, W.K.; Sun, L.; Wang, P. TGF-β1 promotes acinar to ductal metaplasia of human pancreatic acinar cells. Sci Rep 2016, 6, 30904. [Google Scholar] [CrossRef]
- Wagner, M.; Lührs, H.; Klöppel, G.; Adler, G.; Schmid, R.M. Malignant transformation of duct-like cells originating from acini in transforming growth factor transgenic mice. Gastroenterology 1998, 115, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Neuhöfer, P.; Roake, C.M.; Kim, S.J.; Lu, R.J.; West, R.B.; Charville, G.W.; Artandi, S.E. Acinar cell clonal expansion in pancreas homeostasis and carcinogenesis. Nature 2021, 597, 715–719. [Google Scholar] [CrossRef]
- Wang, L.; Yang, H.; Zamperone, A.; Diolaiti, D.; Palmbos, P.L.; Abel, E.V.; Purohit, V.; Dolgalev, I.; Rhim, A.D.; Ljungman, M.; et al. ATDC is required for the initiation of KRAS-induced pancreatic tumorigenesis. Genes Dev 2019, 3, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Ilan, N.; Sanderson, R.D. Forty years of basic and translational heparanase research. Adv Exp Med Biol 2020, 1221, 3–59. [Google Scholar] [CrossRef]
- Levy-Adam, F.; Feld, S.; Cohen-Kaplan, V.; Shteingauz, A.; Gross, M.; Arvatz, G.; Naroditsky, I.; Ilan, N.; Doweck, I.; Vlodavsky, I. Heparanase 2 interacts with heparan sulfate with high affinity and inhibits heparanase activity. J Biol Chem 2010, 285, 28010–28009. [Google Scholar] [CrossRef] [PubMed]
- Kayal, Y.; Barash, U.; Naroditsky, I.; Ilan, N.; Vlodavsky, I. Heparanase 2 (Hpa2)- a new player essential for pancreatic acinar cell differentiation. Cell Death Dis 2023, 14, 465. [Google Scholar] [CrossRef]
- Tan, M.; Chang, Y.; Liu, X.; Li, H.; Tang, Z.; Nyati, M.K.; Sun, Y. The Sag-Shoc2 axis regulates conversion of mPanINs to cystic lesions in Kras pancreatic tumor model. Cell Rep 2022, 41, 111837. [Google Scholar] [CrossRef] [PubMed]
- Radyk, M.D.; Nelson, B.S.; Halbrook, C.J.; Wood, A.; Lavoie, B.; Salvatore, L.; Corfas, G.; Colacino, J.A.; Shah, Y.M.; Crawford, H.C.; et al. Glucose-6-phosphate dehydrogenase deficiency accelerates pancreatic acinar-to-ductal metaplasia. bioRxiv 2023, 2023.11.06.565895. [Google Scholar] [CrossRef]
- Ding, L.; Liou, G.Y.; Schmitt, D.M.; Storz, P.; Zhang, J.S.; Billadeau, D.D. Glycogen synthase kinase-3β ablation limits pancreatitis-induced acinar-to-ductal metaplasia. J Pathol 2017, 243, 65–77. [Google Scholar] [CrossRef]
- Grisan, F.; Spacci, Paoli, C.; Costamagna, A.; Fantuz, M.; Miriam Martini, M.; Lefkimmiatis, K.; Carrer, A. Cholesterol Activates Cyclic AMP Signaling in Metaplastic Acinar Cells. Metabolites 2021, 11, 141. [CrossRef] [PubMed]
- Tekin, C.; Scicluna, B.P.; Lodestijn, S.C.; Shi, K.; Bijlsma, M.F.; Spek, C.A. Protease-activated receptor 1 drives and maintains ductal cell fates in the premalignant pancreas and ductal adenocarcinoma. Mol Oncol 2021, 15, 3091–3108. [Google Scholar] [CrossRef]
- Liou, G.Y.; Döppler, H.; Necela, B.; Krishna, M.; Crawford, H.C.; Raimondo, M.; Storz, P. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-κB and MMPs. J Cell Biol 2013, 202, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, T.; Hong, D.; Dong, B.; Wang, Y.; Huang, H.; Zhang, W.; Lian, B.; Ji, B.; Shi, H. Single-cell transcriptional regulation and genetic evolution of neuroendocrine prostate cancer. iScience 2022, 25, 104576. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.F.; Chen, Y.A.; Hernandez, E.; Pong, R.C.; Ma, S.; Hofstad, M.; Kapur, P.; Zhau, H.; Chung, L.W.; Lai, C.H.; et al. The central role of Sphingosine kinase 1 in the development of neuroendocrine prostate cancer (NEPC): A new targeted therapy of NEPC. Clin Transl Med, 2022, 12, e695. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liang, Y.; Chang, W.; Hu, B.; Zhang, Y. Triple Negative breast cancer depends on sphingosine kinase 1 (SphK1)/sphingosine-1-phosphate (S1P)/sphingosine 1-phosphate receptor 3 (S1PR3)/notch signaling for metastasis. Med Sci Monit 2018, 24, 1912–1923. [Google Scholar] [CrossRef]
- Lee, S.H.; Kwon, H.J.; Park, S.; Kim, C.I.; Ryu, H.; Kim, S.S.; Park, J.B.; Kwon, J.T. Macrophage migration inhibitory factor (MIF) inhibitor 4-IPP downregulates stemness phenotype and mesenchymal trans-differentiation after irradiation in glioblastoma multiforme. PLoS One 2021, 16, e0257375. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Z.; Feng, Y.; Gao, J.; Wang, B.; Lian, C.; Diao, B. Integration analysis of single-cell and spatial transcriptomics reveal the cellular heterogeneity landscape in glioblastoma and establish a polygenic risk model. Front Oncol 2023, 13, 1109037. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




