1. Introduction
Despite progress in water access, 27% of the global population, particularly in low and middle-income countries (LMIC), lacks access to clean drinking water [
1]. According to the World Health Organization (WHO), water-related diseases account for an estimated 3.4 million deaths per year globally [
2]. Microbiologically contaminated drinking water can transmit diseases such as diarrhea, cholera, dysentery, typhoid, and polio and is estimated to cause approximately 505,000 diarrheal deaths each year [
1]. To effectively manage many of these diseases, safe and efficient use of antimicrobial drugs is crucial for treatment and prevention [
3]. However, this may exacerbate antimicrobial resistance (AMR). The misuse and overuse of antimicrobials in humans, animals, and plants are the primary factors driving the development of drug-resistant pathogens [
3,
4]. Moreover, the inadequate water, sanitation, and hygiene (WASH) increase the spread of AMR in communities [
5]. AMR poses a significant global threat to clinical treatment in human and animal health and overall human development. In 2019, bacterial AMR directly caused 1.27 million deaths worldwide and contributed to an additional 4.95 million deaths [
6]. This surpasses the death toll from COVID-19, which stands at about seven million deaths over the four years from 2019 to 2023 [
7].
In order to tackle the spread of infectious disease and AMR, regular and reliable epidemiological surveillance is crucial to timely detect and monitor the emergence, and thereafter guide resource allocation [
8,
9]. Surveillance methods can vary widely based on the pathogen, AMR targets, disease, and resource availability. They evaluate pathogen diversity, variants, and outbreak impact to inform mitigation strategies. Traditional disease surveillance relies on clinical testing and reporting [
10,
11,
12], involving the collection and analysis of individual test reports over time to confirm and assess spatial and temporal outbreak patterns [
11,
13]. But clinical testing tends to focus on symptomatic cases in severe clinical stages, potentially underestimating morbidity from asymptomatic or self-limiting infections, and can under-report cases in areas with limited healthcare access [
9,
14,
15,
16].
Wastewater and Environmental Surveillance (WES) is an emerging approach for monitoring multiple pathogens and AMR targets that complements clinical surveillance. WES provides nearly real-time evidence or in many cases can provide early warning signals for the disease outbreak at a population level [
17,
18,
19,
20]. There is a growing focus on enhancing disease surveillance by integrating WES into existing clinical surveillance systems [
17,
21,
22,
23]. The WES can detect pathogens and AMR targets during the early stage of infection, enabling timely interventions, and taking actions to mitigate potential outbreaks [
19,
24,
25,
26]. It yields results with minimal personal intervention, reduced ethical challenges, and independence from healthcare facility availability or individuals' willingness to undergo testing [
12,
27]. The approach informs evidence-based public health responses, specifically focusing on high-risk areas, and optimizes resource allocation to enhance the effectiveness of disease control measures [
28,
29].
In LMIC, many infected individuals may not seek clinical testing due to several reasons such as limited access to healthcare services, and limitation of diagnostic tools [
14]. In such settings WES can be an effective alternative monitoring approach for accurate estimation of the burden of disease outbreaks at the community level [
14,
15]. When effectively implemented, WES can offer additional evidence of potential infection outbreaks, enabling timely and appropriate responses to emerging public health issues [
30,
31,
32]. However, its adoption still lags, particularly in LMICs [
33,
34,
35,
36].
This study is a part of ODIN consortium project, aiming to improve WES of waterborne pathogens and AMR in sub-Saharan Africa [
37]. Also, the project aims to strengthen genomics and bioinformatics capacities in sub-Saharan countries, foster international multidisciplinary research for reducing disease and illness cases resulting from contaminated drinking water in the region. Within the project scope, this paper compiles data on potential pathogens for WES and the existing surveillance network. The present study explores clinical and environmental surveillance for communicable disease epidemiology in three Sub-Saharan countries (Tanzania, Burkina Faso, and the Democratic Republic of the Congo - DRC). It evaluates existing clinical and environmental surveillance systems and assesses the potential for establishing a new WES system in the region. Additionally, to identify priority pathogens for WES, this study examines recent infectious disease outbreaks in sub-Saharan Africa, focusing specifically on transmission of waterborne pathogens and AMR in the environment.
2. Methodology
We arranged stakeholder workshops (November-December 2023), in each country (Tanzania, Burkina Faso, and the DRC), covering four key themes: (a) current clinical surveillance, (b) existing environmental surveillance, (c) challenges in current clinical surveillance, and (d) potential of wastewater and environmental surveillance. The participants in the workshops comprised distinguished experts in clinical and environmental fields, including academicians, researchers, policymakers, healthcare authorities, and representatives from national and international, governmental, and non-governmental organizations (NGOs), such as the West African Health Organization, WHO country office, National Office for Water and Sanitation, Water Analysis Laboratories, and experts affiliated with WHO Polio Eradication Program (Table 1). Before the main surveys, the questionnaire was piloted to ensure comprehensiveness in terminologies across participating countries.
Among the participants, we conducted two surveys. In both surveys, responses were collected in English in Tanzania and French in Burkina Faso and the DRC. The
Mentimeter.com platform was used for mapping priority pathogens and AMR targets for creating the new WES system in each country. In the Mentimeter survey, priority pathogens were grouped into three categories: (a) waterborne pathogens requiring enhanced public health surveillance, (b) other than waterborne pathogens suitable for monitoring through WES, and (c) antimicrobial resistance (AMR) targets requiring increased surveillance and response for public health significance. A predetermined list of pathogens and AMR targets, based on local epidemiological evidence and prior knowledge of regional outbreaks, was provided to unify the options. Also, a free-text field was included in the survey after each category, allowing respondents to prioritize other pathogens not covered by the survey organizers.
The second survey was conducted covering four key themes, as same as the workshop themes: (a) current clinical surveillance, (b) existing WES, (c) challenges in current clinical surveillance, and (d) potential of WES. The second survey, utilizing the Webropol platform, featured both open-ended and multiple-choice questions in the questionnaire.
For analyzing the survey responses from the three separate country-specific workshops,
Mentimeter.com data were transferred to Microsoft Excel (MS Excel) and visualized using OriginPro software. Quantitative analyses of Webropol responses were also conducted in MS Excel. To provide a comprehensive sub-Saharan overview, responses from all three countries (Tanzania, Burkina Faso, and the DRC) were aggregated. Textual questionnaire responses were organized in MS Word thematically categorized and marked with expertise fields. We employed thematic analysis for qualitative data analysis.
Table 1.
National-level workshop participants.
Table 1.
National-level workshop participants.
| Tanzania |
Burkina Faso |
DRC |
| Academicians and researchers |
Academicians, Researchers |
Academicians and researchers |
| Policymakers and representatives from ministry of Health, and National Public Health Laboratory |
Policymakers and representatives from five- ministries of the One health program:
- -
Health and public hygiene - -
Ministry of the Environment, Energy, Water and Sanitation - -
Livestock - -
Agriculture - -
High education, research and innovation
|
Policymakers and representatives from ministries of Health and Environment |
| Healthcare providers and authorities (local) |
Head of the Nanoro District
Head of the CMA Saint Camille de Nanoro |
- |
| Representatives from national and international non-governmental organizations and community organizations such as healthcare providers and regional water and sewerage authorities (local) |
WaterAid, Directorate-General for Water Resources, the National Office for Water and Sanitation (ONEA), Director General of Wastewater and Excreta Disposal, and Water and environment quality analysis laboratory |
Water Analysis Infrastructure (REGIDESO), and the National Institute of Biomedical Research |
3. Results
3.1. Priority pathogens
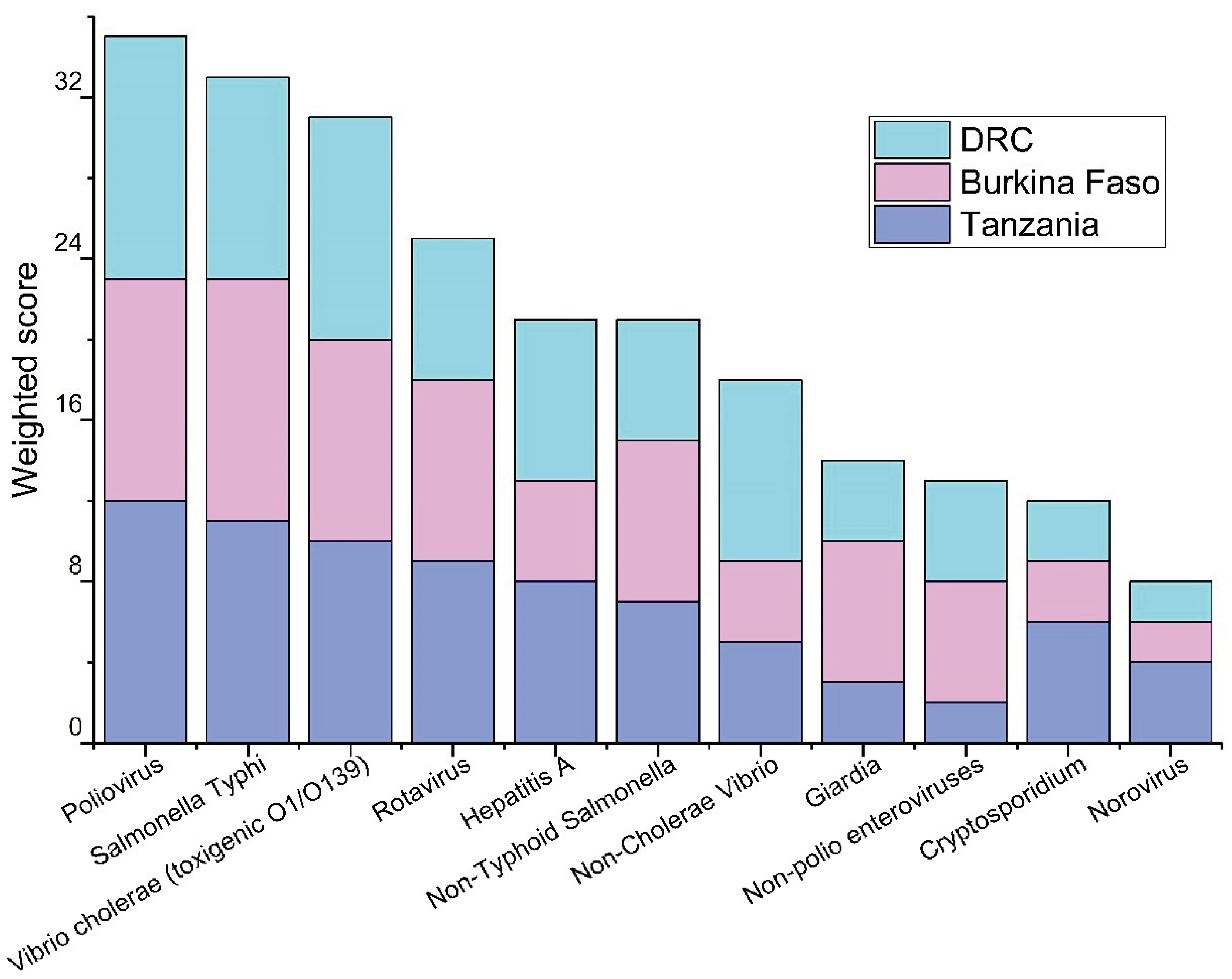
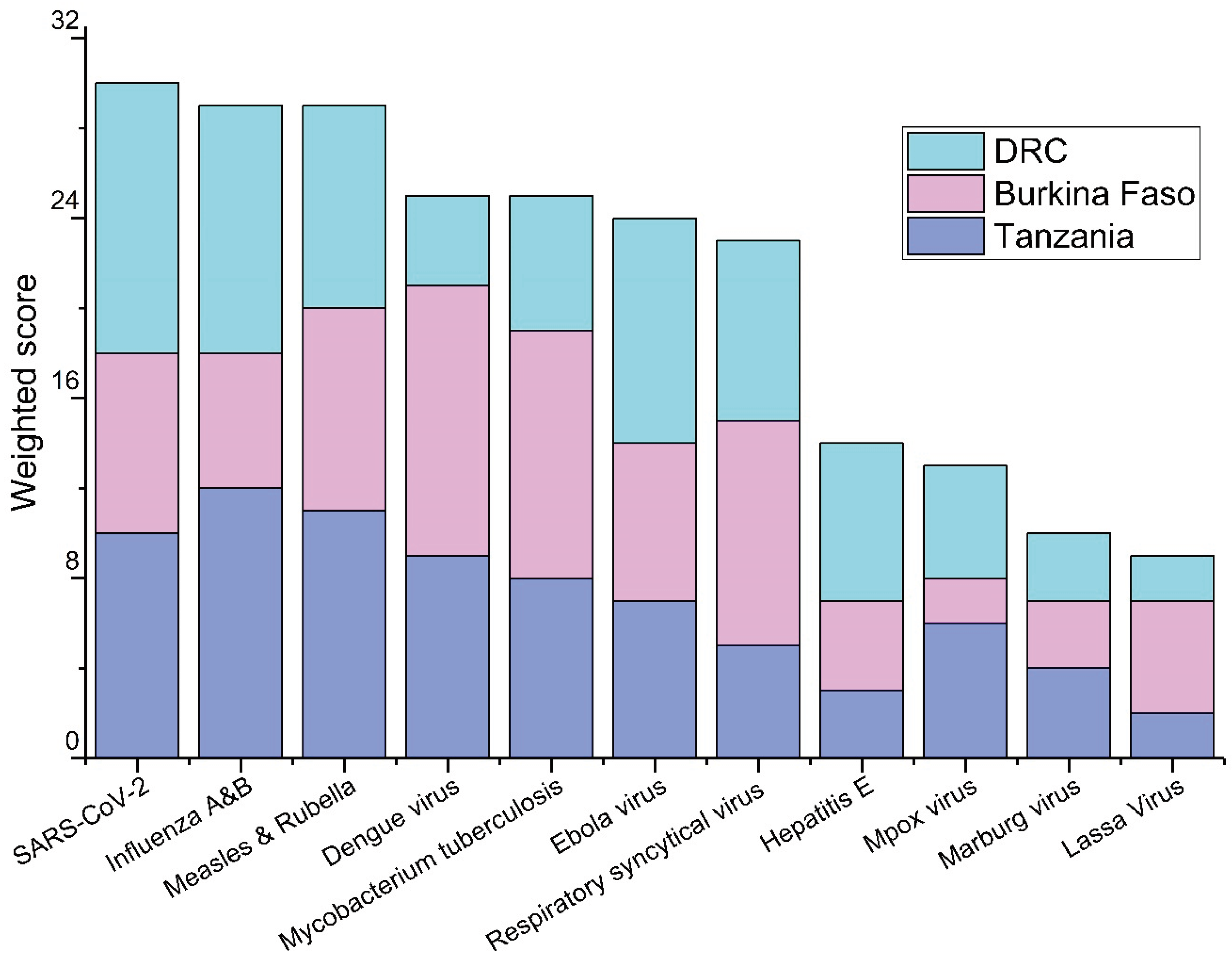
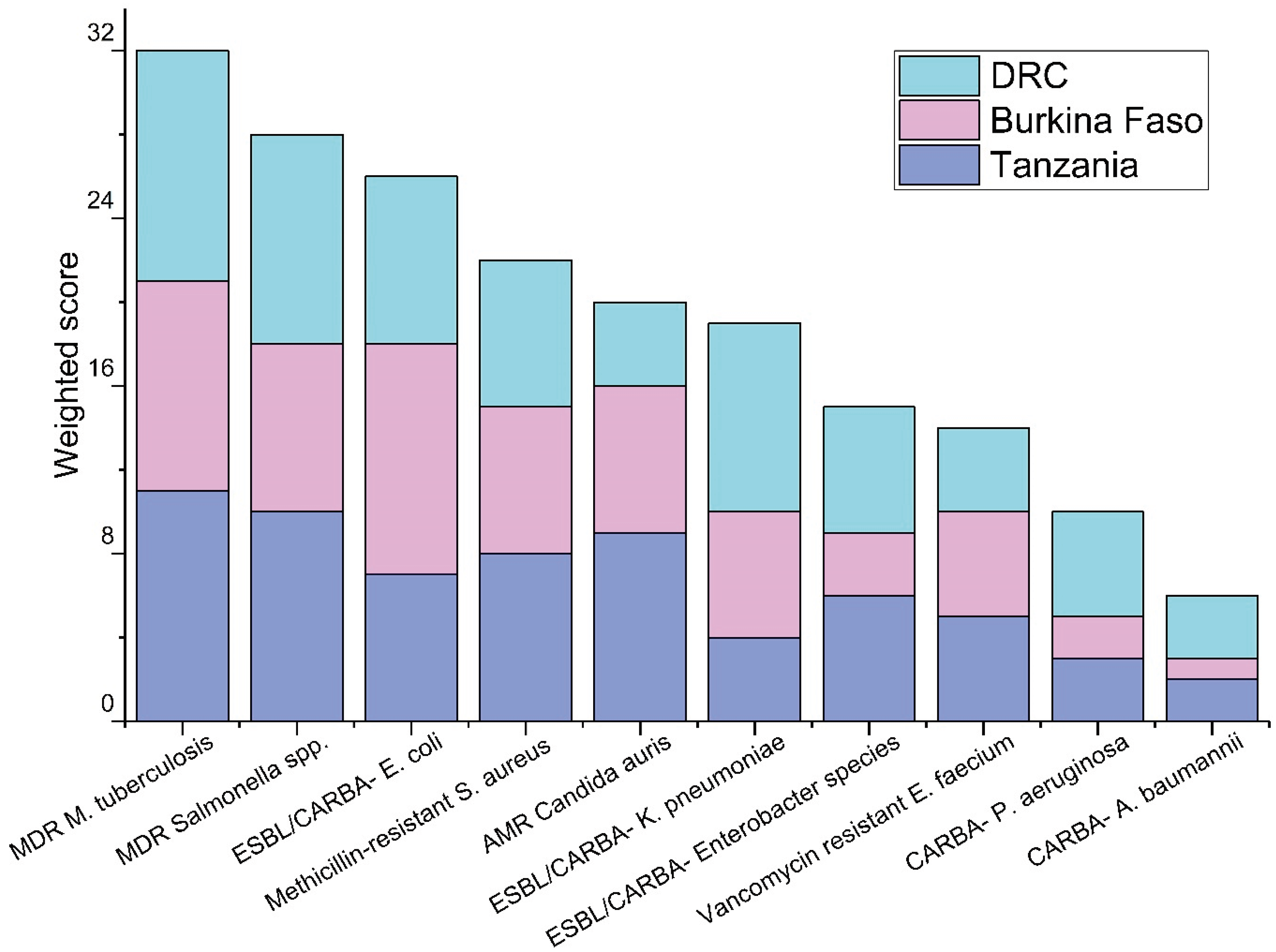
The prioritization of the pathogens based on the responses from the workshop participants are shown in
Figure 1,
Figure 2 and
Figure 3. Among waterborne pathogens, poliovirus,
Salmonella Typhi, and
Vibrio cholerae (toxigenic O1/O139) were rated as the top three priority pathogens in all three countries (
Figure 1). There was no such consensus when other than waterborne pathogens were considered. Of other than waterborne pathogens, influenza A&B, and SARS-CoV-2 were among the top priorities in Tanzania and the DRC, and dengue virus,
Mycobacterium tuberculosis, and respiratory syncytial virus (RSV) were considered as the priority pathogens in Burkina Faso (
Figure 2). Among AMR targets, multidrug-resistant
Mycobacterium tuberculosis, multidrug-resistant
Salmonella spp., and extended-spectrum beta-lactam (ESBL) pathogens including carbapenem-resistant Enterobacteriaceae (
E. coli, and
Klebsiella pneumoniae) had the highest priority by respondents in all three countries (
Figure 3). Based on local outbreak evidence, Burkina Faso survey used additionally multidrug-resistant
Streptococcus pneumoniae as a predetermined option, where it was prioritized as high as the 3rd priority.
3.2. Existing clinical and epidemiological surveillance
The number of participants responding to the Webropol questionnaire in Tanzania's workshop was 20, 24 in Burkina Faso, and 16 in the DRC (
Table 2). Notably, as most of the questions were optional, not all respondents answered every question. The response rates varied, and the percentage shows the lowest response rate with 90% (18/20) in Tanzania, 88% (21/24) in Burkina Faso, and 88% (14/16) in the DRC.
In all three countries, the respondents indicated that the primary approach for gathering clinical information involves various electronic systems like Integrated Disease Surveillance and Response (IDSR), Health Management Information System (HMIS), and Public Health Emergency Operations Centre (PHEOC) in Tanzania, DHSI2, Health data warehouse (EnDoS), Telegram weekly official letter (TLOH), and System for Tracking Epidemiological Data and Laboratory Specimens(STELaB) in Burkina Faso and demographic and health survey (DHSID), and national health information system (
Système National d’Information Sanitaire, SNIS) in the DRC as the official methods for clinical data collection. DHSI2 is one of the largest health information management systems in the world, commonly used in LMICs (
https://dhis2.org/). In the DRC, the reports are entered into the SNIS, the DRC’s national DHIS2 platform for health information management, in the health zones (
https://dhis2.org/drc-data-use/). Additionally, the option to gather information through Excel and paper-based registries was reported to be available in all these countries for cases where electronic systems are not feasible.
Considering the clinical diagnostic methods used within the clinical surveillance, all three countries acknowledged capacity for conventional PCR and quantitative PCR, virus culture, serology, microscopy, rapid diagnostic tests, bacterial culture, and Sanger and Next Generation Sequencing (NGS).
Table 2.
Fields of expertise among the respondents of Webropol questionnaire for clinical and environmental surveillance themes. .
Table 2.
Fields of expertise among the respondents of Webropol questionnaire for clinical and environmental surveillance themes. .
| Characteristic |
Tanzania
Number (%)
Total number of respondents, N=20 |
Burkina Faso
Number (%)
Total number of respondents, N=24 |
DRC
Number (%)
Total number of respondents, N=16* |
| Clinical surveillance/epidemiologist |
6 (30 %) |
4 (17 %) |
1 (6 %) |
| Environmental surveillance/water supply |
6 (30 %) |
3 (13 %) |
3 (19 %) |
| Healthcare professional |
2 (10 %) |
9 (38 %) |
3 (19 %) |
| International organization |
1 (5 %) |
2 (8 %) |
- |
| Non-governmental organization (NGO)/civil society organization (CSO) |
1 (5 %) |
1 (4 %) |
- |
| Researcher |
3 (15 %) |
4 (17 %) |
5 (31 %) |
| Other |
1 (5 %) |
1 (4 %) |
3 (19 %) |
3.3. Existing environmental surveillance
All three countries operate an existing wastewater-based poliovirus surveillance system. Moreover, all three countries conduct limited AMR surveys for academic monitoring. In Tanzania, there is limited SARS-CoV-2 WES initiated in Dar es Samlaam under the National Public Health Laboratory and in Burkina Faso respondents mentioned WES for other pathogens like enterovirus. Respondents from the DRC mentioned WES for SARS-CoV-2 and cholera. There was uncertainty among respondents about key WES practical aspects, such as sampling locations, frequency, analyzing laboratories, and sharing analyzed results. In Tanzania, the Ministry of Health manages an infectious disease database, which includes WES of poliovirus. In Burkina Faso, respondents did not specify entities for WES operations. The Institute of National Research Biomedical (INRB) is handling wastewater database in the DRC. For collecting supplementary environmental information, such as climatic data, the Tanzania Meteorological Authority (TMA) database and The Météo database maintained by the National Geographic Institute for the DRC were mentioned by multiple respondents. One respondent from Burkina Faso suggested Station Synoptique for such purposes in Burkina Faso.
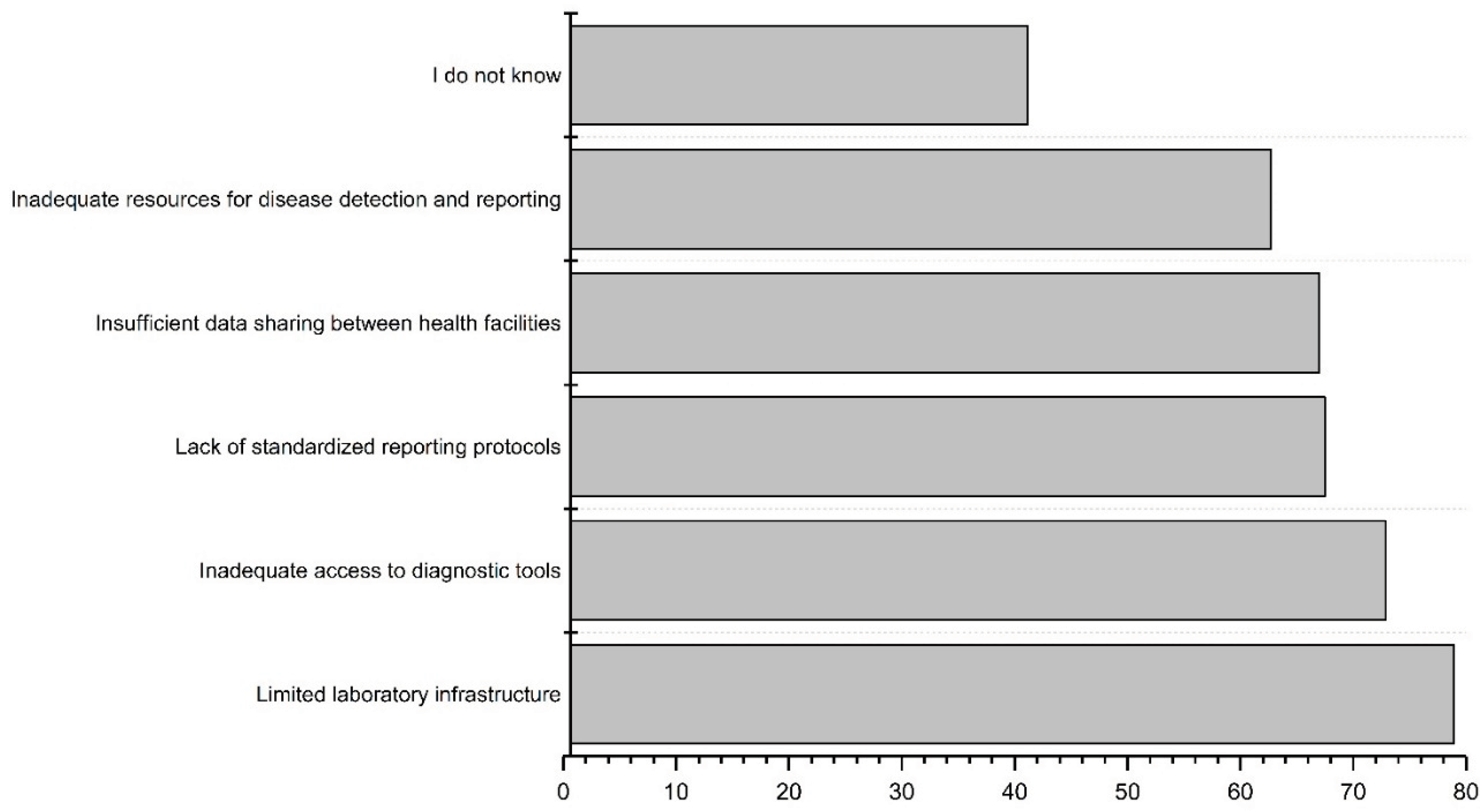
3.4. Challenges and limitations in the current surveillance systems
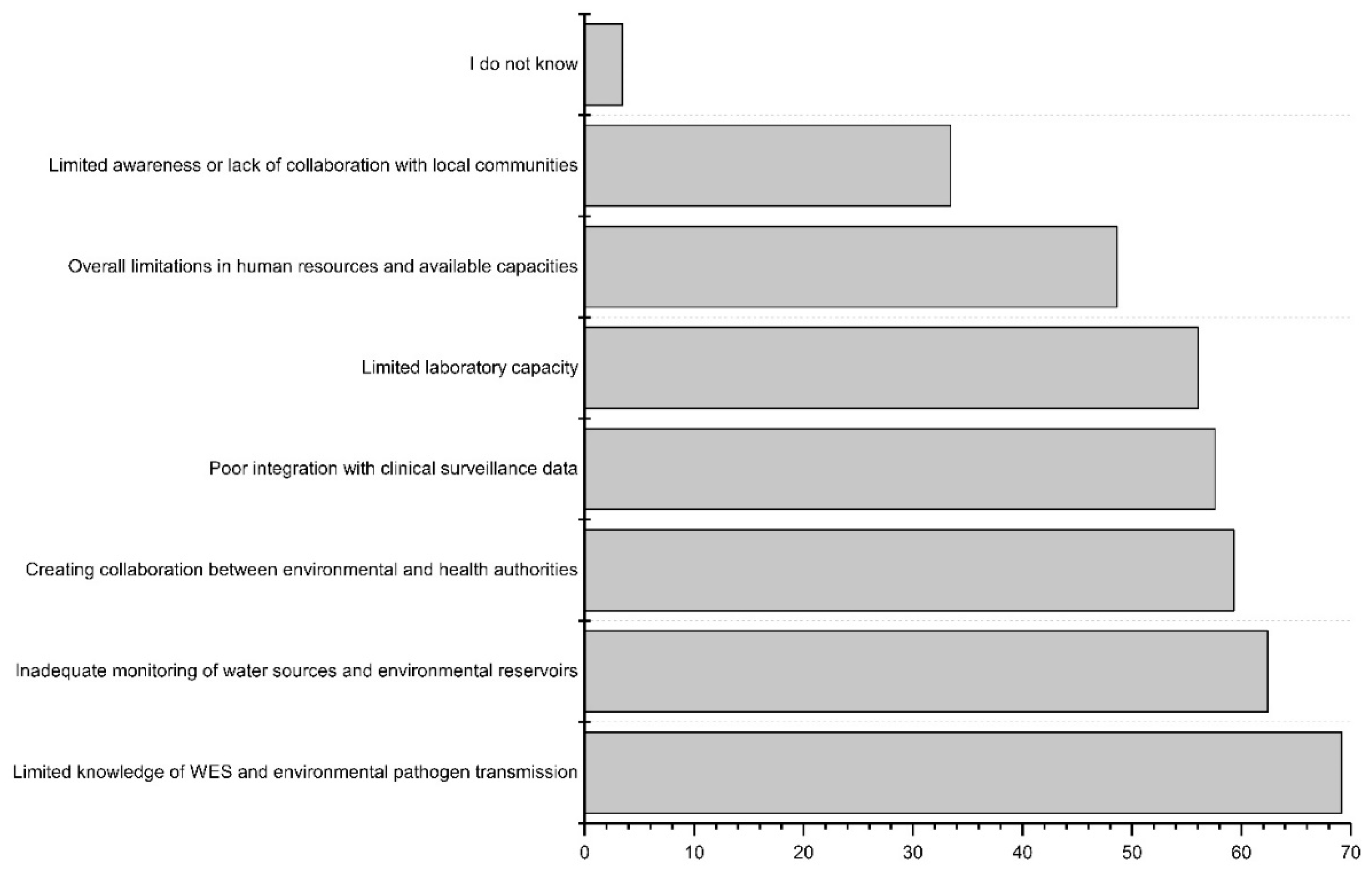
The workshop participants were asked to prioritize the challenges and limitations within the current clinical surveillance systems. The limited laboratory infrastructure was ranked as the biggest challenge, followed by inadequate access to diagnostic tools (
Figure 4). Open-ended responses from all three countries emphasized resource constraints, including a shortage of trained healthcare professionals, inadequate training, and insufficient financial resources. Communication issues between stakeholders and surveillance systems were mentioned, including concerns about the poor linkage between laboratory and surveillance data, and a lack of data sharing or linking among existing systems. In the DRC, ensuring the cold chain for laboratory samples was noted as a challenge. The identified major challenges in all three countries for detecting infectious disease outbreaks and AMR spread were infrastructure deficiencies (e.g., laboratory resources), absence of electronic databases, and insufficient human resources. Open-ended responses for Tanzania and Burkina Faso underscored the importance of written regulations and policies during outbreaks.
There is limited existing WES system working in these countries, except wastewater-based poliovirus surveillance, so the respondents were asked to prioritize the major infrastructural weaknesses in their countries’ current water systems to have information for creating the new WES. Limited sanitation and limited access to clean water supply were chosen as the top weaknesses (
Figure 5). In the open-ended responses, respondents from all three countries emphasized the issue of limited coverage in the sewerage system, both in rural and urban settings, along with constrained laboratory capacity for analyzing environmental samples.
Considering the actual implementation of the new WES of infectious diseases and AMR, challenges for limited knowledge of opportunities in WES or on the environmental transmission of pathogens, as well as inadequate monitoring practices of water sources and environmental reservoirs, such as data or sample collection from the area, stood out among other options (
Figure 5). The open-ended responses highlighted the same concern about limited awareness of WES, which can affect funding opportunities and resource allocation. Considering the limited laboratory capacity, concerns about the quality and management of the samples and data were mentioned. Also, as WES involves multiple stakeholders, including public health and environmental experts, concerns about the effective communication between the different entities were presented. Respondents from the DRC raised a concern about the lack of legal regulation in this new environmental surveillance field.
3.5. Potential of wastewater and environmental surveillance systems
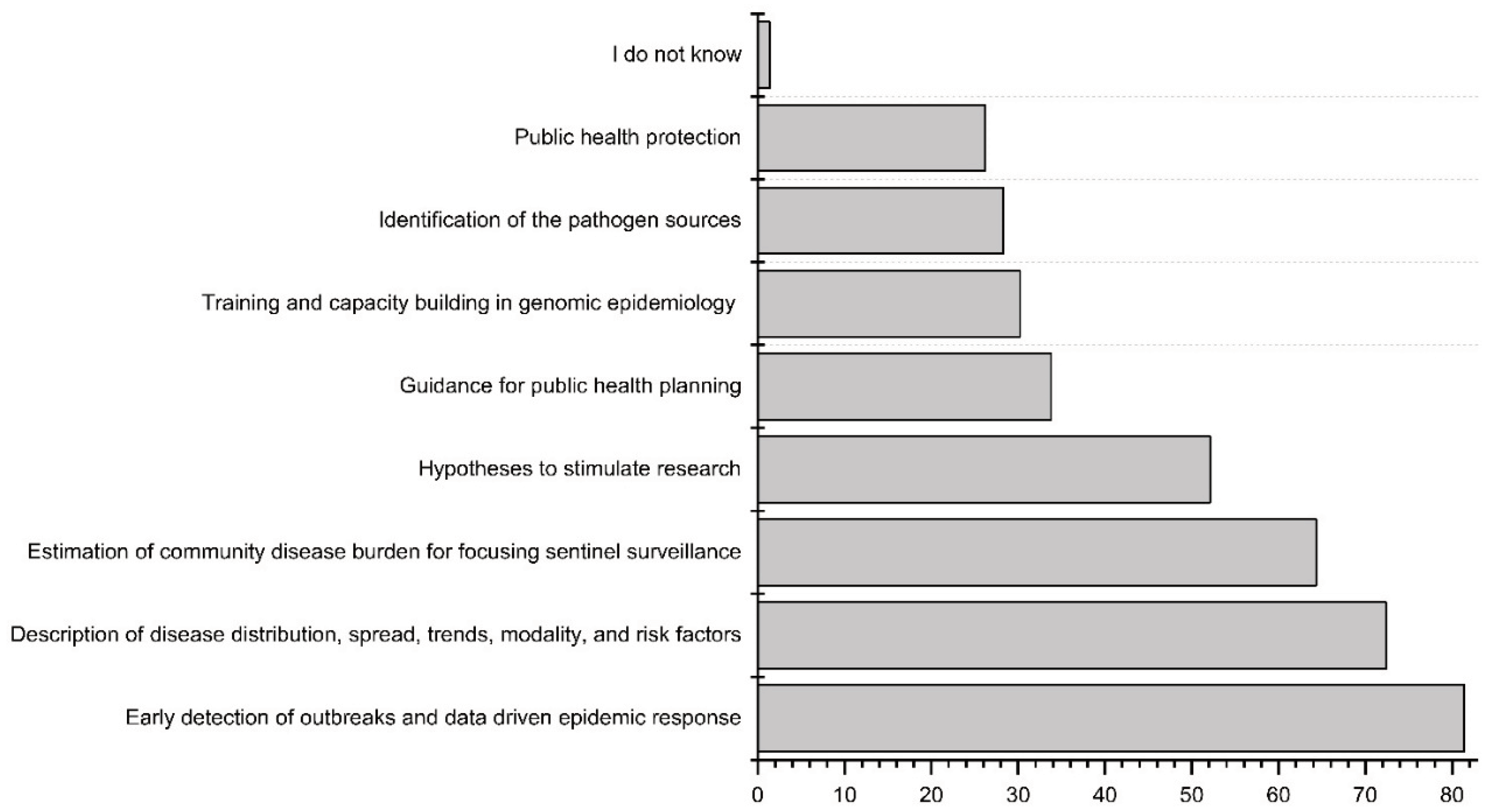
The workshop participants were asked to prioritize the potential benefits of the new WES system (
Figure 6). The foremost benefit, considered by respondents, was the early detection of outbreaks and data-driven epidemic response, with the potential for outbreak prevention through early warning. The second highest considered potential benefit was describing disease distribution, spread, trends, modality, and risk factors, followed by estimating community disease burden and providing early signals for focusing sentinel surveillance, clinical testing, or vaccination.
In the open-ended responses from Tanzania, it was emphasized that the increased awareness brought about by the planning of the new WES could lead to the growth of funding and resources, thus expanding the surveillance system network in sub-Saharan region. Additionally, the respondent from Tanzania underscored that environmental surveillance could provide more accurate data on AMR distribution within sub-Saharan Africa countries. In the responses from Burkina Faso, the knowledge of circulating environmental pathogens was seen as a potential benefit, as well as the research stimulated by the environmental surveillance topic.
4. Discussion
This study evaluates priority pathogens for environmental surveillance and examines existing epidemiological surveillance systems in three sub-Saharan countries: Tanzania, Burkina Faso and the DRC. Among waterborne pathogens, respondents from the three countries selected poliovirus, Salmonella Typhi, and Vibrio cholerae (toxigenic O1 and O139) as priorities for WES. As the survey included only a limited number of pre-chosen pathogens, also the open-ended responses were evaluated. In these responses, participants from all three countries noted the importance of pathogenic E. coli and Campylobacter spp. as waterborne pathogens to be monitored.
WES is an important component of the global poliovirus eradication program, and it has been extensively used [
13,
38,
39,
40,
41]. WES of poliovirus evaluates the effectiveness of mass vaccination campaigns, and the detection of poliovirus in wastewater samples may trigger immediate mass vaccination programs in the community [
38,
41,
42]. While the wastewater monitoring for the poliovirus eradication program has been widely implemented with significant progress in Africa, most countries have not yet tapped on the potential to establish a coordinated system for monitoring and surveillance of other priority pathogens to improve regional preparedness and response to disease outbreaks [
41].
Salmonella Typhi [
43,
44,
45], and toxigenic
Vibrio cholerae (O1 and O139) [
46,
47,
48,
49,
50], are major public health burdens in many LMIC including sub-Saharan African region, and thus potential priority pathogens for WES. Due to the typhoid fever, an illness caused by
Salmonella Typhi, as of 2019, WHO estimated that globally about nine-millions people are ill and about 110 000 people die annually [
51]. Sub-Saharan Africa has one of the highest burdens of typhoid fever in the world with about 762 deaths per 100 000 population each year [
52]. In Tanzania, annually about 79 thousand cases and about 1700 deaths due to typhoid fever were estimated [
53]. Antimicrobial resistance on commonly used antibiotics (amoxicillin, chloramphenicol, and trimethoprim-sulfamethoxazole), is a major concern connected with typhoid cases in recent years, up to 89% of
Salmonella spp. isolates collected from typhoid patients were recorded resistant to antibiotics [
53].
The actual global burden of toxigenic
Vibrio cholerae, serotypes O1 and O139 known as causative agents for cholera, is often underestimated and underreported due to limited epidemiological surveillance and laboratory capacity, along with social, political, and economic disincentives for reporting [
54,
55]. As prioritized in the present study, WES of
V. cholerae O1/O139 can be an effective way for estimating the actual burden at the population level [
56,
57,
58]. From January 1, 2023, to December 18, 2023, there has been reported a total of 879,177 cholera cases, resulting in 5,045 deaths worldwide [
47]. Cholera is still a serious public health concern in sub-Saharan region, both Tanzania and the DRC have reported cholera outbreaks in 2022 [
50]. In 2023, a neighboring country Malawi, declared cholera outbreaks exacerbated by natural disasters (cyclones and floods), resulting in over 59,075 cases and 1,769 deaths as of October 2023 [
59]. In October 2023, Zambia reported a new cholera outbreak that spread in many provinces, putting millions of people at risk [
55,
60]. Also a next East African country, Zimbabwe reported a cholera outbreak in February 2023, with a total number of 6,686 suspected and 1,127 confirmed cases confirmed by November 2023 [
55]. The use of WES for monitoring of toxigenic
V. cholerae is in the early stage of development [
56,
57,
58], and is closely connected with ensuring the safe clean water supply, where together with monitoring of drinking water quality, also Water Safety Planning (WSP) could provide awareness and management tools for outbreak prevention and mitigation of the on-going outbreaks [
61,
62].
Diarrheal diseases pose a significant public health challenge in LMIC, notably contributing to child mortality (around 9% globally) [
63]. The prevalence of pathogens in wastewater can be influenced by ongoing outbreaks. Simultaneously, multiple waterborne outbreaks may be occurring concurrently. In a recent study by Hugho et al. (2023), fecal samples from 146 children (below age 5) with diarrhea were investigated in Moshi, Kilimanjaro region, Tanzania, for finding the causative pathogens [
64]. Predominant clinical symptoms were diarrhea (100%), including vomiting (88.36%) and fever (60.27%). At least one diarrhea-associated pathogen was detected in 80.14% (n = 117) of the children. Major diarrhea-causing pathogens were rotavirus 38.36% (n = 56), adenovirus 40/41 19.86% (n = 29),
Shigella/EIEC 12.33% (n = 18), norovirus GII 11.44% (n = 17), and
Cryptosporidium 9.59% (n = 14) [
64]. In the same way as by studying a huge number of fecal samples from individuals, the use of WES can provide already from a couple of samples representative information of pathogens circulating in the communities. However, the WES samples do not provide information to be used for patient care, as the WES sampling sites represent the population level with a certain coverage depending on the sampling site characteristics.
Among pathogens having other than the waterborne transmission route, respondents in this study prioritized SARS-CoV-2, influenza A&B, measles and rubella, and
Mycobacterium tuberculosis. Of these, the existing use cases of WES have already been reported for SARS-CoV-2 [
28,
65,
66,
67,
68], influenza A&B [
69,
70,
71,
72], and dengue virus [
36,
73,
74]. Measles and rubella, viral diseases, transmitted through respiratory and airborne droplets, are relatively less often monitored in wastewater [
13,
74]. Many infection cases of both measles and rubella may go asymptomatic, and these are detected in urine, so these have high potentiality to be monitored through WES. Rubella is often mild; up to 50% of cases exhibit symptoms [
75]. Measles and rubella viruses have not achieved a 100% vaccination rate in developing countries, which holds significant practical implications for WES. Further, TB is the ninth leading cause of death globally and over 25% of TB deaths occur in the African Region [
76]. The emergence of multidrug-resistant TB (MDR-TB) poses a significant health security threat and jeopardizes progress made in the fight against TB. TB in wastewater is monitored earlier [
77].
Among the group of antimicrobial-resistant pathogens, respondents prioritized multidrug-resistant
Mycobacterium tuberculosis and
Salmonella spp., and extended spectrum beta lactamase (ESBL) and carbapenemase-producing
E. coli as being potentially important to be monitored through WES. Multidrug-resistant
S. pneumoniae was exclusively listed in Burkina Faso's workshop and was ranked there among the top three prioritized AMR pathogens for WES. Antimicrobial resistance poses a serious challenge in LMIC, the situation is exacerbated by poor sanitation [
3,
78]. Poor sanitation practices contribute to the proliferation of pathogens, that need huge amounts of antibiotics for treatment, and ultimately create selective pressure for the development of AMR pathogens [
3]. The synergy between AMR and poor sanitation is particularly concerning, as developing countries often face limited resources, healthcare infrastructure, and education on proper use of antibiotics [
79,
80,
81]. This combination increases the risk of infectious diseases, hampers effective treatment, and compromises public health outcomes. WES can be an effective way for monitoring the situation of AMR in regional and local levels [
25,
79,
80,
81].
4.1. Current clinical surveillance
Disease surveillance involves four essential steps: specimen collection, analysis, result dissemination to stakeholders, and taking appropriate public health actions [
13]. Each of the three countries has a three-level of hospital system: (a) primary level, consisting of dispensary health centers and district hospitals, (b) secondary level, comprising regional referral hospitals, and (c) tertiary level, encompassing zone hospitals and national hospitals. Reference laboratories collect and confirm specimens for major pathogens like yellow fever, tuberculosis, antimicrobial resistance, and typhoid fever. Reference laboratories play a pivotal role in collecting and providing data for reporting, requiring active involvement from clinicians and epidemiologists.
Presently, electronic laboratory information-sharing systems are partially employed in urban hospitals with adequate facilities. In all three countries, the respondents revealed that the primary approach for gathering clinical information involves electronic systems based on DHSI2 (
https://dhis2.org/). DHIS2 has been used globally at least in 80 countries, impacting 3.2 billion individuals (40% of the world's population), including many African countries. However, the prevalence of these modern electronic and automated data collection systems has not yet been fully adopted. Android phone-based apps, viper and WhatsApp could have been widely used for communicating health-related data across different units in these countries.
Health information systems in many developing countries face challenges, whether due to insufficient infrastructure, trained manpower, or lack of hundred percent electrification [
82]. Consequently, information may not always be reported uniformly across all health facilities, with comprehension gaps evident at the community and health center levels [
82]. The workshop participants of this study reported the continued use of paper-based registries and Excel sheets, along with telephone reporting, indicating that these traditional methods persist.
Clinical surveillance is a continuous process for systematically gathering, analyzing, interpreting, and providing feedback on outcome-specific data. In all these countries (Tanzania, Burkina Faso and the DRC), a central body such as the Ministry of Health or the Prime Minister's office oversees the administration of the surveillance system. This includes revising priority pathogens, formulating policies, and collecting data. The African CDC, WHO, and West African Health Organization (WAHO) play a crucial role in providing guidelines to ensure harmonization of surveillance systems at an international level.
In Tanzania, policy guidelines for clinical reporting are governed by the Public Health Act 2009 [
83], and Tanzania Personal Data Protection Law 2022 [
84]. Key stakeholders in clinical surveillance systems include the Ministry of Health, the Prime Minister’s Office- Disasters Management Department (DMD), and local district and regional health management teams and hospitals. In Burkina Faso, the Central National Laboratory, overseen by the Ministry of Health, supervises clinical laboratories and conducts external quality assessments for selected pathogen surveillance. In the DRC, key stakeholders in pathogen surveillance are the Ministry of Health (MoH), local private sector, head physicians in health districts, and professionals at various levels, including the providential chef and Head of Direction Générale de Lutte contre la Maladie. All national hospitals and reference laboratories are interconnected through the Integrated Disease Surveillance and Response (IDSR) framework, guided by the National Plan de Development Sanitaire as the governing legislation for pathogen surveillance and waterborne outbreak monitoring. While both public and private hospitals in districts are mandated to utilize DHIS2 for patient data management, there exist significant gaps in the system's implementation. These challenges impede seamless integration and hinder the optimal utilization of DHIS2 across healthcare facilities.
Despite having infrastructure and disease surveillance systems, highlighted in the responses to the survey conducted during this study, challenges persist due to resource constraints, incomplete electrification, frequent power cuts, and, in some cases, a shortage of skilled human resources. Also, the workshop results highlighted limited resources, including both laboratory infrastructure and human resources, as the main challenges. This emphasizes the necessity for a thorough examination of these aspects in the development of new surveillance systems. This could be facilitated by engaging experts and infrastructures provided by The Africa Center for Disease Control and Prevention (African CDC) that is a regional organization in Africa contributing to the surveillance of pathogens and antimicrobial resistance and responding to outbreaks (
https://africacdc.org/). The primary mission of the African CDC is to ensure public health across the African continent by collaborating with member states to address health challenges and emergencies. Harmonizing disease control and prevention policies and surveillance systems among member states is considered as a key aspect for implementing successful public health actions.
4.2. Wastewater and environmental surveillance
The workshop outcomes reveal that Tanzania, Burkina Faso, and the DRC have each implemented WES to some extent, with a specific focus on poliovirus surveillance. Respondents showed limited awareness of alternative wastewater surveillance initiatives in their countries, reflecting a general lack of knowledge about environmental surveillance opportunities. This underscores the necessity for increased awareness on this subject. Despite this, the recognition of environmental surveillance's potential, especially as an early warning system for outbreak detection and in estimating community disease burden through continuous surveillance, indicates a promising basis for initiating a new environmental surveillance system.
WES have been used for monitoring waterborne, respiratory, multidrug-resistant, and many other pathogens [
9,
13,
26,
80,
85,
86,
87,
88]. However, the choice of pathogens for WES is influenced by their epidemiological relevance, microbiological evidence, and practical feasibility. The surveillance outcomes should be able to trigger an available public health action that could be implemented to mitigate the adverse health effects. Similarly, a prerequisite condition for an effective WES of a pathogen is that the pathogen needs to have a consistent shedding rate to the sewage system from infected individuals. In many nations, WES of health-related pathogens and AMR primarily emphasizes water quality and public health risks assessment. For example, the revised Urban Wastewater Treatment Directive (UWWTD) of the European Union, in Article 17, includes a provision for urban wastewater surveillance [
23]. Member States are mandated to establish a national system facilitating collaboration between health authorities and wastewater treatment authorities. This system aims to identify critical public health parameters, such as SARS-CoV-2 and its variants, poliovirus, influenza virus, emerging pathogens, and any other relevant factors, to be monitored at wastewater treatment plants, primarily in wastewater inlets [
23].
Various human enteric pathogens such as adenovirus, enterovirus, norovirus, and rotavirus, contribute to diarrheal diseases, often undiagnosed based solely on symptoms [
63,
89]. As many infections are asymptomatic or mild, clinical testing is typically sought only in severe conditions. Similarly, diseases like ebolavirus, malaria, cholera, typhoid fever, meningitis, and viral hemorrhagic fevers share clinical symptoms like fever, sore throat, muscle pain, headaches, vomiting, and diarrhea [
90]. Also, three common arboviruses; dengue, zika virus, and chikungunya share similar primary clinical symptoms such as fever, vomiting, severe headache, and muscle and joint pains [
73,
91]. Theoretically, a single wastewater test can confirm the actual agent causing the outbreak and can assist in identifying circulating pathogens in the community. Integrating genomics approaches to WES further allows even more specific implementation of effective monitoring and surveillance strategies and identification of multiple disease pathogens and its evolution, variants of concern, AMR and other emerging infectious threats.
However, establishing WES schemes in developing countries remains challenging also due to lack of centralized sewage systems. Besides wastewater, monitoring hospital wastewater and urban rivers can also provide important sampling materials for environmental surveillance [
36,
92,
93,
94]. Because of the gaps and challenges in implementing effective clinical surveillance systems and effective diagnostic approaches, many disease outbreaks can be highly underreported, based on current surveillance approaches [
44,
45]. Clinical surveillance may not account for cases if individuals do not seek testing or lack access, more likely in many low and middle-income countries, where universal health insurance is not practiced, and testing facilities are scanty. Thus, the combination of environmental surveillance with the current clinical surveillance system may comprehensively account for the spread of pathogens at a community level.
A major implication of current study is its direct benefit for gaining the Sustainable Development Goals (SDGs) [
95]. A previous study indicated that WES for pathogens contributes to more than half of the 17 UN Sustainable Development Goals (SDGs) in the 2030 agenda [
33]. It plays a vital role in disease prevention, water quality assessment, and sanitation improvement—integral elements of sustainable development, addressing social, economic, and environmental challenges. There is growing evidence suggesting a decrease in pathogens incidence, coinciding with the profound impacts on poverty alleviation, economic development of a nation, and overall social and infrastructure development. Waterborne diseases pose a significant global health threat, exerting a substantial toll on morbidity, mortality, and economic well-being.
4.3. Challenges and future directions in WES
Wastewater surveillance, though promising, faces significant developmental challenges, requiring additional empirical evidence to enhance its practicality. Respondents primarily considered deficiencies in infrastructure, coordination gap between WES authorities and epidemiologists, and a lack of human resources as major challenges for WES. In broader context, currently, standard monitoring procedures for numerous pathogens, including wastewater sample collection, population sampling coverage of sample sites, methods for target concentration, nucleic acid extraction, and data interpretation, are lacking [
22,
71,
96,
97]. However, certain pathogens, such as SARS-CoV-2 and poliovirus, are more frequently studied in WES, accumulating practical experiences and evidence compared to other pathogens [
13]. Still, each pathogen's distinct characteristics, such as distribution in wastewater, epidemiology, shedding rate in clinical individuals, and fate and decay rate in the ambient environment, demand specific contexts for sample collection, analysis, and result interpretation [
22,
71,
96,
97]. Respondents primarily considered deficiencies in infrastructure, coordination gap between WES authorities and epidemiologists, and a lack of human resources as major challenges for WES.
One interpretation challenge might arise from the use of live-attenuated vaccines, as observed especially with poliovirus and rotavirus vaccines, which have been linked to enhanced virus shedding in wastewater [
98]. Further, data on pathogen loads in symptomatic and asymptomatic carriers of many pathogens by various individuals, pathogen persistence, fate, and decay kinetics in wastewater and in the environment remain mostly unknown. Understanding that such gaps in data occur is crucial for interpreting WES results. Various factors, such as infection rate, detection limits of the laboratory methods, representativeness and population coverage of the selected WES sampling locations, water flow characteristics, wastewater matrix complexity, and viral shedding, influence the detection of microbial targets in wastewater and from other environmental samples. The persistence of many pathogens in the ambient environment needs more information. A low signal may be attributed to a limited number of cases, underscoring the importance of estimating the minimum number of infected individuals required to detect viral markers in sewage systems. Lastly, ethical considerations, including cultural and community values, are essential before disseminating results to avoid the potential stigmatization of sampled communities.
4.4. One Health perspective for waterborne pathogens
Many of the pathogens included in the current survey for example, bacterial pathogens (e.g.
Salmonella spp., pathogenic
E. coli, Campylobacter spp.), protozoan parasites (e.g.
Cryptosporidium spp.,
Giardia spp.) and other pathogens shedding antimicrobial resistance are zoonotic, i.e. infecting both humans and other animal species [
99,
100,
101]. WES for zoonotic pathogens may require a comprehensive One Health approach in designing the surveillance scheme, interpreting results, and planning management actions [
32,
80,
102]. Some zoonotic pathogens identified through WES could be originated from animal sources. If these pathogens escape into the environment from the wastewater environment, subsequently infecting animal hosts, there is a potential for them to circulate back into human hosts, perpetuating the chain. The monitoring of pathogens in wastewater and environmental waters has potential to reveal deficiencies in the sanitation systems and raise awareness of fecal pathogen distribution and thus could lead to significant public health interventions.
The risks and impact of pathogens depend on factors like persistence, prevalence, dose, invasiveness, virulence, and an individual's immune status, leading to acute and chronic health consequences. Infections can be asymptomatic, with the proportion of carriers varying based on pathogen and population characteristics. Even asymptomatic individuals can contribute to pathogen transmission. Integrated risk management, involving safeguarding drinking water sources, optimizing drinking treatment processes, and effectively managing drinking water distribution systems, is crucial to prevent waterborne outbreaks [
103]. Environmental surveillance of pathogens is vital for protecting drinking water sources and managing outbreaks.
4.5. Limitation of current study
The list of pathogens designed for multiple-choice questionnaires for the workshops relied on microbiological evidence and the epidemiological severity of various pathogens. However, biases in the results gained may arise due to the predetermined lists of pathogens and other answering options provided. Respondent biases could also be present; for example, perceptions of a pathogen may be influenced by its seasonality. Visibility in the media can also shape perceptions of the stakeholders; for example, SARS-CoV-2 received extensive attention during the COVID-19 pandemic, potentially affecting the prioritization. Existing surveillance programs and personal experience may also influence responses, e.g., a poliovirus expert might more likely nominate this pathogen as a priority without further thought. Personal stigma may cause hesitation; for instance, Mpox has been widely communicated as a gay-related disease, and may have caused hesitation in mentioning it [
12,
22]. Pathogens vary in microbiology, incubation period, outbreak progression, infectivity, and fatality rates. Consequently, interpreting pathogen monitoring in wastewater requires careful consideration.
5. Conclusions
This study highlights the importance of customized WES systems in three sub-Saharan countries (Tanzania, Burkina Faso, and the DRC), emphasizing localized approaches for effective monitoring of waterborne pathogens and AMR. Based on various surveys and existing knowledge on pathogens, this study evaluates priority pathogens for WES including, poliovirus, Salmonella Typhi, Vibrio cholerae, influenza A&B, measles and rubella, SARS-CoV-2, and Mycobacterium tuberculosis. Recommended AMR pathogens include drug-resistant Mycobacterium tuberculosis, Salmonella spp., and extended spectrum beta lactamase (ESBL) and carbapenemase-producing E. coli. However, many factors could affect practical use of these pathogens for WES, majorly microbiological evidence and practically feasibility.
Currently these countries employ centralized electronic systems for clinical data collection, complemented by Excel and paper-based registries. However, WES can be a valuable tool for early detection of locally circulating human-derived pathogens, aiding in outbreak detection, data-driven epidemic response, and timely intervention. Integrating WES with the current disease surveillance system can be crucial for monitoring waterborne pathogens, respiratory pathogens, and pathogens with AMR. The WES results can be also important for measuring effectiveness of existing sanitation practices, and safeguarding human, animal, and environmental health. Thus, WES can be an important tool for risk management, reducing waterborne outbreaks, and promptly achieving UN Sustainable Development Goals.
Author Contributions
Ananda Tiwari: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Taru Miller: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Vito Baraka: Writing – review & editing, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization. Marc Christian Tahita: Writing – review & editing, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization. Vivi Maketa: Writing – review & editing, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization. Berenger Kabore: Review & editing, Paul Tunde Kingpriest: Review & editing, Patrick Mitashi: Review & editing, Erick Lyimo: Review & editing, Hillary Sebukoto: Review & editing, Tarja Pitkänen: Writing – review & editing, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization.
Funding
This work was part of the ODIN consortium project ”Strengthening Environmental Surveillance to Advance Public Health Action”. The project is supported by the Commission of the European Communities as part of the Horizon Europe – the Framework Programme for Research and Innovation (Grant Agreement no 101103253) and Global Health EDCTP3.
Acknowledgements
We acknowledge the stakeholder workshop participants and organizers, and all ODIN consortium members.
Conflicts of Interest
The authors declare that they have no competing interests.
Availability of data
The produced data have been processed and made available in the article and supplemental material.
Ethical approval
This study does not require ethical approval.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- WHO Drinking Water. Available online: https://www.who.int/news-room/fact-sheets/detail/drinking-water (accessed on 7 November 2023).
- UNICEF Triple Threat How Disease, Climate Risks, and Unsafe Water, Sanitation and Hygiene Create a Deadly Combination for Children. Available online: https://www.unicef.org/media/137206/file/triple-threat-wash-EN.pdf (accessed on 7 November 2023).
- WHO. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Pieri, A.; Aschbacher, R.; Fasani, G.; Mariella, J.; Brusetti, L.; Pagani, E.; Sartelli, M.; Pagani, L. Country Income Is Only One of the Tiles: The Global Journey of Antimicrobial Resistance among Humans, Animals, and Environment. Antibiotics 2020, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- WHO Key Facts Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 10 January 2024).
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Wordometer Coronavirus Cases. Available online: https://www.worldometers.info/coronavirus/#countries.
- Bishop, R.F.; Kirkwood, C.D. Enteric Viruses. In Encyclopedia of Virology; Elsevier, 2008; pp. 116–123.
- Bisseux, M.; Colombet, J.; Mirand, A.; Roque-Afonso, A.-M.; Abravanel, F.; Izopet, J.; Archimbaud, C.; Peigue-Lafeuille, H.; Debroas, D.; Bailly, J.-L.; et al. Monitoring Human Enteric Viruses in Wastewater and Relevance to Infections Encountered in the Clinical Setting: A One-Year Experiment in Central France, 2014 to 2015. Eurosurveillance 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Matrajt, G.; Lillis, L.; Meschke, J.S. Review of Methods Suitable for Environmental Surveillance of Salmonella Typhi and Paratyphi. Clin. Infect. Dis. 2020, 71, S79–S83. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Cohen, A.L. Infectious Disease Surveillance. In International Encyclopedia of Public Health; Elsevier, 2017; pp. 222–229.
- Bowes, D.A.; Darling, A.; Driver, E.M.; Kaya, D.; Maal-Bared, R.; Lee, L.M.; Goodman, K.; Adhikari, S.; Aggarwal, S.; Bivins, A.; et al. Structured Ethical Review for Wastewater-Based Testing in Support of Public Health. Environ. Sci. Technol. 2023, 57, 12969–12980. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, P.; Hill, D.; Anderson, K.; Collins, M.B.; Green, H.; Kmush, B.L.; Larsen, D.A. Wastewater Surveillance for Infectious Disease: A Systematic Review. Am. J. Epidemiol. 2023, 192, 305–322. [Google Scholar] [CrossRef] [PubMed]
- von Kalckreuth, V.; Konings, F.; Aaby, P.; Adu-Sarkodie, Y.; Ali, M.; Aseffa, A.; Baker, S.; Breiman, R.F.; Bjerregaard-Andersen, M.; Clemens, J.D.; et al. The Typhoid Fever Surveillance in Africa Program (TSAP): Clinical, Diagnostic, and Epidemiological Methodologies. Clin. Infect. Dis. 2016, 62, S9–S16. [Google Scholar] [CrossRef] [PubMed]
- Marks, F.; von Kalckreuth, V.; Aaby, P.; Adu-Sarkodie, Y.; El Tayeb, M.A.; Ali, M.; Aseffa, A.; Baker, S.; Biggs, H.M.; Bjerregaard-Andersen, M.; et al. Incidence of Invasive Salmonella Disease in Sub-Saharan Africa: A Multicentre Population-Based Surveillance Study. Lancet Glob. Heal. 2017, 5, e310–e323. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Phan, N.; Tandukar, S.; Ashoori, R.; Thakali, O.; Mousazadesh, M.; Dehghani, M.H.; Sherchan, S.P. Persistence and Occurrence of SARS-CoV-2 in Water and Wastewater Environments: A Review of the Current Literature. Environ. Sci. Pollut. Res. 2021. [Google Scholar] [CrossRef]
- Lundy, L.; Fatta-Kassinos, D.; Slobodnik, J.; Karaolia, P.; Cirka, L.; Kreuzinger, N.; Castiglioni, S.; Bijlsma, L.; Dulio, V.; Deviller, G.; et al. Making Waves: Collaboration in the Time of SARS-CoV-2 - Rapid Development of an International Co-Operation and Wastewater Surveillance Database to Support Public Health Decision-Making. Water Res. 2021, 199, 117167. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, K.; Du, W.; Ali, W.; Feng, X.; Zhang, H. The Potential of Wastewater-Based Epidemiology as Surveillance and Early Warning of Infectious Disease Outbreaks. Curr. Opin. Environ. Sci. Heal. 2020, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bibby, K.; Bivins, A.; Wu, Z.; North, D. Making Waves: Plausible Lead Time for Wastewater Based Epidemiology as an Early Warning System for COVID-19. Water Res. 2021, 202, 117438. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.; Radniecki, T.S.; Kaya, D.; Alegre, D.; Geniza, M.; Girard, A.-M.; Carter, K.; Dasenko, M.; Sanders, J.L.; Cieslak, P.R.; et al. Detection of SARS-CoV-2 B.1.351 (Beta) Variant through Wastewater Surveillance before Case Detection in a Community, Oregon, USA. Emerg. Infect. Dis. 2022, 28, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sims, N.; Kasprzyk-Hordern, B. Future Perspectives of Wastewater-Based Epidemiology: Monitoring Infectious Disease Spread and Resistance to the Community Level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Adhikari, S.; Kaya, D.; Islam, M.A.; Malla, B.; Sherchan, S.P.; Al-Mustapha, A.I.; Kumar, M.; Aggarwal, S.; Bhattacharya, P.; et al. Monkeypox Outbreak: Wastewater and Environmental Surveillance Perspective. Sci. Total Environ. 2022, 856, 159166. [Google Scholar] [CrossRef]
- EU Regulation 2020/741 Proposal for a DIRECTIVE OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL Concerning Urban Wastewater Treatment (Recast). Off. J. Eur. Union 2022, 0345, 1–68.
- Keshaviah, A.; Diamond, M.B.; Wade, M.J.; Scarpino, S. V; Ahmed, W.; Amman, F.; Aruna, O.; Badilla-Aguilar, A.; Bar-Or, I.; Bergthaler, A.; et al. Wastewater Monitoring Can Anchor Global Disease Surveillance Systems. Lancet Glob. Heal. 2023, 11, e976–e981. [Google Scholar] [CrossRef]
- Tiwari, A.; Krolicka, A.; Tran, T.T.; Räisänen, K.; Ásmundsdóttir, Á.M.; Wikmark, O.-G.; Lood, R.; Pitkänen, T. Antibiotic Resistance Monitoring in Wastewater in the Nordic Countries: A Systematic Review. Environ. Res. 2024, 246, 118052. [Google Scholar] [CrossRef]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.-M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater Surveillance of Antibiotic-Resistant Bacterial Pathogens: A Systematic Review. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef]
- Mansfeldt, C.; Maal-Bared, R.; Kaya, D.; Bowes, D.A.; Keenum, I.; Aggarwal, S.; Tiwari, A.; Hutchison, J.M. Unveiling the Targeted Opportunities and Universal Challenges of Wastewater-Based Surveillance for Public Health. ACS ES&T Water 2023, 3, 1987–1989. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First Detection of SARS-CoV-2 RNA in Wastewater in North America: A Study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Smith, W.J.M.; Metcalfe, S.; Stephens, M.; Jennison, A. V.; Moore, F.A.J.; Bourke, J.; Schlebusch, S.; McMahon, J.; et al. Detection of the Omicron (B.1.1.529) Variant of SARS-CoV-2 in Aircraft Wastewater. Sci. Total Environ. 2022, 820, 153171. [Google Scholar] [CrossRef] [PubMed]
- Santiso-Bellón, C.; Randazzo, W.; Pérez-Cataluña, A.; Vila-Vicent, S.; Gozalbo-Rovira, R.; Muñoz, C.; Buesa, J.; Sanchez, G.; Rodríguez Díaz, J. Epidemiological Surveillance of Norovirus and Rotavirus in Sewage (2016–2017) in Valencia (Spain). Microorganisms 2020, 8, 458. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Malla, B.; Haramoto, E. Estimation of Norovirus Infections in Japan: An Application of Wastewater-Based Epidemiology for Enteric Disease Assessment. Sci. Total Environ. 2024, 912, 169334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shi, J.; Li, X.; Tiwari, A.; Gao, S.; Zhou, X.; Sun, X.; O’Brien, J.W.; Coin, L.; Hai, F.; et al. Wastewater-Based Epidemiology of Campylobacter Spp.: A Systematic Review and Meta-Analysis of Influent, Effluent, and Removal of Wastewater Treatment Plants. Sci. Total Environ. 2023, 903, 166410. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Halden, R.U. Opportunities and Limits of Wastewater-Based Epidemiology for Tracking Global Health and Attainment of UN Sustainable Development Goals. Environ. Int. 2022, 163, 107217. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Adhikari, S.; Zhang, S.; Solomon, T.B.; Lipponen, A.; Islam, M.A.; Thakali, O.; Sangkham, S.; Shaheen, M.N.F.; Jiang, G.; et al. Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants. Water 2023, 15, 1018. [Google Scholar] [CrossRef]
- Islam, M.A.; Rahman, M.A.; Jakariya, M.; Bahadur, N.M.; Hossen, F.; Mukharjee, S.K.; Hossain, M.S.; Tasneem, A.; Haque, M.A.; Sera, F.; et al. A 30-Day Follow-up Study on the Prevalence of SARS-COV-2 Genetic Markers in Wastewater from the Residence of COVID-19 Patient and Comparison with Clinical Positivity. Sci. Total Environ. 2023, 858, 159350. [Google Scholar] [CrossRef]
- Thakali, O.; Raya, S.; Malla, B.; Tandukar, S.; Tiwari, A.; Sherchan, S.P.; Sherchand, J.B.; Haramoto, E. Pilot Study on Wastewater Surveillance of Dengue Virus RNA: Lessons, Challenges, and Implications for Future Research. Environ. Challenges 2022, 100614. [Google Scholar] [CrossRef]
- THL Strengthening Environmental Surveillance to Advance Public Health Action (ODIN). Available online: https://thl.fi/tutkimus-ja-kehittaminen/tutkimukset-ja-hankkeet/strengthening-environmental-surveillance-to-advance-public-health-action-odin- (accessed on 23 January 2023).
- WHO Guidelines for Environmental Surveillance of Poliovirus Circulation; Geneva, Switzerland, 2003.
- Hovi, T.; Stenvik, M.; Partanen, H.; Kangas, A. Poliovirus Surveillance by Examining Sewage Specimens. Quantitative Recovery of Virus after Introduction into Sewerage at Remote Upstream Location. Epidemiol. Infect. 2001, 127, 101–106. [Google Scholar] [CrossRef]
- Lanrewaju, A.A.; Enitan-Folami, A.M.; Sabiu, S.; Edokpayi, J.N.; Swalaha, F.M. Global Public Health Implications of Human Exposure to Viral Contaminated Water. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- WHO Global Polio Surveillance Action Plan 2022–2024 2022.
- WHO Poliomyelitis Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/poliomyelitis#:~:text=Cases due to wild poliovirus,at risk of contracting polio (accessed on 23 January 2023).
- Kim, C.L.; Cruz Espinoza, L.M.; Vannice, K.S.; Tadesse, B.T.; Owusu-Dabo, E.; Rakotozandrindrainy, R.; Jani, I. V; Teferi, M.; Bassiahi Soura, A.; Lunguya, O.; et al. The Burden of Typhoid Fever in Sub-Saharan Africa: A Perspective. Res. Rep. Trop. Med. 2022, Volume 13, 1–9. [Google Scholar] [CrossRef]
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, Clinical Presentation, Laboratory Diagnosis, Antimicrobial Resistance, and Antimicrobial Management of Invasive Salmonella Infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef] [PubMed]
- Dougan, G.; Baker, S. Salmonella Enterica Serovar Typhi and the Pathogenesis of Typhoid Fever. Annu. Rev. Microbiol. 2014, 68, 317–336. [Google Scholar] [CrossRef] [PubMed]
- Madullu, M.T.; Thomas, D.S.K.; Nyanza, E.C.; Seni, J.; Ngallaba, S.E.; Kiluvia, S.; Asori, M.; Kangmennaang, J. Spatial Distribution of Suspected and Confirmed Cholera Cases in Mwanza City, Northern Tanzania. PLOS Glob. Public Heal. 2023, 3, e0001261. [Google Scholar] [CrossRef]
- ECDC Cholera Worldwide Overview. Available online: https://www.ecdc.europa.eu/en/all-topics-z/cholera/surveillance-and-disease-data/cholera-monthly (accessed on 12 January 2024).
- Ali, M.; Nelson, A.R.; Lopez, A.L.; Sack, D.A. Updated Global Burden of Cholera in Endemic Countries. PLoS Negl. Trop. Dis. 2015, 9, e0003832. [Google Scholar] [CrossRef] [PubMed]
- Rajasingham, A.; Hardy, C.; Kamwaga, S.; Sebunya, K.; Massa, K.; Mulungu, J.; Martinsen, A.; Nyasani, E.; Hulland, E.; Russell, S.; et al. Evaluation of an Emergency Bulk Chlorination Project Targeting Drinking Water Vendors in Cholera-Affected Wards of Dar Es Salaam and Morogoro, Tanzania. Am. J. Trop. Med. Hyg. 2019, 100, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- WHO Disease Outbreak News Cholera – Global Situation. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON437 (accessed on 12 January 2024).
- WHO Typhoid. Available online: https://www.who.int/news-room/fact-sheets/detail/typhoid (accessed on 10 January 2024).
- Antillón, M.; Warren, J.L.; Crawford, F.W.; Weinberger, D.M.; Kürüm, E.; Pak, G.D.; Marks, F.; Pitzer, V.E. The Burden of Typhoid Fever in Low- and Middle-Income Countries: A Meta-Regression Approach. PLoS Negl. Trop. Dis. 2017, 11, e0005376. [Google Scholar] [CrossRef] [PubMed]
- Msemo, O.A.; Mbwana, J.; Mahende, C.; Malabeja, A.; Gesase, S.; Crump, J.A.; Dekker, D.; Lusingu, J.P.A. Epidemiology and Antimicrobial Susceptibility of Salmonella Enterica Bloodstream Isolates Among Febrile Children in a Rural District in Northeastern Tanzania: A Cross-Sectional Study. Clin. Infect. Dis. 2019, 68, S177–S182. [Google Scholar] [CrossRef]
- Luhata Lungayo, C.; Burke, R.M.; Cikomola, A.; Mukamba, E.; Burnett, E.; Tate, J.E.; Samuel Otomba, J.; Albert, M.K.; Nimpa, M.M.; Dommergues, M.A.; et al. Epidemiology and Pre-Vaccine Burden of Rotavirus Diarrhea in Democratic Republic of Congo (DRC): Results of Sentinel Surveillance, 2009–2019. Vaccine 2022, 40, 5933–5941. [Google Scholar] [CrossRef]
- WHO Multi-Country Outbreak of Cholera. External Situation Report #4, Published 6 July 2023 2023.
- Zohra, T.; Ikram, A.; Salman, M.; Amir, A.; Saeed, A.; Ashraf, Z.; Ahad, A. Wastewater Based Environmental Surveillance of Toxigenic Vibrio Cholerae in Pakistan. PLoS One 2021, 16, e0257414. [Google Scholar] [CrossRef] [PubMed]
- Chigwechokha, P.; Nyirenda, R.L.; Dalitsani, D.; Namaumbo, R.L.; Kazembe, Y.; Smith, T.; Holm, R.H. Vibrio Cholerae and Salmonella Typhi Culture-Based Wastewater or Non-Sewered Sanitation Surveillance in a Resource-Limited Region. J. Expo. Sci. Environ. Epidemiol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Bwire, G.; Debes, A.K.; Orach, C.G.; Kagirita, A.; Ram, M.; Komakech, H.; Voeglein, J.B.; Buyinza, A.W.; Obala, T.; Brooks, W.A.; et al. Environmental Surveillance of Vibrio Cholerae O1/O139 in the Five African Great Lakes and Other Major Surface Water Sources in Uganda. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- UNICEF Malawi Appeal Humanitarian Action for Children. Available online: https://www.unicef.org/appeals/malawi (accessed on 23 January 2023).
- WHO Zambia Races to Curb Fast-Spreading Cholera Outbreak. Available online: https://www.afro.who.int/countries/zambia/news/zambia-races-curb-fast-spreading-cholera-outbreak#:~:text=The government declared a new,the epicentre of the outbreak (accessed on 23 January 2024).
- WHO Guidelines for Drinking-Water Quality: First Addendum to the Fourth Edition. Geneva: World Health Organization; 2017; Vol. 109; ISBN 9789241550017.
- WHO Water Safety Plan Manual: Step-by-Step Risk Management for Drinking-Water Suppliers. 2023.
- UNICEF Diarrhoea Remains a Leading Killer of Young Children, despite the Availability of a Simple Treatment Solution. Available online: https://data.unicef.org/topic/child-health/diarrhoeal-disease/.
- Hugho, E.A.; Kumburu, H.H.; Amani, N.B.; Mseche, B.; Maro, A.; Ngowi, L.E.; Kyara, Y.; Kinabo, G.; Thomas, K.M.; Houpt, E.R.; et al. Enteric Pathogens Detected in Children under Five Years Old Admitted with Diarrhea in Moshi, Kilimanjaro, Tanzania. Pathogens 2023, 12, 618. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Lipponen, A.; Hokajärvi, A.-M.; Luomala, O.; Sarekoski, A.; Rytkönen, A.; Österlund, P.; Al-Hello, H.; Juutinen, A.; Miettinen, I.T.; et al. Detection and Quantification of SARS-CoV-2 RNA in Wastewater Influent in Relation to Reported COVID-19 Incidence in Finland. Water Res. 2022, 215, 118220. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First Environmental Surveillance for the Presence of SARS-CoV-2 RNA in Wastewater and River Water in Japan. Sci. Total Environ. 2020, 737, 140405. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Joshi, M.; Jiang, G.; Yamada, R.; Honda, R.; Srivastava, V.; Mahlknecht, J.; Barcelo, D.; Chidambram, S.; Khursheed, A.; et al. Response of Wastewater-Based Epidemiology Predictor for the Second Wave of COVID-19 in Ahmedabad, India: A Long-Term Data Perspective. Environ. Pollut. 2023, 337, 122471. [Google Scholar] [CrossRef] [PubMed]
- Hokajärvi, A.M.; Rytkönen, A.; Tiwari, A.; Kauppinen, A.; Oikarinen, S.; Lehto, K.M.; Kankaanpää, A.; Gunnar, T.; Al-Hello, H.; Blomqvist, S.; et al. The Detection and Stability of the SARS-CoV-2 RNA Biomarkers in Wastewater Influent in Helsinki, Finland. Sci. Total Environ. 2021, 770, 145274. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Hughes, B.; Duong, D.; Chan-Herur, V.; Buchman, A.; Wolfe, M.K.; White, B.J. Wastewater Concentrations of Human Influenza, Metapneumovirus, Parainfluenza, Respiratory Syncytial Virus, Rhinovirus, and Seasonal Coronavirus Nucleic-Acids during the COVID-19 Pandemic: A Surveillance Study. The Lancet Microbe 2023, 4, e340–e348. [Google Scholar] [CrossRef]
- Lehto, K.-M.; Hyder, R.; Länsivaara, A.; Luomala, O.; Lipponen, A.; Hokajärvi, A.-M.; Heikinheimo, A.; Pitkänen, T.; Oikarinen, S.; Group, W.S. Wastewater-Based Surveillance Is an Efficient Monitoring Tool for Tracking Influenza A Virus in the Community. medRxiv 2023, 2008–2023. [Google Scholar]
- Ahmed, W.; Smith, W.J.M.; Tiwari, A.; Bivins, A.; Simpson, S.L. Unveiling Indicator, Enteric, and Respiratory Viruses in Aircraft Lavatory Wastewater Using Adsorption-Extraction and Nanotrap® Microbiome A Particles Workflows. Sci. Total Environ. 2023, 2, 165007. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Stephens, M.; Metcalfe, S.; Smith, W.J.M.; Sirikanchana, K.; Kitajima, M.; Simpson, S.L. Occurrence of Multiple Respiratory Viruses in Wastewater in Queensland, Australia: Potential for Community Disease Surveillance. Sci. Total Environ. 2023, 864, 161023. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.K.; Paulos, A.H.; Zulli, A.; Duong, D.; Shelden, B.; White, B.J.; Boehm, A.B. Wastewater Detection of Emerging Arbovirus Infections: Case Study of Dengue in the United States. Environ. Sci. Technol. Lett. 2024, 11, 9–15. [Google Scholar] [CrossRef]
- Gentry, Z.; Zhao, L.; Faust, R.A.; David, R.E.; Norton, J.; Xagoraraki, I. Wastewater Surveillance beyond COVID-19: A Ranking System for Communicable Disease Testing in the Tri-County Detroit Area, Michigan, USA. Front. Public Heal. 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Mawson, A.R.; Croft, A.M. Rubella Virus Infection, the Congenital Rubella Syndrome, and the Link to Autism. Int. J. Environ. Res. Public Health 2019, 16, 3543. [Google Scholar] [CrossRef] [PubMed]
- WHO Tuberculose (TB). 2019.
- Mtetwa, H.N.; Amoah, I.D.; Kumari, S.; Bux, F.; Reddy, P. Wastewater-Based Surveillance of Antibiotic Resistance Genes Associated with Tuberculosis Treatment Regimen in KwaZulu Natal, South Africa. Antibiotics 2021, 10, 1362. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global Trends in Antimicrobial Resistance in Animals in Low- and Middle-Income Countries. Science 2019, 365. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Bortolaia, V.; Tate, H.; Tyson, G.H.; Aarestrup, F.M.; McDermott, P.F. Using Genomics to Track Global Antimicrobial Resistance. Front. Public Heal. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic Resistance in European Wastewater Treatment Plants Mirrors the Pattern of Clinical Antibiotic Resistance Prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef]
- Markkanen, M.A.; Haukka, K.; Pärnänen, K.M.M.; Dougnon, V.T.; Bonkoungou, I.J.O.; Garba, Z.; Tinto, H.; Sarekoski, A.; Karkman, A.; Kantele, A.; et al. Metagenomic Analysis of the Abundance and Composition of Antibiotic Resistance Genes in Hospital Wastewater in Benin, Burkina Faso, and Finland. mSphere 2023, 8. [Google Scholar] [CrossRef]
- Diallo, C.O.; Schiøler, K.L.; Samuelsen, H.; Drabo, K.M. Information System as Part of Epidemic Management in Burkina Faso: From Plan to Reality (Field Findings). BMC Public Health 2022, 22, 1726. [Google Scholar] [CrossRef]
- PUBLIC HEALTH ACT THE PUBLIC HEALTH ACT, ACT NO. 1 OF 2009 2009.
- Government Notice The Personal Data Protection Act, 2022 2023.
- Faleye, T.O.C.; Bowes, D.A.; Driver, E.M.; Adhikari, S.; Adams, D.; Varsani, A.; Halden, R.U.; Scotch, M. Wastewater-Based Epidemiology and Long-Read Sequencing to Identify Enterovirus Circulation in Three Municipalities in Maricopa County, Arizona, Southwest United States between June and October 2020. Viruses 2021, 13, 1803. [Google Scholar] [CrossRef] [PubMed]
- Dehghan Banadaki, M.; Torabi, S.; Rockward, A.; Strike, W.D.; Noble, A.; Keck, J.W.; Berry, S.M. Simple SARS-CoV-2 Concentration Methods for Wastewater Surveillance in Low Resource Settings. Sci. Total Environ. 2024, 912, 168782. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the Moment: Now Is the Time for Integrated Global Surveillance of Antimicrobial Resistance in Wastewater Environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Berglund, F.; Flach, C.-F.; Kristiansson, E.; Larsson, D.G.J. Predicting Clinical Resistance Prevalence Using Sewage Metagenomic Data. Commun. Biol. 2020, 3, 711. [Google Scholar] [CrossRef] [PubMed]
- George, C.M.; Perin, J.; Parvin, T.; Bhuyian, M.S.I.; Thomas, E.D.; Monira, S.; Zohura, F.; Hasan, M.T.; Alam, M.; Tofail, F. Diarrhea Prevalence and Child Growth Faltering Are Associated with Subsequent Adverse Child Developmental Outcomes in Bangladesh (CHoBI7 Program). Am. J. Trop. Med. Hyg. 2022, 106, 233–238. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; Falzarano, D.; Onyango, C.; Rosenke, K.; Marzi, A.; Ochieng, M.; Juma, B.; Fischer, R.J.; Prescott, J.B.; Safronetz, D.; et al. The Merits of Malaria Diagnostics during an Ebola Virus Disease Outbreak. Emerg. Infect. Dis. 2016, 22. [Google Scholar] [CrossRef] [PubMed]
- WHO Chikungunya Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/chikungunya.
- Shrestha, S.; Yoshinaga, E.; Chapagain, S.K.; Mohan, G.; Gasparatos, A.; Fukushi, K. Wastewater-Based Epidemiology for Cost-Effective Mass Surveillance of COVID-19 in Low- and Middle-Income Countries: Challenges and Opportunities. Water 2021, 13, 2897. [Google Scholar] [CrossRef]
- Joshi, M.; Kumar, M.; Srivastava, V.; Kumar, D.; Rathore, D.S.; Pandit, R.; Graham, D.W.; Joshi, C.G. Genetic Sequencing Detected the SARS-CoV-2 Delta Variant in Wastewater a Month Prior to the First COVID-19 Case in Ahmedabad (India). Environ. Pollut. 2022, 310, 119757. [Google Scholar] [CrossRef]
- Uprety, S.; Sherchan, S.P.; Narayanan, P.; Dangol, B.; Maggos, M.; Celmer, A.; Shisler, J.; Amarasiri, M.; Sano, D.; Nguyen, T.H. Microbial Assessment of Water, Sanitation, and Hygiene (WaSH) in Temporary and Permanent Settlements Two Years after Nepal 2015 Earthquake. Sci. Total Environ. 2023, 877, 162867. [Google Scholar] [CrossRef]
- UN Transforming Our World: The 2030 Agenda for Sustainable Development. Available online: https://sdgs.un.org/2030agenda (accessed on 7 January 2024).
- Ahmed, W.; Bivins, A.; Korajkic, A.; Metcalfe, S.; Smith, W.J.M.; Simpson, S.L. Comparative Analysis of Adsorption-Extraction (AE) and Nanotrap® Magnetic Virus Particles (NMVP) Workflows for the Recovery of Endogenous Enveloped and Non-Enveloped Viruses in Wastewater. Sci. Total Environ. 2023, 859, 160072. [Google Scholar] [CrossRef]
- Zhang, S.; Sharma, E.; Tiwari, A.; Chen, Y.; Sherchan, S.P.; Gao, S.; Zhou, X.; Shi, J.; Jiang, G. The Reduction of SARS-CoV-2 RNA Concentration in the Presence of Sewer Biofilms. Water 2023, 15, 2132. [Google Scholar] [CrossRef]
- Armas, F.; Chandra, F.; Lee, W.L.; Gu, X.; Chen, H.; Xiao, A.; Leifels, M.; Wuertz, S.; Alm, E.J.; Thompson, J. Contextualizing Wastewater-Based Surveillance in the COVID-19 Vaccination Era. Environ. Int. 2023, 171, 107718. [Google Scholar] [CrossRef] [PubMed]
- Dobrowsky, P.H.; De Kwaadsteniet, M.; Cloete, T.E.; Khan, W. Distribution of Indigenous Bacterial Pathogens and Potential Pathogens Associated with Roof-Harvested Rainwater. Appl. Environ. Microbiol. 2014, 80, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Pitkänen, T. Review of Campylobacter Spp. in Drinking and Environmental Waters. J. Microbiol. Methods 2013, 95, 39–47. [Google Scholar] [CrossRef]
- Zahedi, A.; Monis, P.; Deere, D.; Ryan, U. Wastewater-Based Epidemiology—Surveillance and Early Detection of Waterborne Pathogens with a Focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitol. Res. 2021, 120, 4167–4188. [Google Scholar] [CrossRef]
- Xiao, K.; Zhang, L. Wastewater Pathogen Surveillance Based on One Health Approach. The Lancet Microbe 2023, 4, e297. [Google Scholar] [CrossRef]
- WHO Guidelines for Drinking-Water Quality: Fourth Edition Incorporating the First and Second Addenda; Geneva, Switzerland, 2022.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).