1. Introduction
According to the World Health Organization (WHO), glaucoma, diabetic retinopathy (DR), and age-related macular degeneration are among the most common causes of visual impairment [
1]. These diseases can be identified and/or monitored using a non-invasive procedure called retinal fundus imaging. Retinal blood vessels, the primary anatomical structures visible in color fundus images, are crucial for diagnosing these conditions, especially in patients with diabetes who are at a heightened risk of DR due to retinal blood vessel damage. Early diagnosis, treatment, and detection can effectively decrease the disease's severity and slow its progression [
2]. Artificial intelligence employs image processing and deep learning models to accurately detect and classify cases of DR [
3]. The integration of artificial intelligence techniques into retinal vascular segmentation has emerged because of the progressive advancement and expansion of this field [
4]. However, traditional methods for diagnosing DR, primarily involving manual segmentation by ophthalmologists, are labor-intensive, time-consuming, and subject to variability in results [
5].
Patients with diabetes are primarily affected by diabetic retinopathy (DR), which is the leading cause of vision loss. Patients with diabetes are more likely to experience DR due to retinal blood vessel damage. Therefore, it is essential to identify and separate the retinal blood vessels to diagnose DR, which prevents diabetic patients from losing their vision earlier. A precise segmented image is essential to the screening of fundus diseases, the subsequent diagnosis, and the treatment of them. In addition to being time-consuming, arduous, and requiring expert techniques, manual segmentation is also labor-intensive. A variation in the segmentation results between different ophthalmologists will result in inaccurate final segmentation results [
6,
7]. As a result of the automatic segmentation of retinal vessels, the diagnostic burden is alleviated for ophthalmologists and those with limited experience can effectively address issues. Automatic detection and segmentation of retinal blood vessels are essential to the detection of DR [
8]. Nevertheless, the accuracy of blood vessel detection and the selection of abnormal features play a major role in detecting retinal disease DR, which still offers researchers the opportunity to investigate this condition. Therefore, the programmed division of retinal vessels is essential for clinical diagnosis and treatment of ophthalmic infections.
Herein lies the novelty of our approach: We propose a groundbreaking method utilizing a hybrid Convolutional Neural Network-Radial Basis Function (CNN-RBF) classifier combined with Multi-Scale Discriminative Robust Local Binary Pattern (MS-DRLBP) features. This advanced technique aims to automate the segmentation of retinal blood vessels, overcoming the limitations of manual segmentation and enhancing the accuracy and efficiency of DR diagnosis. Unlike existing methods, our approach leverages the strengths of both deep learning and pattern recognition algorithms to achieve a more precise and reliable diagnosis of DR, thereby offering a significant advancement in the field of retinal disease screening.
In this paper, we detail the development and implementation of this novel method, demonstrating its superior performance in classifying retinal diseases compared to traditional techniques. Our method not only paves the way for more accurate and efficient DR detection but also has the potential to be extended to other retinal diseases, revolutionizing the landscape of ophthalmic diagnostics.
In the literature, several studies have been published on the use of artificial intelligence (AI) to diagnose retinal diseases such as DR, glaucoma, and macular degeneration. Indeed, local binary pattern (LBP)-based feature extraction was combined with conventional machine learning (Support Vector Machine (SVM), K Nearest Neighbour (KNN), Random Forest (RF), Decision Tree (DT), etc.) and deep learning (CNN) for retinal disease classification. Although LBP is a computationally inexpensive and straightforward method of feature extraction, the features extracted in the local region may not be powerful enough to discriminate between classes. Additionally, CNN requires more annotated datasets and high computation power to achieve efficient classification. Nevertheless, there is a need to improve diagnostic rates and algorithms for accurate detection and classification of retinal diseases, thus allowing researchers to expand their research in this specialized field. Hence, the proposed research work has developed an automated threshold algorithm based on blood vessel segmentation for DR diagnosis. In addition, multi-scale discriminative robust LBP, together with a CNN-RBF classifier, is used for the classification of retinal diseases to overcome the limitations of earlier works.
In moving with discriminative robust local binary patterns (DRLBP), edges and textures can be extracted from objects by using a variety of processes. Due to its emphasis on pixel distinctions, it is effective for brightening and contrasting different kinds of images. Images are stored with their contrast information using the proposed method. The data contains both edge and texture information, making them useful for recognizing objects. In DRLBP, an object is segregated according to its texture and shape, framed by its limits. A radial basis function is one of the best candidates for pattern classification and regression and is widely used in classical machine learning applications. In this study, CNN-RBF classifiers have been used for deep learning applications and have proved to perform well compared to state-of-the-art techniques. This research work uses the MS-DRLBP and RBF architectures to create unique opportunities for using CNN models with MS-DRLBP. By using blood vessel segmented features of the fundus images, a new training process for RBFs provides the opportunity to use labeled or unlabeled data for better classification.
In this paper, the following contributions are made:
LBP is used to extract more discriminative features to classify retinal diseases using a multi-scale discriminative feature extraction approach.
Deep learning techniques were used to develop a new classifier using CNNs and RBFs
In comparison with the state-of-the-art methods reported in the literature, we achieved better results in retinal disease classification.
Despite their benefits, RBFs are difficult to integrate into contemporary CNN architectures due to two aspects of the architecture and training process. Firstly, RBFs don't allow efficient gradient flow due to their nonlinear activations and computational charts. In addition, RBFs assume that the MS-DRLBP features are fixed, and the cluster centers are introduced initially. The rest of the paper is organized as follows: This paper includes a review of related works in
Section 2, followed by a description of the materials and method, and a proposal of the method in
Section 3.
Section 4 presents the experimental outcomes of applying CNN-RBF architectures along with various CNN backbones to benchmark datasets. The conclusion is presented in
Section 5.
2. Related Works
The retinal vascular organization reflects the health of the retina, which is a useful analytic sign. As a result, segmenting retinal blood vessels can be an extremely useful tool for diagnosing vascular diseases (Eladawiet al. 2017) [
9]. A generalized Gauss-Markov random field (GGMRF) was employed for noise reduction, and a joint Markov-Gibbs random field (MGRF) for blood vessel segmentation. In 2015, Hassan et al. [
10] presented a mathematical morphological and K-means clustering method for segmenting blood vessels. In their study, Fraz et al. (2012) [
11] clearly stated that automated blood vessel segmentation is the key to a retinal disease screening system and that this has been examined in two-dimensional retinal images. In a study by Roychowdhury et al. (2015) [
12], a three-stage segmentation algorithm involving green channel extraction, high pass filtering, and Gaussian mixture models (GMMs) was presented. In their study [
13], Barkana et al. (2017) segmented retinal vessels and extracted descriptive statistics from images of different retinal diseases. Using machine learning algorithms (fuzzy logic, artificial neural networks, support vector machines) and classifier fusion methods, these features are classified to diagnose different diseases.
Based on the Digital Retinal Images for Vessel Extraction (DRIVE) and Structured Analysis of the Retina (STARE) datasets, Marin et al. (2011) proposed a new supervised learning-based neural network scheme for retinal vessel segmentation and classification using a 7-D vector composed of gray level and moment invariant features [
14]. By preprocessing retinal vessel images with a Gaussian filter and segmenting them through curvelet transform, Kavitha et al. (2014) evaluated the method of diagnosing diabetes using retinal vessel images [
15]. Based on Digital Retinal Images for Vessel Extraction (DRIVE), Structured Analysis of the Retina (STARE), and Child Heart and Health Study in England (CHASE) databases, Liskowski et al. (2016) developed a supervised approach to classify retinal images using deep neural networks [
16].
By combining preprocessing, feature extraction, and classification, Vasanthi and Wahida Banu (2014) extracted blood vessels from retinal images [
17]. An Adaptive Neuro Fuzzy Inference System (ANFIS) with an Extreme Learning Machine (ELM) is used to classify images as normal or abnormal. Morales et al. (2015) examined texture analysis's discrimination capabilities to differentiate pathologic images from healthy images [
18,
19]. A texture discriminator called Local Binary Patterns (LBP) is used to distinguish DR from AMD. An intelligent vessel segmentation model was developed by Sangeethaa and Uma Maheswari (2018) [
20] using morphological processing, thresholding, edge detection, and adaptive histogram equalization. In a study of mammograms, Mohammed Alkhaleefah and Chao-Cheng Wu (2018) developed a hybrid classifier using CNN and RBF-based SVM for breast cancer classification [
21]. By using transfer learning, knowledge from deep neural networks can be taken and applied to different tasks. In 2021, Kishore Balasubramanian & Ananthamoorthy [
22] investigated mean-oriented superpixel segmentation with CNN features and binary SVM classifiers to classify vessel and non-vessel regions using blood vessel segmentation. The early detection of DR and timely treatment are possible through regular retinal screening.
To simplify the screening process, Dai et al (2021) developed a deep learning system, named DeepDR, which can identify diabetic retinopathy from the early stages to late stages [
23]. DeepDR is trained for real-time image quality assessment, lesion detection, and grading. As a result of deep learning algorithms, there are significant improvements in screening, recognition, segmentation, prediction, and classification applications in different domains of healthcare. Nadeem et al (2022) [
24] conducted and presented a comprehensive review of deep learning developments in the field of diabetic retinopathy (DR) analysis, such as screening, segmentation, prediction, classification, and validation.
A study by Radha et al (2023) [
25] clearly states that diabetic retinopathy (DR) may develop due to damage to the retina's blood vessels. Ophthalmologists can manually recognize and mark these vessels based on some clinical and geometrical features, but it is time-consuming. The extraction and segmentation of a vessel's anatomy is crucial for distinguishing between a healthy vessel and one that has just developed abnormally. The identification, extraction, and examination of blood vessels in the retina is an intricate process. S. S. Prabha and colleagues (2022) investigated that diabetic retinopathy is a medical condition in which high blood sugar levels in the body begin to affect the retinal blood vessels [
26]. A contrast-limited adaptive histogram equalization has been implemented to enhance base contrast and cancel noise. Segmentation is further divided into two steps, and the first step consists of Fuzzy C-Means clustering to locate the coarse vessels in the retina. Second, the region-based active contour is used to highlight the blood vessels in the region of interest.
In the study by Sivapriya et al. (2024) [
27], a ResEAD2Net design was proposed, which has the potential to accurately separate imperceptible micro-vessels. The self-attention mechanism employed in the subsequent decoding stage of the network is utilized to extract more advanced semantic features, hence enhancing the network's ability to distinguish between different classes and accumulate information within the same class. The network undergoes testing using the publicly available DRIVE, STARE, and CHASE_DB1 datasets. Jaspreet and Prabhpreet (2022) [
28] employed image preprocessing to eliminate undesirable pixels, the optic disc, and blood vessels from the retinal images. Also, they used KNN classifier on Diabetic Retinopathy Database (DIARETDB1) dataset. In 2021, Li et al. [
29] introduces a network architecture combining U-Net with DenseNet model to improve micro vessel segmentation accuracy and completeness. Retinal vascular segmentation was done on the public DRIVE dataset.
Different features of microaneurysm and hemorrhage (area, major and minor axis, perimeter, etc.) are extracted after removing morphological features from the fundus images by the study done by Kumar et al., (2020) [
30]. They stated that their technique has improved sensitivity and specificity for detecting DR based on DIARETDB1 data. Kamran et al. (2021) [
31] asserted that segmentation techniques based on autoencoding are unable to restore retinal microvascular structure due to the loss of resolution during encoding and the subsequent inability to recover it during decoding. They introduced a generative architecture called RV-GAN to address this issue by segmenting retinal vascular tissue at many scales. Their model was based on DRIVE, STARE, and CHASE datasets. Similarly, for these three public datasets, Zhendi et al., (2024) [
32] employed U-Net model with decoder fusion module (DFM) and context squeeze and excitation (CSE) module to successfully blends multi-scale elements and gathers information from multiple levels. Thus, improving segmentation of the fundus images.
Numerous eye health problems can be identified using retinal blood vessels. Yubo et al., (2024) [
33] focuses on visual cortex cells and orientation selection processes in their study. They utilized DRIVE, CHASE, High-Resolution Fundus (HRF), and STARE datasets for the model analysis, which was based on W-shaped Deep Matched Filtering (WS-DMF). By utilizing the CHASE, DRIVE, and STARE datasets, the study by Xialan et al., (2024) [
34] presents a revised technique for segmenting retinal blood vessels using a network-based approach. This method extracts various features of the blood vessels at different scales and segments them in a continuous manner. Low contrasts in retinal fundus images hinder segmentation. Soomro et al., (2018) [
35] accessed independent component analysis (ICA) structures using STARE and DRIVE datasets to fix image segmentation issues caused by uneven contrast.
3. Materials and Methods
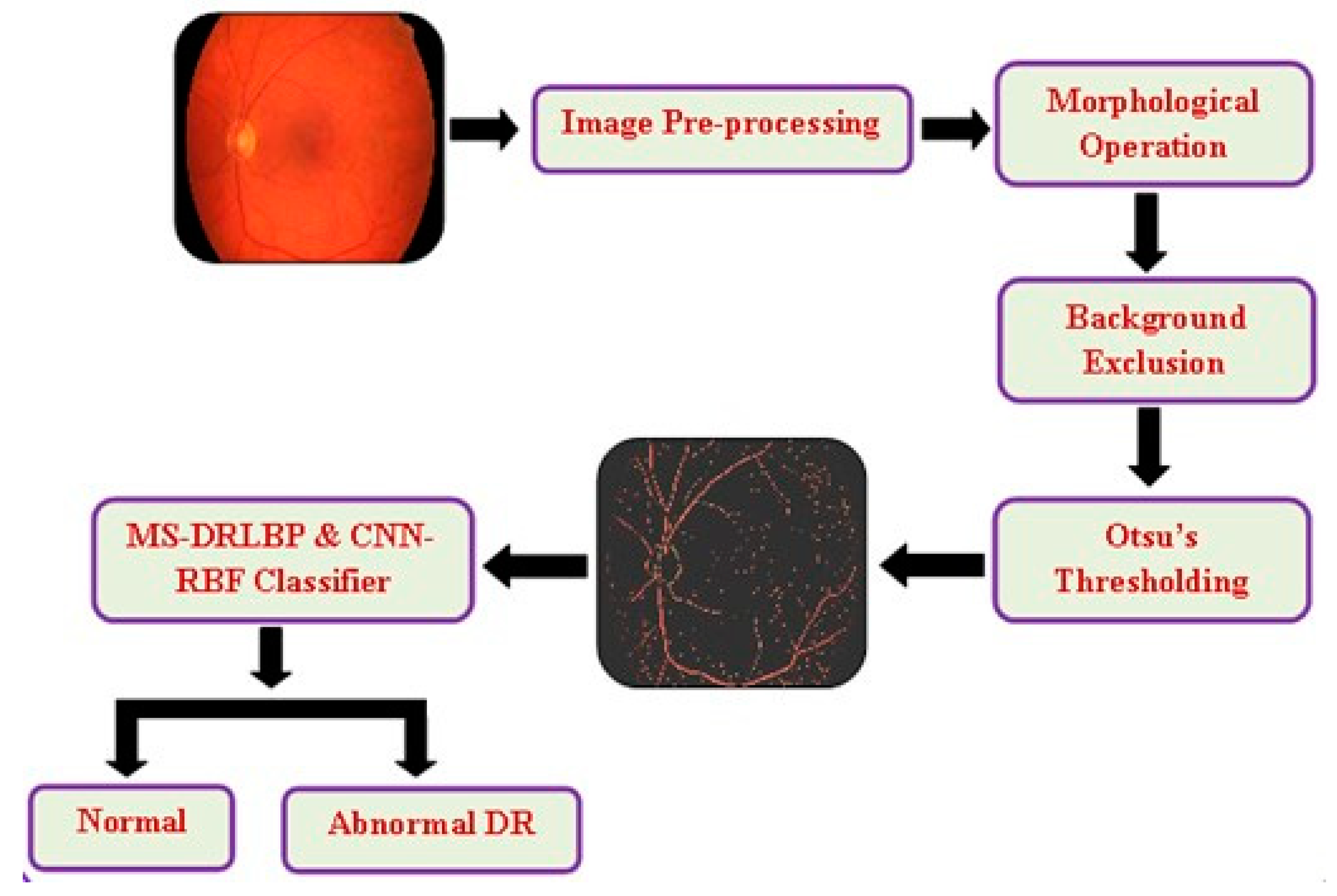
A literature review indicates that blood vessels play an important role in screening for retinal diseases, including diabetic retinopathy, which can lead to loss of vision if not detected in time. Therefore, image preprocessing algorithms developed for noise reduction and enhancement are combined with morphological operation, background exclusion, and Otsu's thresholding to perform the segmentation of blood vessels. Finally, the CNN-RBF classifier and MS-DRLBP features are used to screen for retinal disease DR.
Figure 1 illustrates the proposed approach. This study used three publicly available datasets, including Structured Analysis of the Retina (STARE), High-Resolution Fundus (HRF), and Fundus Fluorescein Angiography (FFA) for retinal disease classification [
36,
37,
38]. There are 90 images in the STARE dataset (38 Normal and 52 DR images), 30 images in the HRF dataset (15 Normal and 15 DR images), and 70 images in the FFA dataset (30 Normal and 40 DR images). The combined datasets for the work comprise 83 normal images and 107 DR images, totaling 190 images. There are four types of retinal diseases such as DME, DR, AMD, and CNV. In the above datasets, there are few images in each class. Hence, we combined all the four classes (DME, DR, AMD, and CNV) and referred to abnormal DR and the healthy control images as normal DR.
3.1. Preprocessing
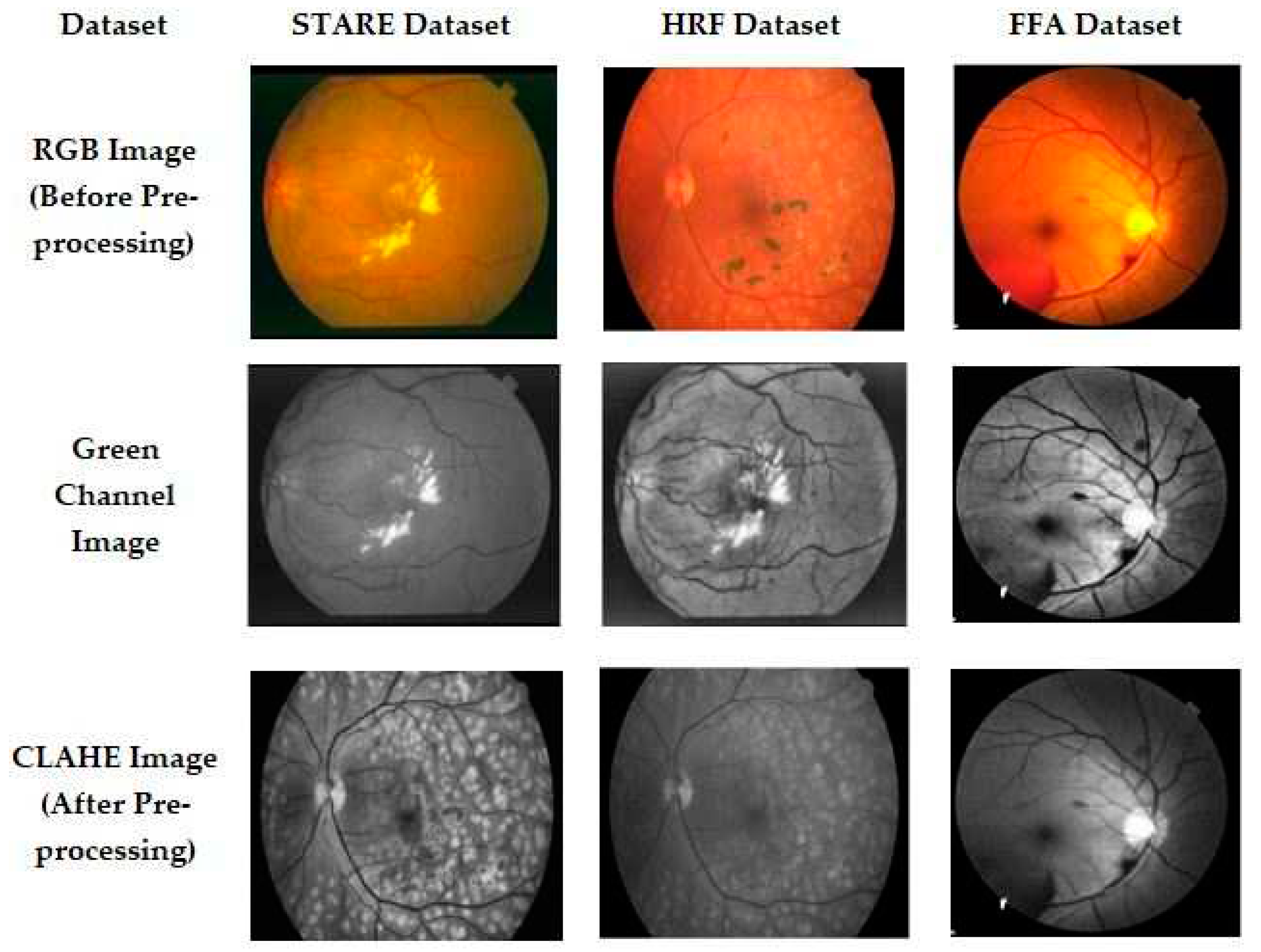
Preprocessing is essential to improve the image quality, which can increase the success rate of the proposed method. As shown in
Figure 1, we used the model developed by Sangeethaa & Uma Maheswari (2018) and performed five steps, including the extraction of green channels, contrast enhancement, morphological operations, background exclusion, and Otsu's thresholding [
20]. The preprocessed images are illustrated in
Figure 2. Due to the noise in the red and blue channels, the green channel has been extracted from the RGB image. The extracted green channel image is then subjected to Contrast Limited Adaptive Histogram Equalization (CLAHE), which splits the total area into a small, tiny area, thereby improving the contrast for better blood vessel information.
3.1.1. Morphological Operations
In retinal images, morphological operations are used to brighten small or tortuous blood vessels. A pre-processed image is morphologically opened with a structuring element (Se) using dilation, erosion, opening, and closing operations.
3.1.2. Background Exclusion
Every image captured has its own background part, which may replicate some sort of information that leads to incorrect diagnosis. Due to this, it is important to remove the background so that the foreground objects can be easily analyzed. CLAHE image enhancement is used in this work to exclude background by subtracting the morphological image.
3.1.3. Otsu’s Thresholding
There are several global thresholding methods employed in earlier works for blood vessel segmentation from retinal images. However, Otsu's thresholding gives the most effective segmentation performance due to its ease of implementation and robustness. During the iterative process, the algorithm determines the threshold that minimizes the within-class variance, calculated by summing the variances of the two classes (background and foreground). Greyscale colors generally range from 0-255 (0-1 if they are floats). If we choose a threshold of 100, then all pixels with values less than 100 become the background, and all pixels with values greater than or equal to 100 become the foreground.
The formula for finding the within-class variance at any threshold t is given by Equation (1).
Where ωbg(t) and ωfg(t) represents the probability of number of pixels for each class at threshold t and σ2 represents the variance of color values.
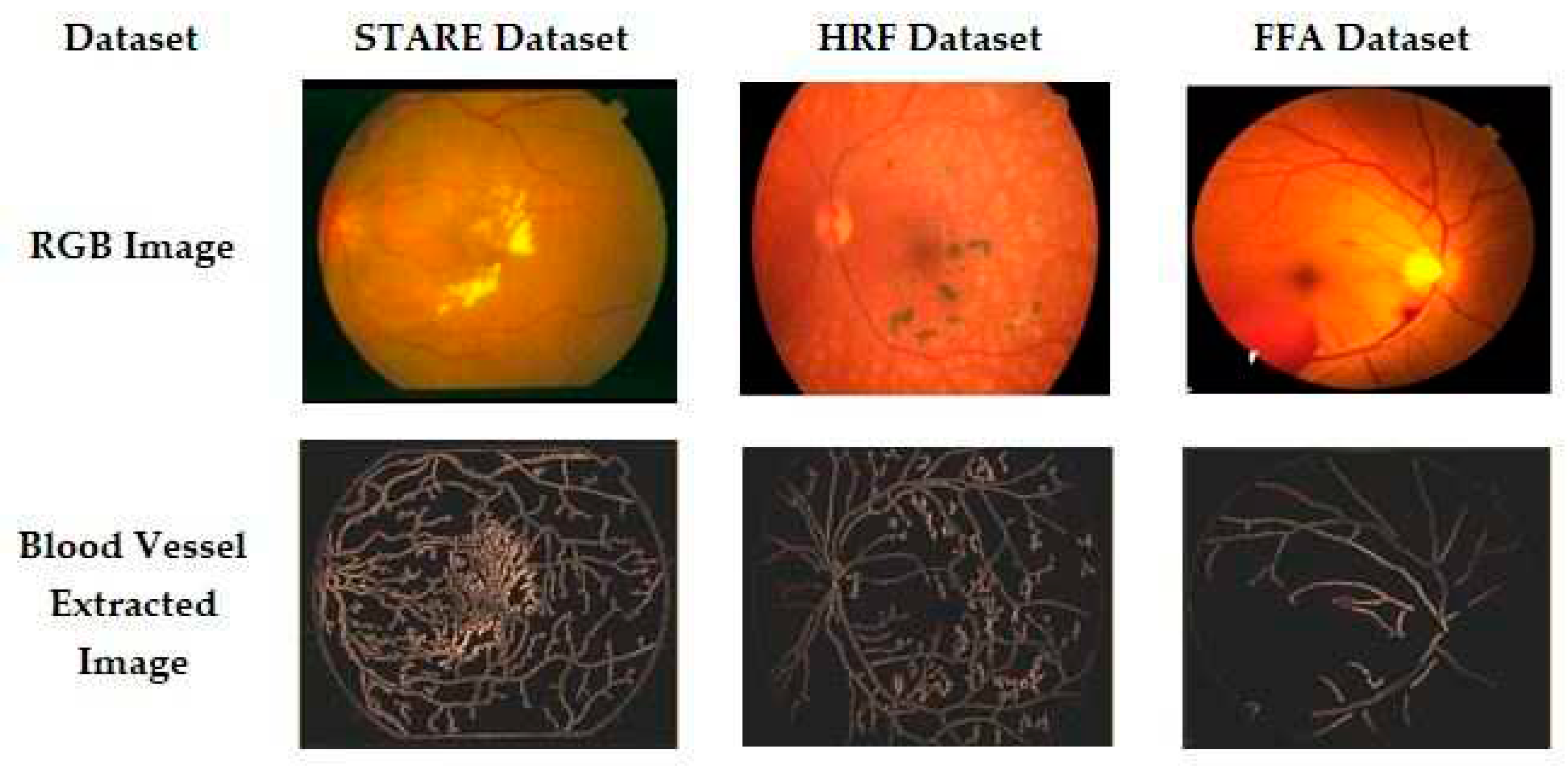
Figure 3 shows an example of the resultant input and output images for better understanding and clarity. There is good accuracy in the segmentation of blood vessels using the proposed methodology.
3.2. Network Model and Training
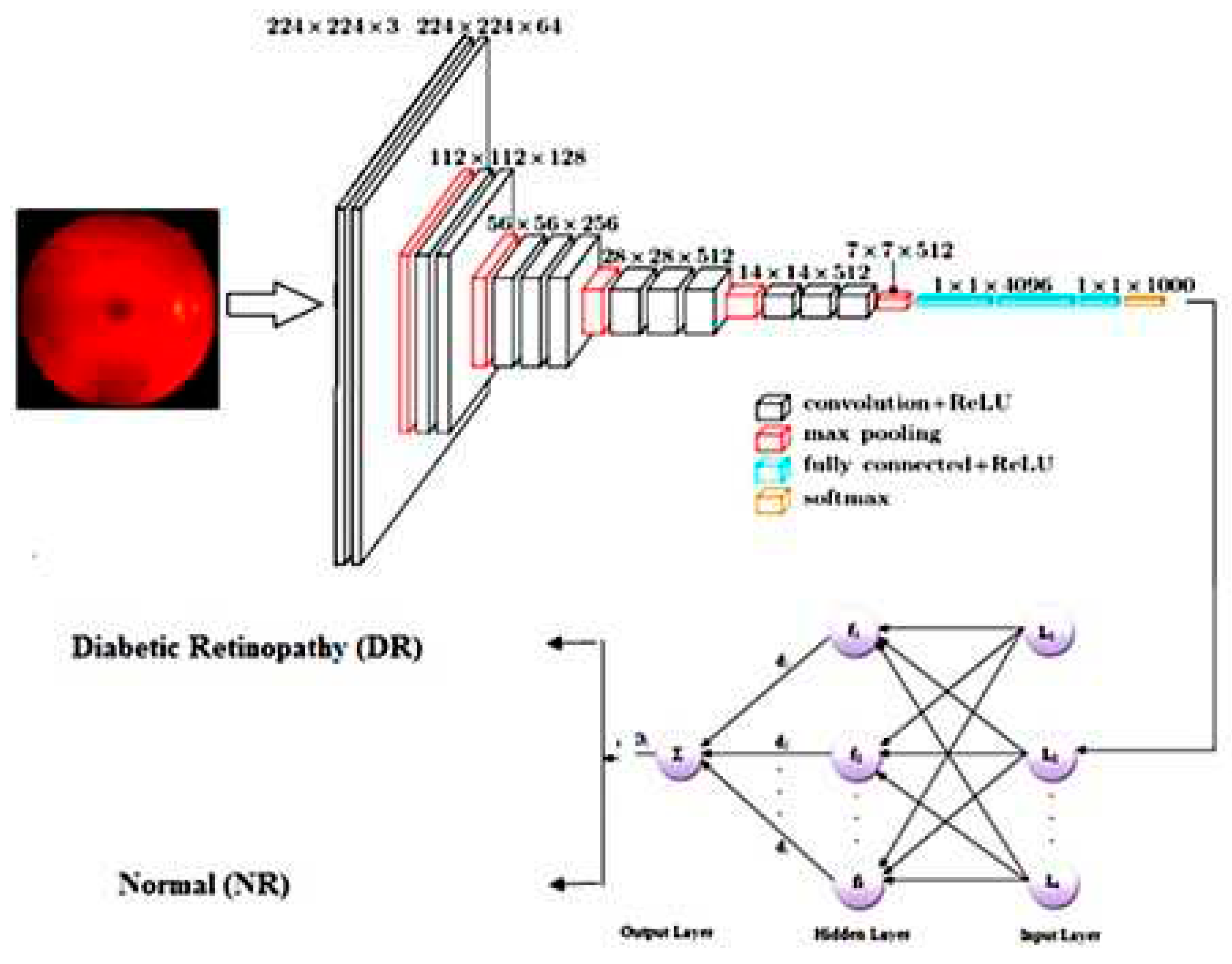
CNN is a deep-learning algorithm. This is where individual neurons are arranged in tiles so that they can relate to a given area in a visual field. In the proposed work, the segmented RBV is verified by calculating the value of the percentage of RBV pixels accurately classified as RBV pixel P, the percentage of background pixels correctly classified as background pixel N, and the percentage of pixels classified T. A segmented blood vessel image is then presented to MS-DRLBP and CNN-RBF classifiers for classification as normal or DR.
Figure 4 shows the CNN-RBF classifier network model. The proposed deep neural network has the following hyperparameter values, maximum number of epochs is 100, learning rate is 0.001, momentum factor is 0.9, batch size has been tested with 8, 16, and 32. Finally, the batch size of 32 is selected for achieving higher accuracy in retinal image classification. We used the stochastic gradient descent with momentum optimizer and cross-entropy loss function in our network. All these hyperparameters are selected based on heuristic approach to achieve the higher classification rate in detecting retinal diseases.
4. Results and Discussion
This section presents the experimental results of retinal image classification and discusses the results with state-of-the-art publications with their advantages and disadvantages. We conducted the simulation on a Desktop Computer (Intel i7 processor) with Windows 11 operating system running at 3.20 GHz with 16 GB RAM. We developed and tested the code for the proposed retinal image classification algorithm in MATLAB software. We consider 90 images from the STARE database, 30 images from the HRF database, and 70 images from the FFA database for evaluating the performance of the proposed system. Here, the DR class includes the images of all four different types of retinal diseases such as DR, AMD, DME, and
Table 1 shows descriptions of the datasets.
Table 2 shows that images are separated into training and testing types based on their classes.
Here, we compare the proposed CNN-RBF classifier with existing CNN, RBF, Adaptive Neuro Fuzzy Inference System (ANFIS), Nearest Neighbor (NN), Naive Bayes (NB), and Support Vector Machine (SVM) classifiers in terms of precision, recall, F-Score, sensitivity, specificity, and accuracy. According to the Equations 2-5, the True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN) values were used to calculate the four performance measures, namely, precision (P), sensitivity (Se), specificity (Sp), and accuracy (Acc).
True positive (TP): DR image correctly identified as DR image.
False positive (FP): Normal image (NR) incorrectly identified as DR image.
True negative (TN): Normal image (NR) correctly identified as Normal image (NR).
False negative (FN): DR image incorrectly identified as Normal image (NR).
Table 3 shows the results of the proposed CNN-RBF classifier in retinal image classification using STARE dataset. The maximum sensitivity, specificity, accuracy of 96.49%, 100%, and 97.22% is achieved compared to other conventional machine learning (ANFIS, NN, NB and SVM) and deep learning (CNN) algorithms. In contrast to the standalone classifiers such as CNN and RBF, the fusion of CNN-RBF reported higher accuracy in retinal image classification.
The confusion matrix of STARE dataset based retinal image classification is given in
Table 4. From
Table 4, we could observe the efficiency of the proposed classifier in detecting individual class of DR from the STARE dataset.
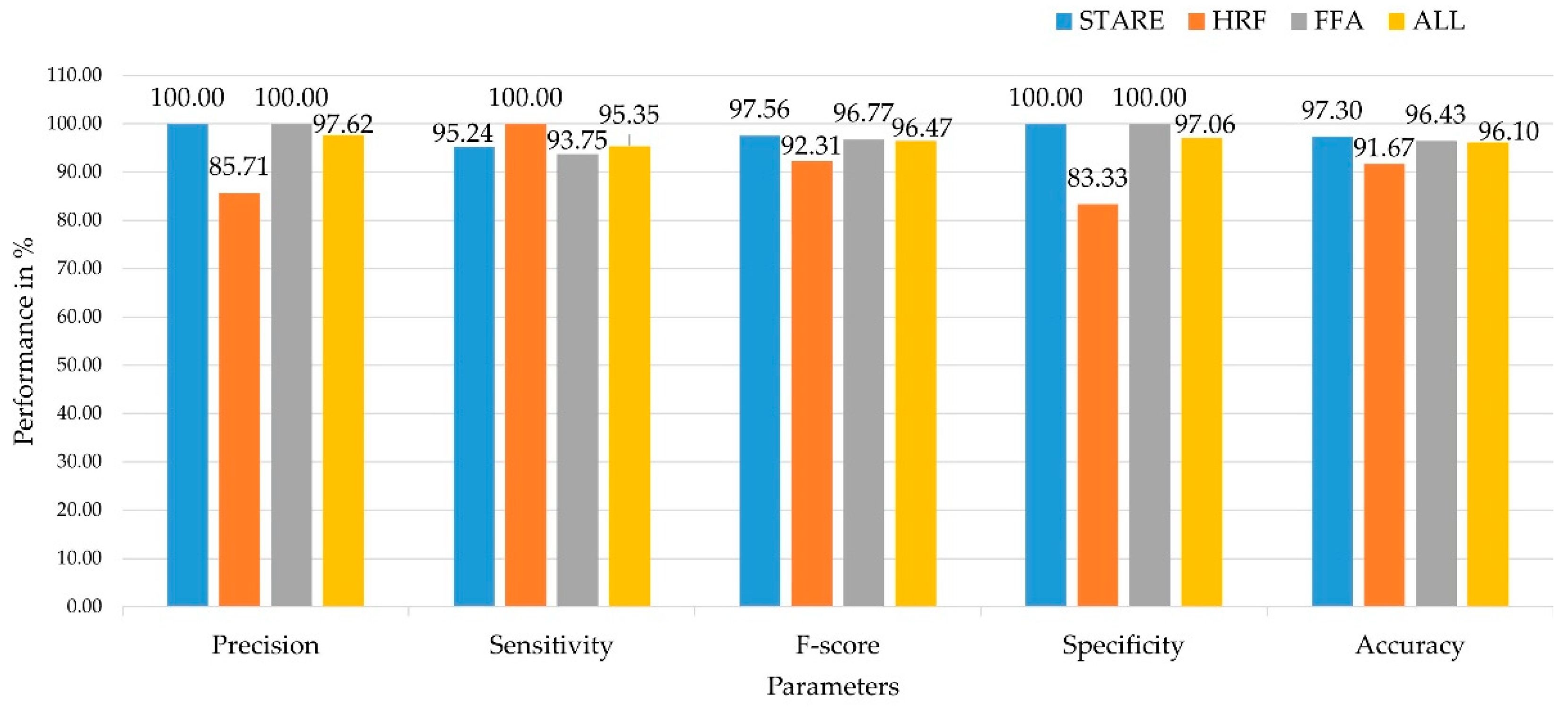
Table 5 shows the performance of the proposed and the conventional classifiers on three different publicly available datasets, namely, STARE, HRF, PPA, and ALL (combining all the three datasets).
Among the three different datasets and the combination of all datasets, the proposed CNN-RBF classifier performed well compared to other conventional machine and deep learning algorithms. Also, the maximum accuracy of 97.30%, 91.67%, 96.43%, and 96.10% is achieved in DR classification using STARE, HRF, FFA, and ALL datasets, respectively. In addition, the deep learning algorithms (CNN-RBF and CNN) reported higher accuracy compared to machine learning algorithms such as Naïve Bayes, Nearest Neighbour, Support Vector Machine and ANFIS. This shows that the deep learning algorithms extract more meaningful and useful information from the retinal image and accurately identified the retinal diseases from the input fundus image. Among the different machine learning algorithms, NB reported the lower accuracy in retinal image classification.
Figure 5 summarizes the maximum performance achieved in retinal image classification across different datasets. All performance measures such as precision, sensitivity, specificity, and accuracy showed that the proposed CNN-RBF classifier performed at least 90% better than the baseline classifier. Compared to other state-of-the-art methods in retinal image classification, the proposed classifier is simple, efficient, and achieves maximum performance across all three datasets. The comparison of previous studies is represented in
Table 6.
4.1. Limitation and Future Works
The proposed methodology performed well in retinal image classification with a limited number of features and with less computational complexity, but it has a few major limitations:
The proposed methodology has been developed and tested using only three open-source or publicly available datasets. Consequently, the results may not be more robust if they are tested with unknown datasets. A larger number of images from a variety of datasets needs to be tested to validate and generalize the proposed methodology.
The present work only considered binary classification due to the limited number of images in each class of retinal diseases (DME, DR, CNV, AMD). The proposed methodology, however, must be trained with many multi-class data for a better clinical interpretation.
5. Conclusions
This research marks a significant advancement in the field of retinal disease detection, introducing pivotal architectural modifications that integrate Radial Basis Function (RBF) with Convolutional Neural Networks (CNNs). Our innovative approach centers on a multi-scale Local Binary Pattern (LBP) feature-based CNN-RBF classifier, which has demonstrated superior performance in classifying retinal images compared to existing methods. The results conclusively show that our CNN-RBF classifier outshines conventional deep learning models, such as standalone CNNs, and other machine learning algorithms like Support Vector Machines (SVM), Neural Networks (NN), Naive Bayes (NB), and Adaptive Neuro Fuzzy Inference Systems (ANFIS) in terms of accuracy and reliability.
A key innovation of our study is the development of a training and activation process that seamlessly integrates with any CNN architecture. This includes complex models featuring inception blocks and residual connections, thereby broadening the applicability of our method in various diagnostic scenarios. The application of the CNN-RBF classifier for screening Diabetic Retinopathy (DR) using retinal blood vessel analysis represents a substantial leap forward in ophthalmic diagnostics.
Uniquely, this study utilized three different publicly available datasets to train and test our hybrid Deep Neural Network (DNN) classifier for DR screening, a first in this domain to our knowledge. The proposed method achieved outstanding classification rates, with a maximum mean accuracy of 97.30% in the STARE dataset, 91.67% in the HRF dataset, 96.43% in the FFA dataset, and 96.10% when combining all three datasets. These results not only validate the effectiveness of our algorithm but also demonstrate its superiority over existing state-of-the-art solutions.
In conclusion, our research offers a highly accurate, efficient, and adaptable tool for the early detection of retinal diseases, particularly DR, which has significant implications for improving patient outcomes and reducing the burden of vision loss globally.
Author Contributions
Conceptualization, H.G.R., A.M. and M.M.; methodology, H.G.R., and P.N.B; software, H.G.R. and M.M.; validation, A.M. resources, H.G.R.; data curation, P.N.B.; writing—original draft preparation, M.M.; writing—review and editing, A.M. and M.M.; project administration, A.M.; funding acquisition, A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Kuwait University Research Grant No. EO04/18.
Ethical Statement
The authors declare that the study was conducted with the publicly available datasets and therefore there is no violation of ethics towards publication of this manuscript.
Data Availability Statement
The authors declare that the study was conducted with publicly available datasets.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organization. Blindness and vision impairment. Available online: https://www.who.int/news-room/fact-sheets/detail/blindness-and-visual-impairment (accessed on 28 December 2023).
- Murugappan, M.; Prakash, N.; Jeya, R.; Mohanarathinam, A.; Hemalakshmi, G. A Novel Attention Based Few-shot Classification Framework for Diabetic Retinopathy Detection and Grading. Measurement 2022, 111485. [Google Scholar] [CrossRef]
- Mutawa, A.M.; Alnajdi, S.; Sruthi, S. Transfer Learning for Diabetic Retinopathy Detection: A Study of Dataset Combination and Model Performance. Applied Sciences 2023, 13. [Google Scholar] [CrossRef]
- Qin, Q.; Chen, Y. A review of retinal vessel segmentation for fundus image analysis. Engineering Applications of Artificial Intelligence 2024, 128, 107454. [Google Scholar] [CrossRef]
- Kar, S.S.; Maity, S.P. Automatic Detection of Retinal Lesions for Screening of Diabetic Retinopathy. IEEE Transactions on Biomedical Engineering 2018, 65, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Huang, L.; Pan, X.; Zheng, J.; Chen, S. Optimization of deep convolutional neural network for large scale image retrieval. Neurocomputing 2018, 303, 60–67. [Google Scholar] [CrossRef]
- Khan, M.B.; Ahmad, M.; Yaakob, S.B.; Shahrior, R.; Rashid, M.A.; Higa, H. Automated Diagnosis of Diabetic Retinopathy Using Deep Learning: On the Search of Segmented Retinal Blood Vessel Images for Better Performance. Bioengineering 2023, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Marín, D.; Aquino, A.; Gegundez-Arias, M.E.; Bravo, J.M. A New Supervised Method for Blood Vessel Segmentation in Retinal Images by Using Gray-Level and Moment Invariants-Based Features. IEEE Transactions on Medical Imaging 2011, 30, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Eladawi, N.; Elmogy, M.; Helmy, O.; Aboelfetouh, A.; Riad, A.; Sandhu, H.; Schaal, S.; El-Baz, A. Automatic blood vessels segmentation based on different retinal maps from OCTA scans. Comput Biol Med 2017, 89, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Hassan, G.; El-Bendary, N.; Hassanien, A.E.; Fahmy, A.; Abullah M, S.; Snasel, V. Retinal Blood Vessel Segmentation Approach Based on Mathematical Morphology. Procedia Computer Science 2015, 65, 612–622. [Google Scholar] [CrossRef]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. Blood vessel segmentation methodologies in retinal images--a survey. Comput Methods Programs Biomed 2012, 108, 407–433. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. Blood Vessel Segmentation of Fundus Images by Major Vessel Extraction and Subimage Classification. IEEE J Biomed Health Inform 2015, 19, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Barkana, B.D.; Saricicek, I.; Yildirim, B. Performance analysis of descriptive statistical features in retinal vessel segmentation via fuzzy logic, ANN, SVM, and classifier fusion. Knowledge-Based Systems 2017, 118, 165–176. [Google Scholar] [CrossRef]
- Marin, D.; Aquino, A.; Gegundez, M.; Bravo, J.M. A New Supervised Method for Blood Vessel Segmentation in Retinal Images by Using Gray-Level and Moment Invariants-Based Features. IEEE Transactions on Medical Imaging 2011, 30, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.; Palani, S. Blood Vessel, Optical Disk and Damage Area-Based Features for Diabetic Detection from Retinal Images. Arabian Journal for Science and Engineering 2014, 39, 7059–7071. [Google Scholar] [CrossRef]
- Liskowski, P.; Krawiec, K. Segmenting Retinal Blood Vessels With Deep Neural Networks. IEEE Trans Med Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Vasanthi, S.; Banu, R.W. Automatic segmentation and classification of hard exudates to detect Macular Edema in fundus images. Journal of Theoretical and Applied Information Technology 2014, 66, 684–690. [Google Scholar]
- Morales, S.; Engan, K.; Naranjo, V.; Colomer, A. Retinal Disease Screening Through Local Binary Patterns. IEEE J Biomed Health Inform 2017, 21, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Morales, S.; Naranjo, V.; Angulo, J.; Fuertes, J.; Alcañiz Raya, M. Segmentation and analysis of retinal vascular tree from fundus image processing. In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2012) 2012; 2012; pp. 321–324. [Google Scholar] [CrossRef]
- Sangeethaa, S.N.; Uma Maheswari, P. An Intelligent Model for Blood Vessel Segmentation in Diagnosing DR Using CNN. J Med Syst 2018, 42, 175. [Google Scholar] [CrossRef]
- Alkhaleefah, M.; Wu, C.-C. A Hybrid CNN and RBF-Based SVM Approach for Breast Cancer Classification in Mammograms. 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), 2018; 894–899. [Google Scholar] [CrossRef]
- Balasubramanian, K.; Ananthamoorthy, N. Robust retinal blood vessel segmentation using convolutional neural network and support vector machine. Journal of Ambient Intelligence and Humanized Computing 2021, 12. [Google Scholar] [CrossRef]
- Dai, L.; Wu, L.; Li, H.; Cai, C.; Wu, Q.; Kong, H.; Liu, R.; Wang, X.; Hou, X.; Liu, Y.; et al. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat Commun 2021, 12, 3242. [Google Scholar] [CrossRef]
- Nadeem, M.W.; Goh, H.G.; Hussain, M.; Liew, S.-Y.; Andonovic, I.; Khan, M.A. Deep Learning for Diabetic Retinopathy Analysis: A Review, Research Challenges, and Future Directions. Sensors (Basel, Switzerland) 2022, 22. [Google Scholar] [CrossRef] [PubMed]
- Kanagaraj, R.; Karuna, Y. Retinal vessel segmentation to diagnose diabetic retinopathy using fundus images: A survey. International Journal of Imaging Systems and Technology 2023, 34, n/a-n/a. [Google Scholar] [CrossRef]
- Prabha, S.; Sasikumar, S.; Leela Manikanta, C. Diabetic Retinopathy Detection Using Automated Segmentation Techniques. Journal of Physics: Conference Series 2022, 2325. [Google Scholar] [CrossRef]
- Sivapriya, G.; Manjula Devi, R.; Keerthika, P.; Praveen, V. Automated diagnostic classification of diabetic retinopathy with microvascular structure of fundus images using deep learning method. Biomedical Signal Processing and Control 2024, 88, 105616. [Google Scholar] [CrossRef]
- Kaur, J.; Kaur, P. Automated Computer-Aided Diagnosis of Diabetic Retinopathy Based on Segmentation and Classification using K-nearest neighbor algorithm in retinal images. The Computer Journal 2022, 66, 2011–2032. [Google Scholar] [CrossRef]
- Li, Z.; Jia, M.; Yang, X.; Xu, M. Blood Vessel Segmentation of Retinal Image Based on Dense-U-Net Network. Micromachines 2021, 12, 1478. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Adarsh, A.; Kumar, B.; Singh, A.K. An automated early diabetic retinopathy detection through improved blood vessel and optic disc segmentation. Optics & Laser Technology 2020, 121, 105815. [Google Scholar] [CrossRef]
- Kamran, S.A.; Hossain, K.F.; Tavakkoli, A.; Zuckerbrod, S.L.; Sanders, K.M.; Baker, S.A. RV-GAN: Segmenting Retinal Vascular Structure in Fundus Photographs Using a Novel Multi-scale Generative Adversarial Network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, Cham; 2021; pp. 34–44. [Google Scholar]
- Ma, Z.; Li, X. An improved supervised and attention mechanism-based U-Net algorithm for retinal vessel segmentation. Computers in Biology and Medicine 2024, 168, 107770. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Yang, K.-F.; Zhao, S.-X.; Wang, J.; Liu, L.; Li, Y.-J. Deep matched filtering for retinal vessel segmentation. Knowledge-Based Systems 2024, 283, 111185. [Google Scholar] [CrossRef]
- He, X.; Wang, T.; Yang, W. Research on Retinal Vessel Segmentation Algorithm Based on a Modified U-Shaped Network. Applied Sciences 2024, 14, 465. [Google Scholar] [CrossRef]
- Soomro, T.A.; Khan, T.M.; Khan, M.A.U.; Gao, J.; Paul, M.; Zheng, L. Impact of ICA-Based Image Enhancement Technique on Retinal Blood Vessels Segmentation. IEEE Access 2018, 6, 3524–3538. [Google Scholar] [CrossRef]
- STructured Analysis of the Retina. Available online: https://cecas.clemson.edu/~ahoover/stare/ (accessed on 2 June 2023).
- High-Resolution Fundus (HRF) Image Database. Available online: https://www5.cs.fau.de/research/data/fundus-images/ (accessed on 3 July 2023).
- Fundus Photography for Health Technicians Manual. Available online: https://wwwn.cdc.gov/nchs/data/nhanes3/manuals/fundus.pdf (accessed on 10 July 2023).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).