1. Introduction
The management of water resources constitutes one of central issues of the sustainable development for the environment and human health. The responsible use of water resources relies on understanding the complex and interrelated processes that affect the quantity and quality of water available for human needs, economic activities, and ecosystems. Global demand for freshwater continues to increase at a rate of 1% per year since 1980 driven by population growth and socioeconomic changes. Simultaneously, the increase in evaporation caused by rising temperatures leads to a decrease in streamflow volumes in many areas of the world, which already suffer from water scarcity problems [
1,
2]. Achieving socioeconomic and environmental sustainability under such challenging conditions will require the application of innovative technologies, capable of measuring hydrological characteristics at a range of spatial and temporal scales [
3].
Traditional surface water management practices are primarily based on data collected from networks of in situ hydrometric gauges. Point measurements do not provide sufficient spatial resolution to fully characterize river networks. Moreover, the decline of existing measurement networks is being observed all over the world and many developing regions lack them altogether [
4]. Remote sensing methods are considered a solution to cover data gaps specific to point measurement networks [
5]. A leading example of remote sensing are measurements made from satellites. However, due to the low spatial resolution, satellite data is suitable only for studying the largest rivers. For example, the SWOT mission will allow only observation of rivers of width greater than 50-100 m [
6]. Small surface streams of the first and second order (according to Strahler's classification [
7]) constitute 70%-80% of the length of all rivers in the world. Small streams play a significant role in hydrological systems and provide an ecosystem for living organisms [
8]. In this regard, measurement techniques based on unmanned aerial vehicles (UAVs) are promising in many key aspects, as they provide observations in high spatial and temporal resolution, their deployment is simple and fast, and can be used in inaccessible locations [
9]. One of the most important stream characteristics is spatially distributed Water Surface Elevation (WSE), as it is used for the validation and calibration of hydrologic, hydraulic, or hydrodynamic models to make hydrological forecasts, including predicting dangerous events such as floods and droughts [
10,
11,
12,
13,
14].
To date, remote sensing methods for measuring WSE in small streams mainly rely on the use of UAVs with various types of sensors attached. A clear comparison in this matter was made by Bandini et al. [
15] where UAV based methods using RADAR, LIDAR and photogrammetry were compared on the same case study. As a result, the method using RADAR with root mean-square error (RMSE) of 3 cm proved to be far superior to methods based on photogrammetry and LIDAR with RMSEs of 16 cm and 22 cm respectively. Besides its high accuracy, the advantage of a radar-based solution is the short processing time. Nonetheless, this approach necessitates non-standard UAV instrumentation and the requisite knowledge for its configuration.
Photogrammetric structure-from-motion (SfM) algorithms are able to generate orthophotos and digital surface models (DSMs) of terrain from multiple aerial photographs. Photogrammetric DSMs are precise in determining the elevation of solid surfaces to within a few centimeters [
16,
17], but water surfaces are usually falsely stated. This is related to the fact that the general principle of SfM algorithms is based on the automatic search for distinguishable and static terrain points that appear in several images showing these points from different perspectives. The surface of water lacks such points as it is uniform, transparent, and in motion. The transparency of the water makes the surface level of the stream depicted on the photogrammetric DSM lower than in reality. The stream bottom is represented by photogrammetric DSMs for clear and shallow streams [
18]. Photogrammetric DSMs for opaque water bodies are affected by artifacts brought on by lack of distinguishable key points [
19]. The above factors make the measurement of WSE by direct DSM sampling yield results with high uncertainty. Some studies report that it is possible to read the WSE from a photogrammetric DSM near streambank where the stream is shallow and there are no undesirable effects associated with light penetration below the water surface [
19,
20,
21]. However, this method gives satisfactory results only for unvegetated and smoothly sloping streambanks where the boundary line between water and land is easy to define [
15]. For this reason, this method is not suitable for many streams that do not meet these conditions.
The exponentially growing interest [
22] and the promising results of machine learning algorithms in various fields offer prospects for the application of this technology for estimation of stream WSE. The topic still remains insufficiently explored. There are only a few loosely related studies on the subject. Convolutional neural networks (CNNs) were used to estimate water surface elevation in laboratory conditions using high speed camera recordings of water surface waves [
23]. In another study several machine learning approaches were tested to extract a flood water depth based on synthetic aperture radar and DSM data [
24]. In the context of using photogrammetric DSM to estimate river water levels, artificial intelligence appears to be a promising tool. Thanks to its flexibility, it can potentially take into account a number of the adverse factors mentioned above and make a more accurate estimate of the WSE compared to direct DSM sampling.
The objective of this study is to assess the capability of CNNs in handling the disturbances in water areas present in photogrammetric DSMs of small streams, with the aim of accurately estimating the WSE.
2. Materials and Methods
2.1. Case study site
Photogrammetric data and WSE observations were obtained for Kocinka – a small lowland stream (length 40 km, catchment area ) located in the Odra River basin in southern Poland. Data were collected on two stream stretches with similar hydromorphological characteristics and different water transparency:
-
About 700 m stretch of the Kocinka stream located near Grodzisko village (50.8744N, 18.9711E). This stretch has a water surface width of about 2 m. There are no trees in close proximity to the stream. The streambed is made up of dark silt and the water is opaque. The banks and the streambed are overgrown with rushes that protrude above the water surface. The banks are steeply sloping at angles of about 50° to 90° relative to the water surface. There are marshes nearby, with stream water flowing into them in places. Data from this stretch were collected on the following days:
- o
December 19, 2020. Total cloud cover was present during the measurements. Due to the winter season, the foliage was reduced. Samples obtained from this survey are labeled with the identifier "GRO20".
- o
July 13, 2021. There was no cloud cover during the measurements. The rushes were high and the water surface was densely covered with Lemna plants. Samples obtained from this survey are labeled with the identifier "GRO21".
-
About 700 m stretch of the Kocinka stream located near Rybna village (50.9376N, 19.1143E). This stretch has a water surface width of about 3 m and is overhung by sparse deciduous trees. There is a pale, sandy streambed that is visible through the clear water. There are no rushes that emerge from the streambed. The banks slope at angles of about. 20° to 90° relative to the water surface. Data from this stretch were collected on the following days:
- o
December 19, 2020. Total cloud cover was present during the measurements. Due to the winter season, the trees were devoid of leaves and the grasses were reduced. Samples obtained from this survey are labeled with the identifier "RYB20".
- o
July 13, 2021. There was no cloud cover during the measurements. The streambank grasses were high. With good lighting and exceptionally clear water, the streambed was clearly visible through the water. The samples obtained from this survey are labeled with the identifier "RYB21".
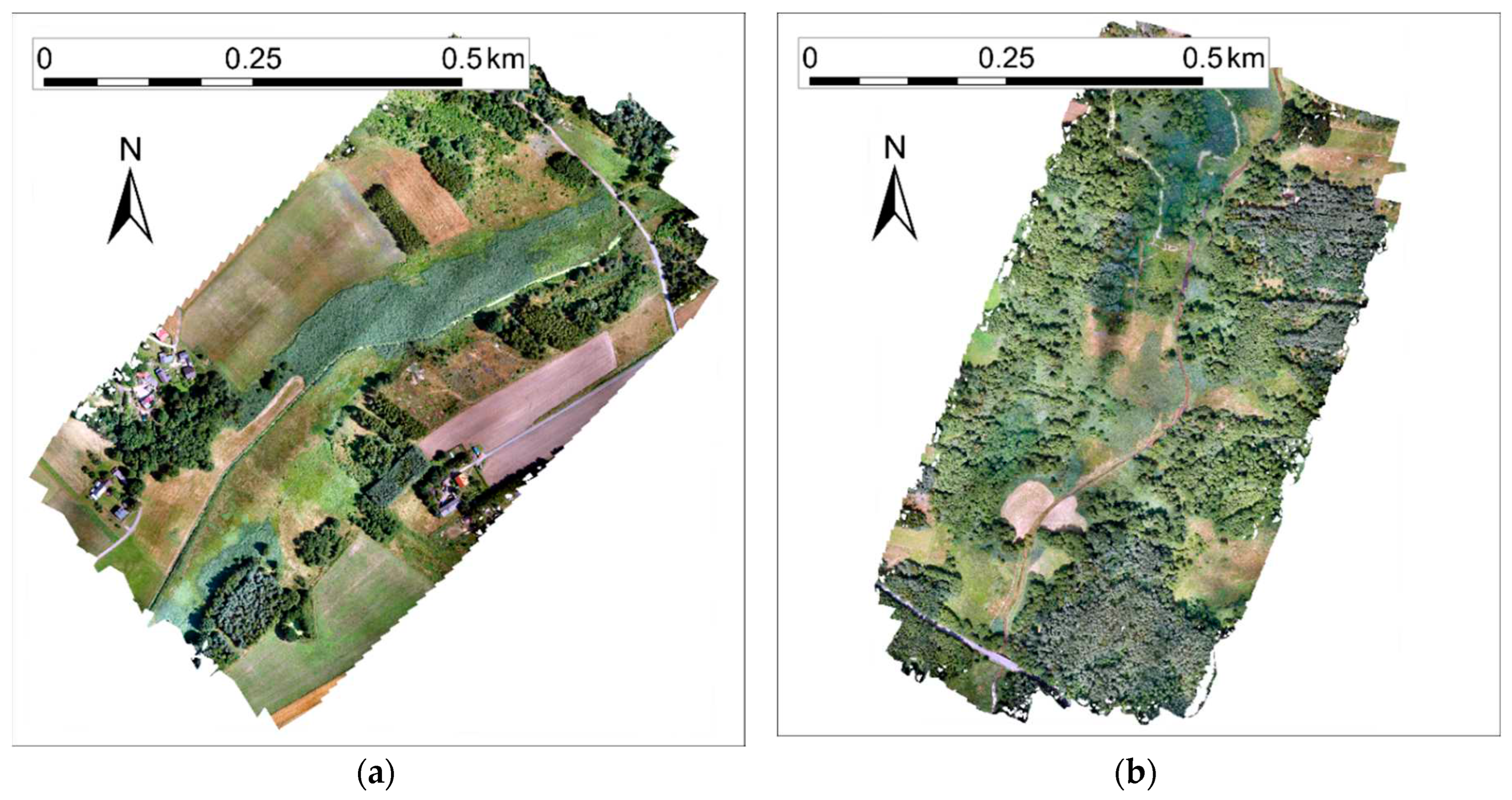
The orthophotos of the Grodzisko and Rybna case studies are shown in
Figure 1. The photo of part of the Rybna case study is shown in
Figure 2.
Furthermore, the data set was supplemented with data from surveys conducted by Bandini et. al [
25] over approximately 2.3 km stretch of the stream Åmose Å (Denmark) on November 21, 2018. The stream is channelized and well maintained. The banks are overgrown with low grass and the neighboring few trees are devoid of leaves due to winter. Further details about this case study can be found in related study, where current state-of-the-art methods to measure stream WSE with UAVs using RADAR, LIDAR and photogrammetry were tested [
15]. The supplemented data is therefore a comparative benchmark to evaluate the proposed method against existing ones. The samples obtained from this survey are labeled in our data set with the identifier "AMO18".
2.2. Field surveys
During the survey campaigns, photogrammetric measurements were conducted over the stream area. Aerial photos were taken from a DJI S900 UAV using a Sony ILCE a6000 camera with a Voigtlander SUPER WIDE HELIAR VM 15 mm f/4.5 lens. The flight altitude was approximately 77 m AGL, resulting in a 20 mm terrain pixel. The front overlap was 80%, and side overlap was 60%.
During the flights, the camera was oriented towards nadir. Some studies propose conducting multiple flights at various altitudes and camera position angles to effectively capture areas obscured by inclined vegetation and steep terrain [
15]. However, in this study, the adoption of such techniques was omitted, considering time efficiency and the recognition that for the objectives of the deep learning solution employed, the three-dimensional photogrammetric model is ultimately transformed into its two-dimensional representation in the form of an orthographic DSM raster, effectively presenting view solely from the nadir perspective.
In addition to drone flights, ground control points (GCPs) were established homogeneously in the area of interest using a Leica GS 16 RTN GNSS receiver. Ground-truth WSE point measurements were also made using an RTN GNSS receiver. They were carried out along the stream every approximately 10-20 meters on both banks.
2.3. Data processing
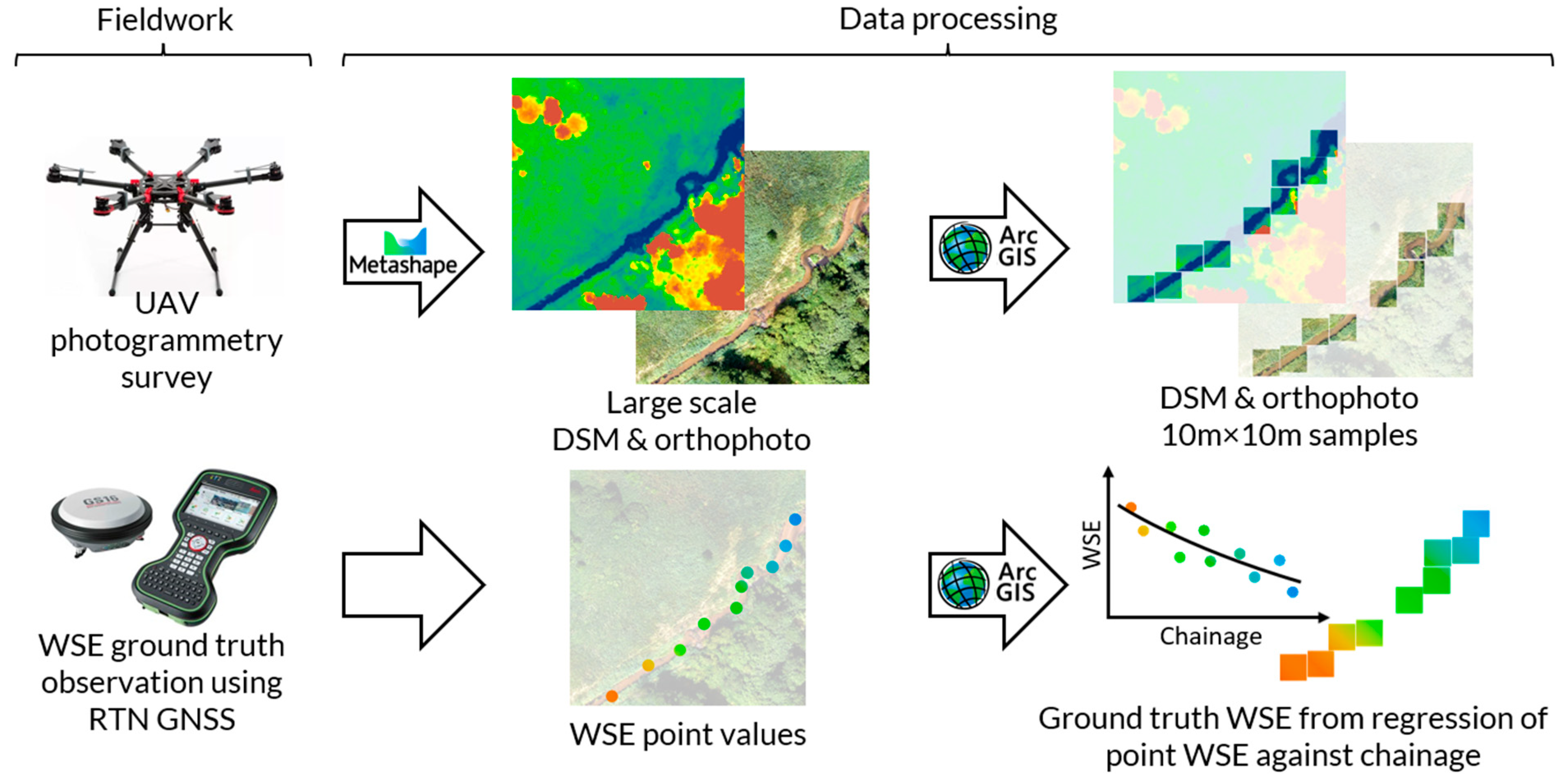
Orthophoto and DSM raster files were generated using Agisoft Metashape photogrammetric software. GCPs were used to embed rasters in a geographic reference system of latitude, longitude, and elevation. Further data processing was performed using ArcGIS ArcMap software. Each of the obtained rasters had the width and height of several tens of thousands of pixels and represented a part of a basin area exceeding 30 ha. For use in the Machine Learning algorithm, samples representing 10m x 10m areas of the terrain were manually extracted from large-scale orthophoto and DSM rasters. Each sample contains areas of water and adjacent land. The samples do not overlap.
The point measurements of ground-truth WSE were interpolated using polynomial regression as a function of chainage along the stream centerline. Where beaver dam caused an abrupt change in the WSE, regressions were made separately for the sections upstream and downstream of a dam. The WSE values interpolated by regression analysis were assigned to the raster samples according to the geospatial location (an average WSE from a stream centerline segment located within the sample area was assigned to the sample as ground-truth WSE). The standard error of estimate metric [
26] was used to determine the accuracy of ground-truth data. It was calculated using the formula:
where
– number of WSE point measurements,
– measured WSE value,
– WSE from regression analysis.
The results of the standard error of estimate examination are included in
Table 1, revealing that the ground-truth WSE error extends up to 2 cm.
Figure 3 shows a data set preparation workflow that include both fieldwork and data processing.
2.4. Machine learning data set structure
The machine learning data set comprises 322 samples. For details on the number of samples in each subset, see
Table 1. Every sample includes the data described below.
Photogrammetric orthophoto. A square crop of an orthophoto representing area, containing the water body of a stream and adjacent land. Grayscale image represented as a array of integer values from to (1-channel image of pixels).
Photogrammetric DSM. A square crop of the DSM representing the same area as the orthophoto sample described above. Stored as array of floating point numbers containing elevations of pixels expressed in m MSL.
Water Surface Elevation. Ground-truth WSE of the water body segment included in orthophoto and DSM sample. Represented as a single floating point value expressed in m MSL.
-
Metadata. The following additional information is stored for each sample:
- o
DSM statistics. Mean, standard deviation, minimum, and maximum values of the photogrammetric DSM sample array, which can be used for standardization or normalization. Represented as floating point values expressed in m MSL.
- o
Centroid latitude and longitude. WGS-84 geographical coordinates of the centroid of the shape of the sample area. Represented as floating-point numbers.
- o
Chainage. Sample position expressed using a chainage relative for a given stream section.
- o
Subset ID. Text value that identifies the survey subset to which the sample belongs. Available values: "GRO21", "RYB21", "GRO20", "RYB20, "AMO18". For additional information about case studies, see section 2.1.
2.5. DSM-WSE relationship
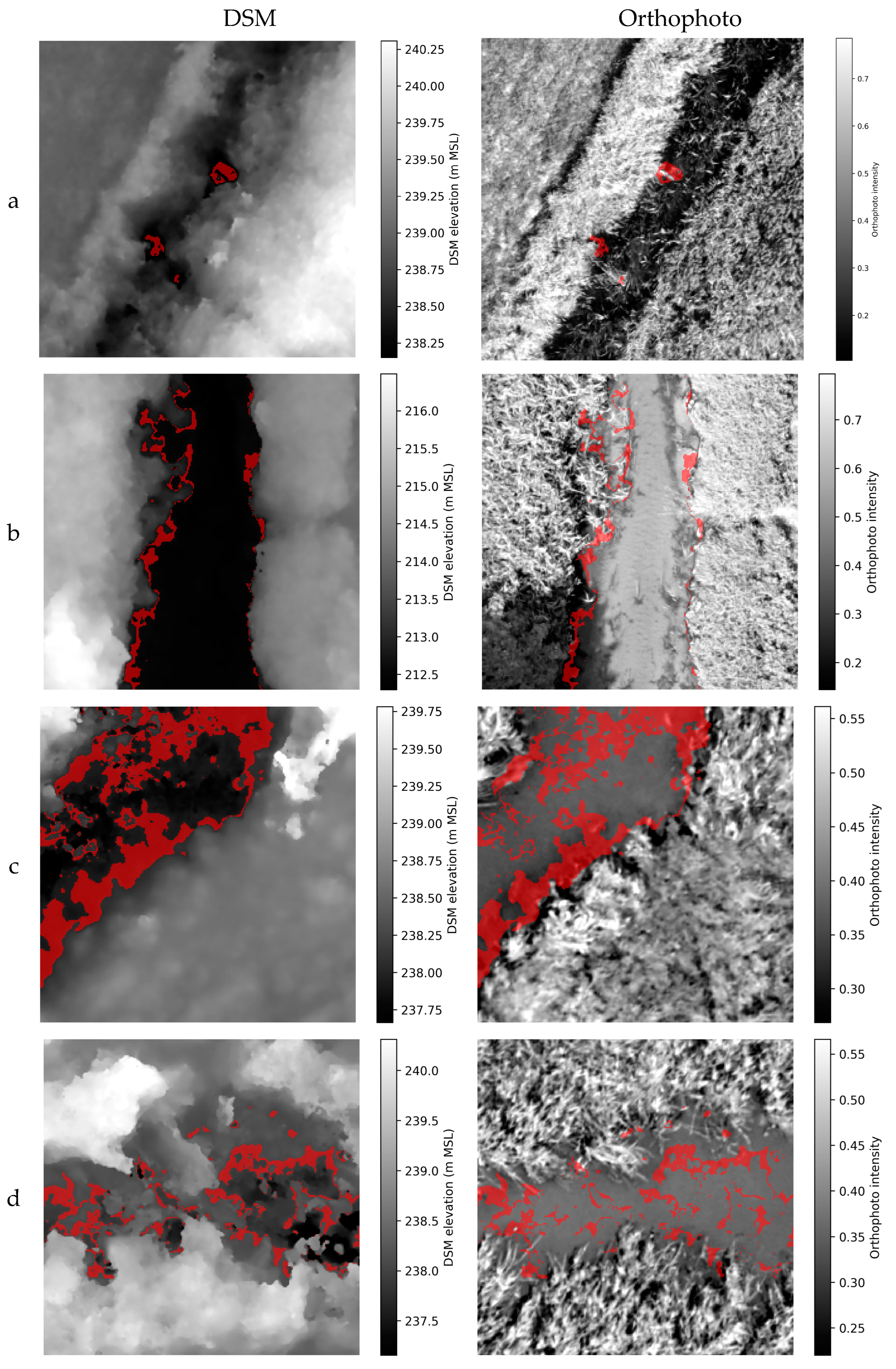
Figure 5 shows example dataset samples with marked areas where the DSM equals the actual WSE ±5cm. It can be seen that the patterns are not straightforward and in places do not meet the rule saying that the water level read from the DSM at the streambank corresponds to the WSE.
Figure 4.
Example DSM and orthophoto dataset samples (a-d) with marked areas where the DSM equals the actual WSE ±5 cm (red color).
Figure 4.
Example DSM and orthophoto dataset samples (a-d) with marked areas where the DSM equals the actual WSE ±5 cm (red color).
2.6. Deep learning framework
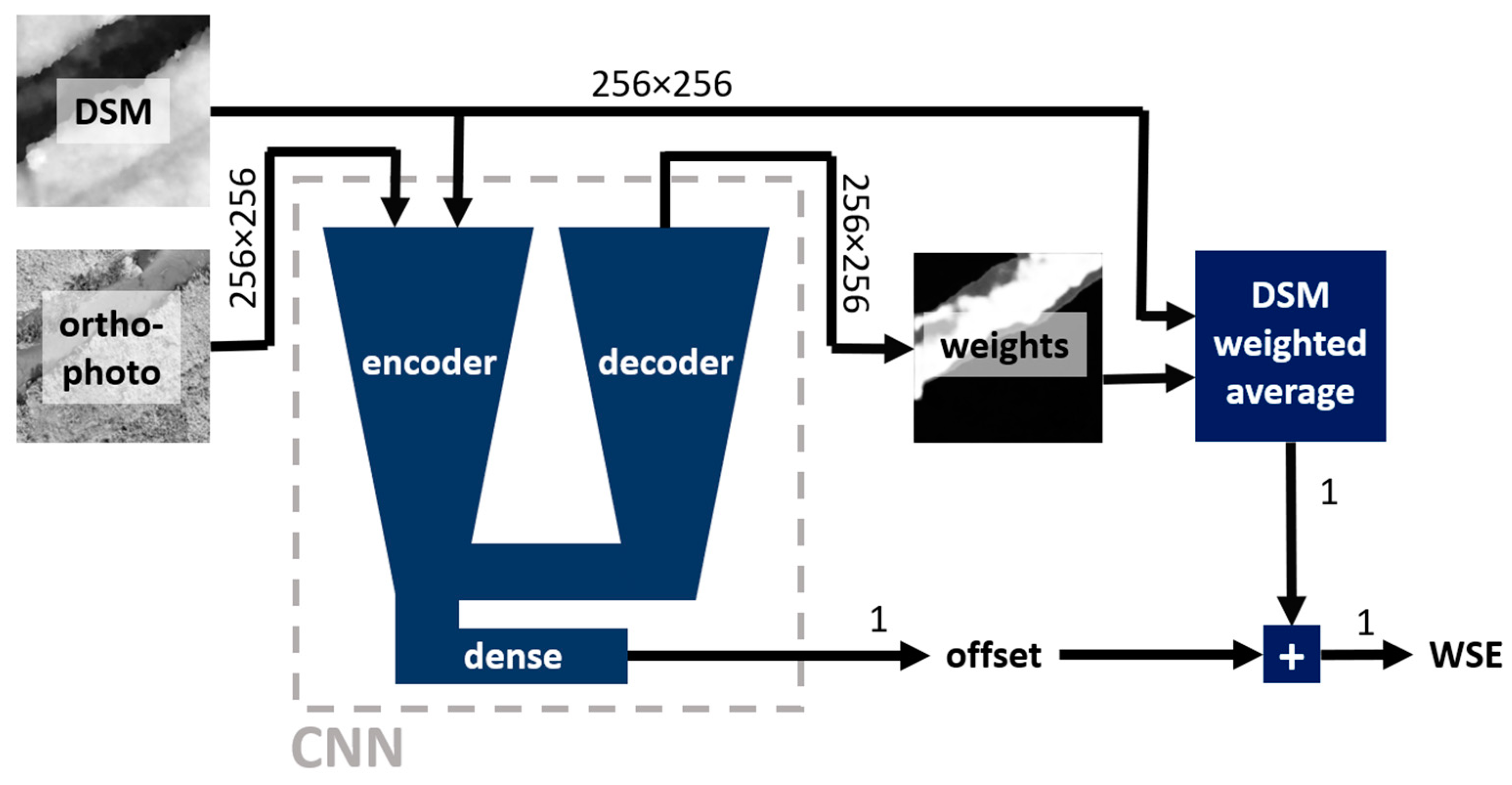
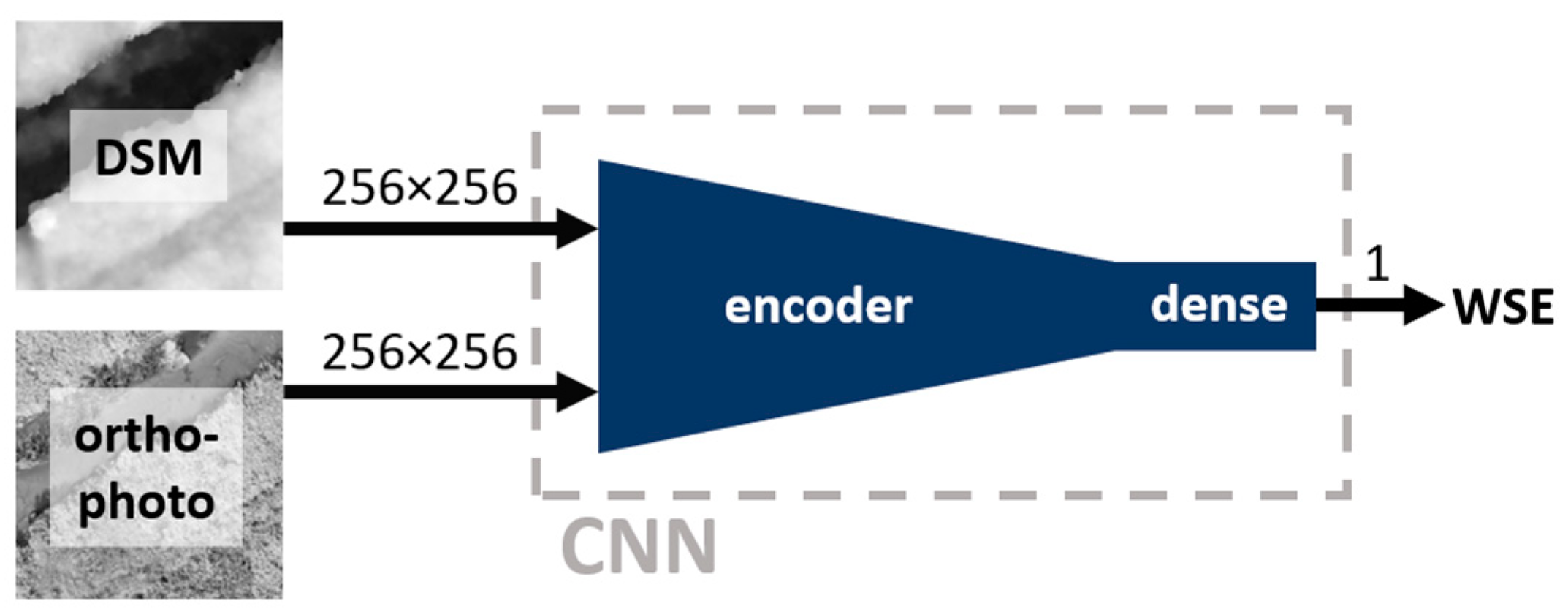
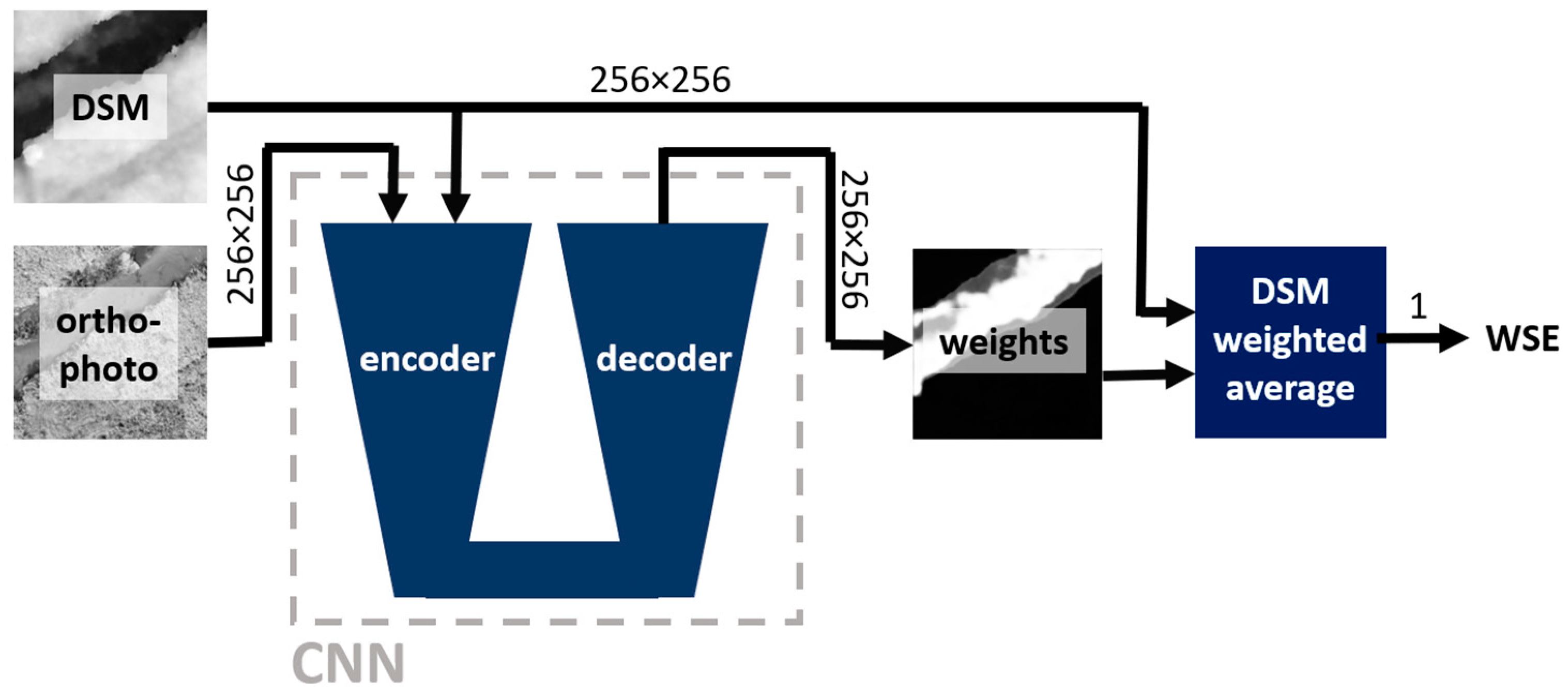
In this study, a deep learning (DL) convolutional neural network (CNN) is utilized to estimate a WSE from a DSM and an orthophoto. Two approaches are tested: direct regression of WSE using an encoder and solution based on weighted average of the DSM using a weight mask predicted by an encoder-decoder network. Note that the proposed approaches will be referred to hereafter as "direct regression", "mask averaging".
Figure 5 and
Figure 6 depict schematic representations of the proposed approaches.
All CNN models used in this study were configured to incorporate two input channels (DSM and grayscale orthophoto). In all approaches, training is conducted using mean squared error (MSE) loss. This implies that in mask averaging approach, no ground truth masks were employed for training. Instead, the network autonomously learns to determine the optimal weight mask through the optimization of the MSE loss. CNN architectures originally designed for semantic segmentation were employed to generate weight masks. They were configured to generate single-channel predictions. By employing the sigmoid activation function at the output of the model, the model generates weight masks with values ranging between 0 and 1. All of the architectures used in this study were sourced from the Segmentation Models Pytorch library [
27].
All the training was performed with a learning rate of 10
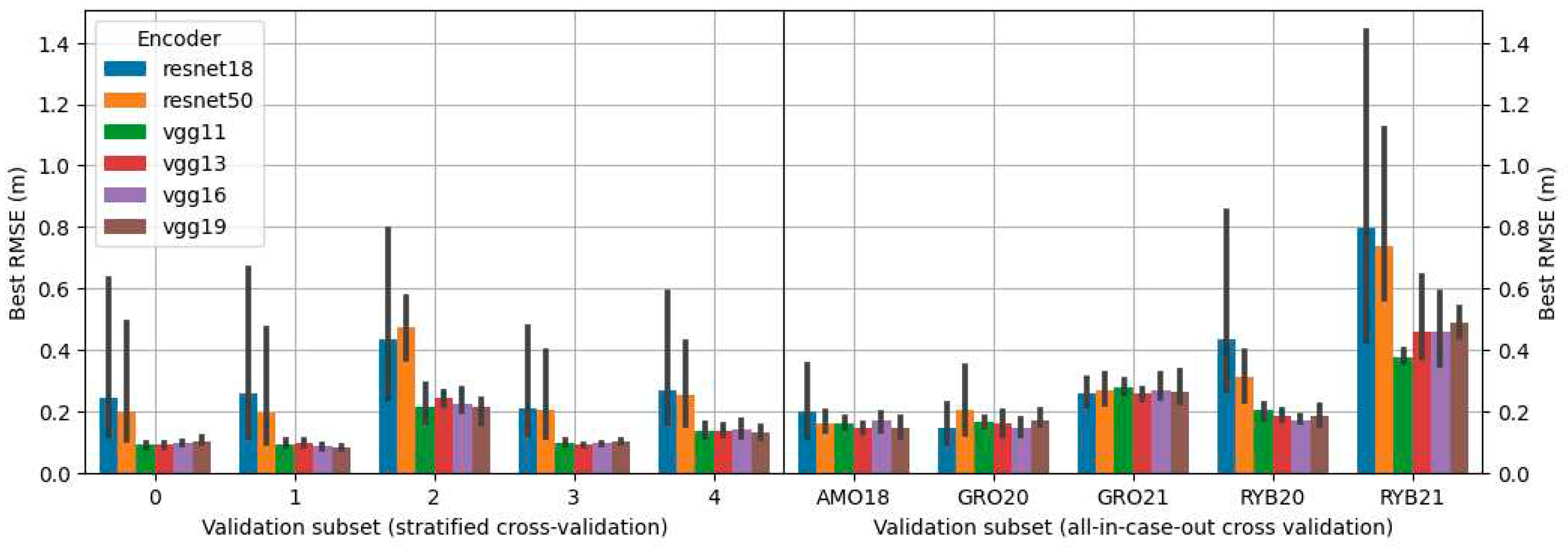
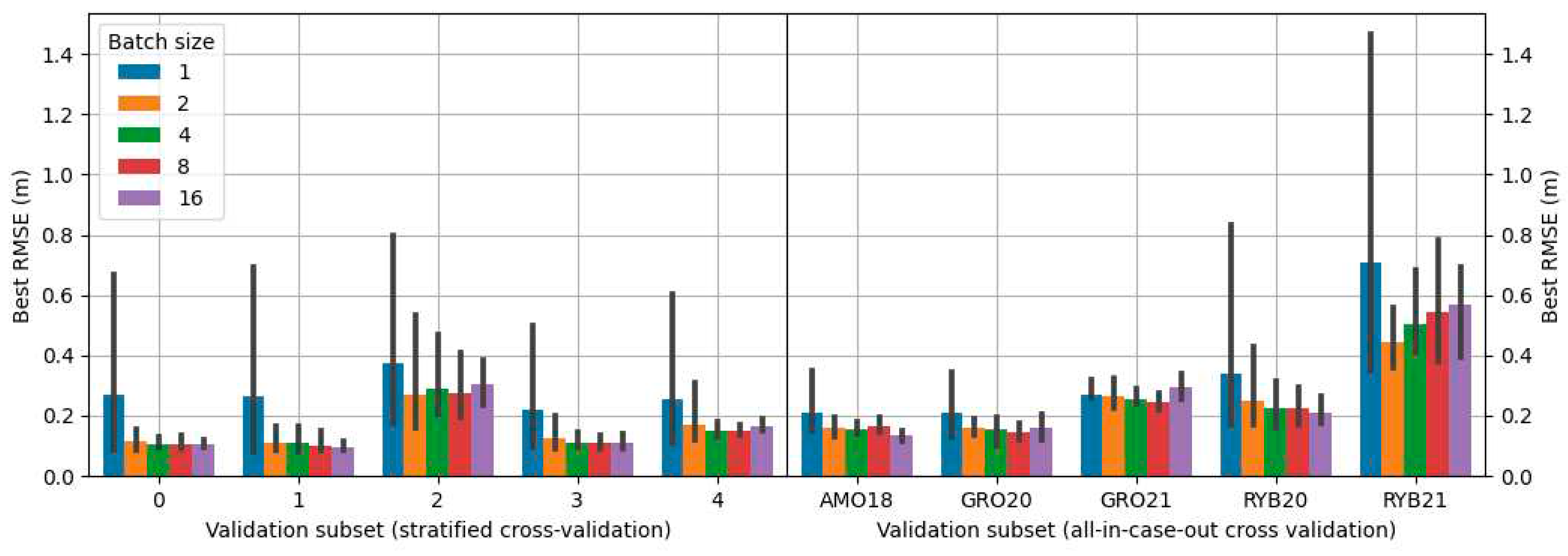
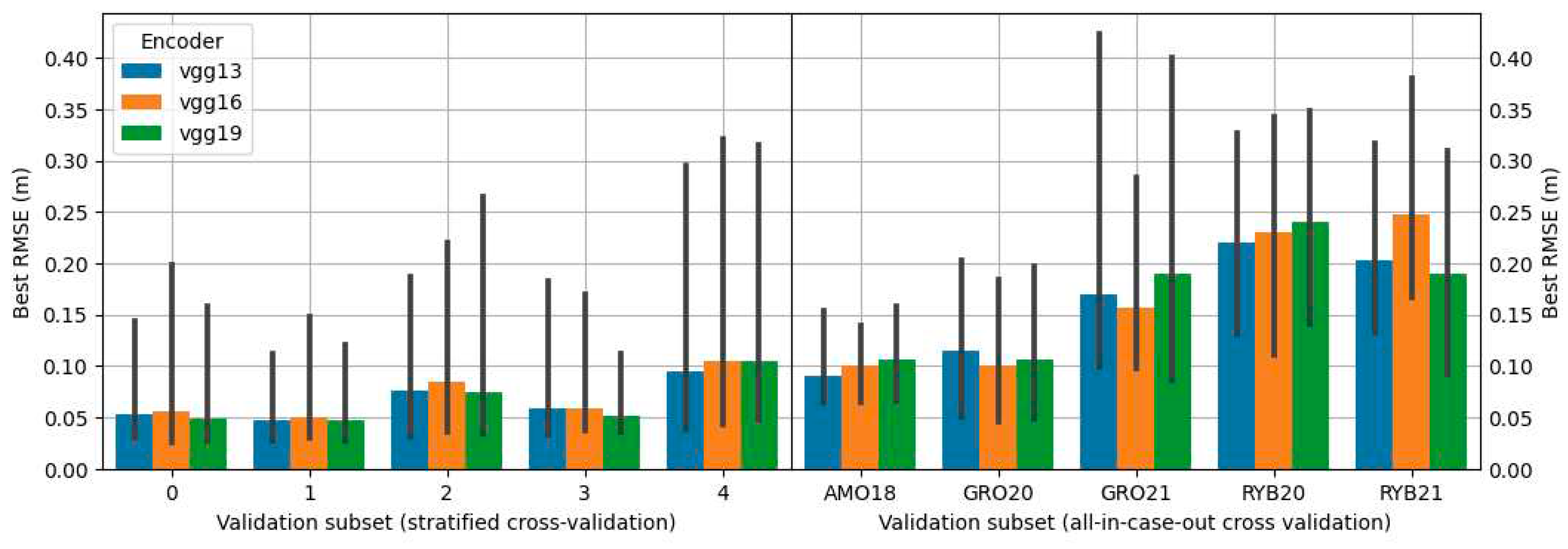
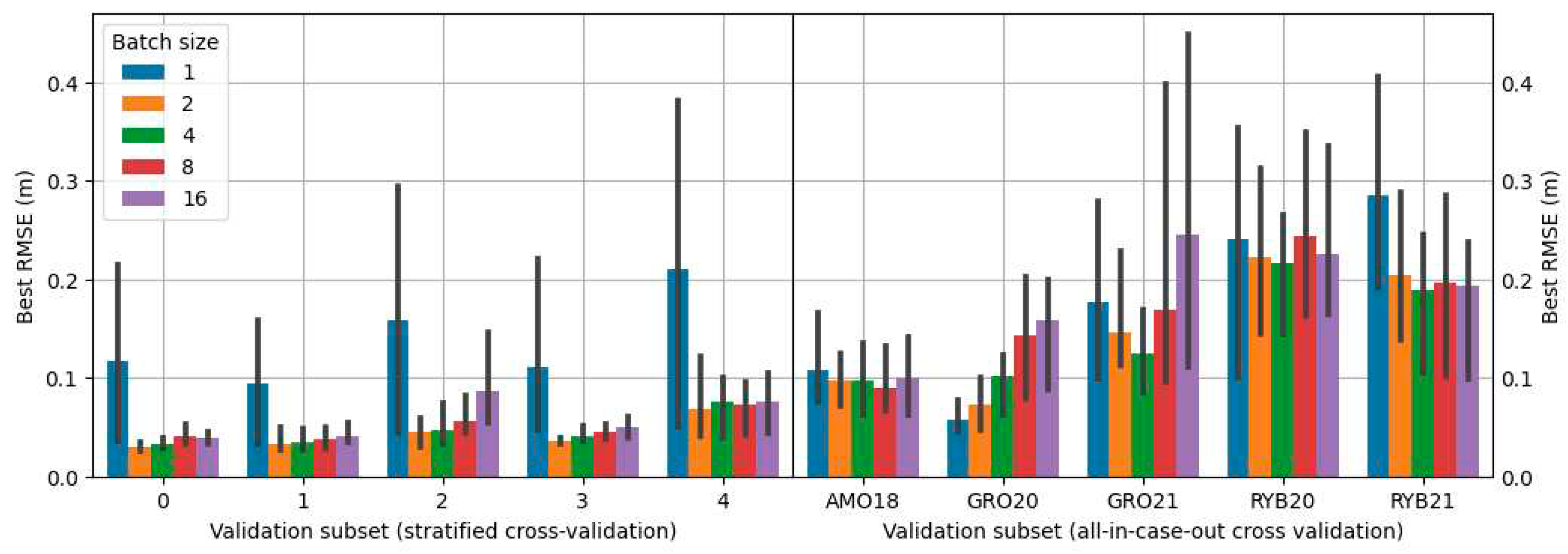
-5 using the Adam optimizer. Training was undertaken until the RMSE on the validation subset showed no further reduction for the next 20 learning epochs. Given that batch size has a notable impact on accuracy, various values of it were tested during the exploration for the optimal model during grid search (refer to
Section 2.10).
2.7. Standardization
As the DSM and orthophoto arrays have values from different ranges and distributions, they are subjected to feature scaling before they are fed into the CNN model in order to ensure proper convergence of the gradient iterative algorithm during training [
28]. The DSMs were standardized according to the equation:
where:
– standardized sample DSM 2D array with values centered around 0,
– raw sample DSM 2D array with values expressed in ,
– mean DSM value of a sample ,
– standard deviation of DSM arrays pixel values for the entire data set.
This method of standardization has two advantages. Firstly, by subtracting the average value of a sample, standardized DSMs are always centered around zero, so the algorithm is insensitive to absolute altitude differences between case studies. Actual WSE relative to mean sea level can be recovered by inverse standardization. Secondly, dividing all samples by the same value of the entire data set ensures that all standardized samples are scaled equally. It was experimentally found during preliminary model tests that multiplying the denominator by 2 results in better model accuracy compared to standardization that does not include this factor.
Orthophotos were standardized using ImageNet [
29] data set mean and standard deviation according to the equation:
where:
– standardized 1-channel orthophoto gray-scale image (2D array) with values centered around 0,
– 1-channel orthophoto gray-scale image (2D array) represented with values from the range [0,1],
– mean value of ImageNet data set red, green and blue channel values means (0.485, 0.456, 0.406),
– mean value of ImageNet data set red, green and blue channel values standard deviations (0.229, 0.224, 0.225).
2.8. Augmentation
In order to increase the size of the training data set and therefore improve prediction generalization, each sample array used to train the model was subjected to the following augmentation operations: i) rotation of 0°, 90°, 180° or 270°, ii) no inversion, inversion in the x-axis, inversion in the y-axis, or inversion both in the x-axis and the y-axis. This gives a total of 16 permutations, which makes the training data set 16 times larger.
2.9. Cross validation
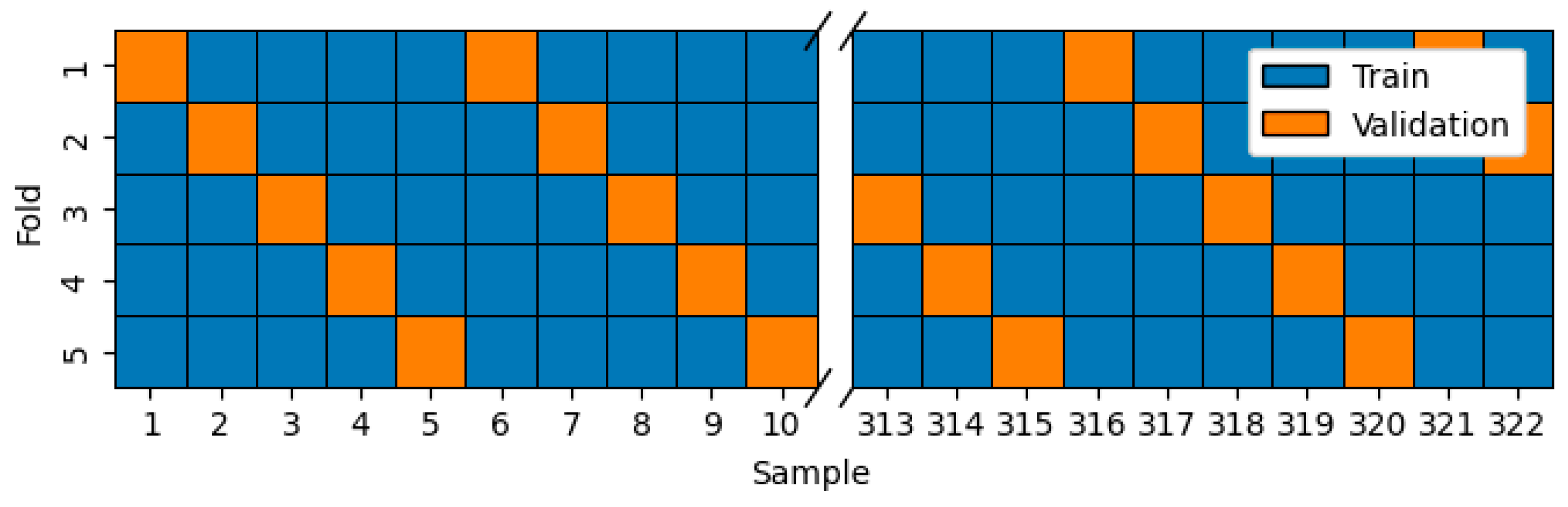
Two variations of k-fold cross-validation methods were employed: one with stratified folds of mixed samples from each case study and another with all-in-case-out folds of isolated samples for each case study.
Stratified folds were generated by selecting for validation samples at intervals of every fifth element from the entire dataset. 5 folds were created. The validation subset in each fold contained a comparable number of samples representing each of the case studies. The illustration in
Figure 7 highlights the selection of validation subsets for each of the 5 folds.
In the all-in-case-out variant of k-fold cross-validation, 5 folds were also created. However, in this scenario, a validation subset for each fold contained samples exclusively from one case study, while the remaining samples from the other 4 case studies were utilized for training. This method of cross-validation assesses the model's ability to generalize, i.e., its capacity to predict from data outside the training data distribution.
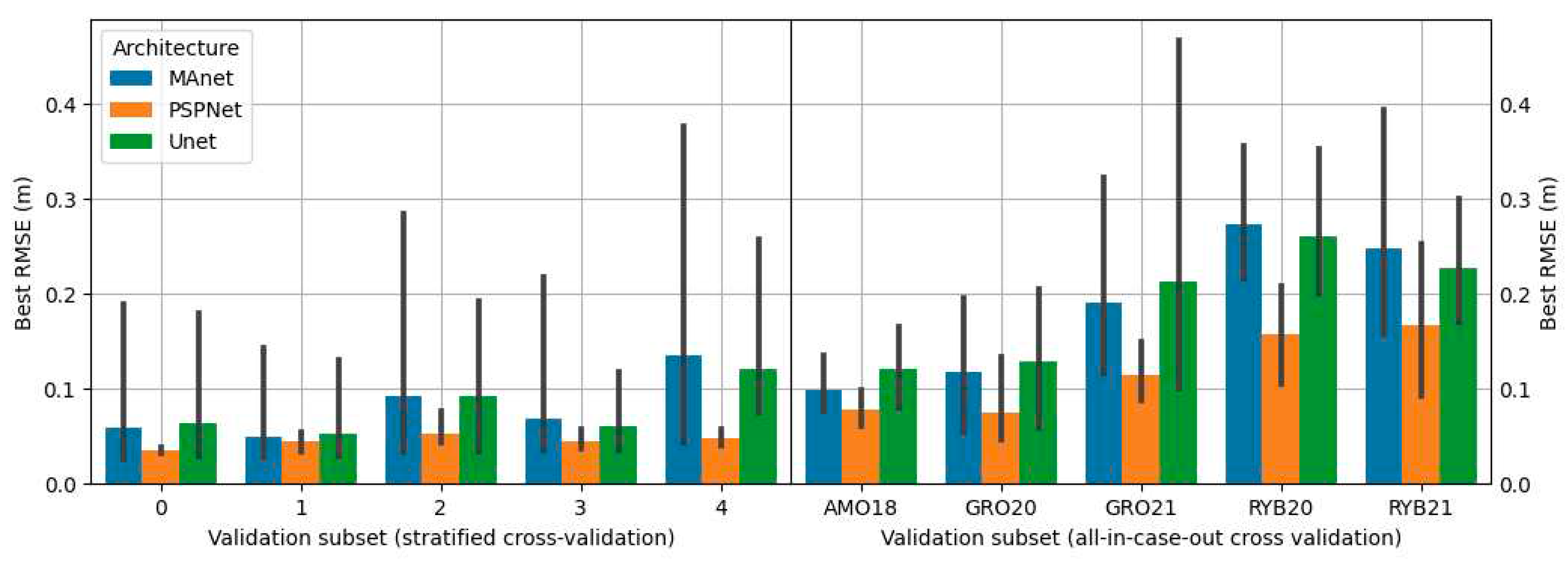
2.10. Grid search
The search for the best configuration of the proposed solutions was carried out using grid search in which all possible combinations of proposed parameters were tested. Propositions of configurations depended on the approach variant. The combinations included different types of encoders, architectures, and batch sizes. The architectures tested were: U-Net [
30], MA-Net [
31] and PSP-Net [
32]. Encoders tested were various depths of the VGG [
33] and ResNet [
34] encoders. Details on configurations used for each approach are presented in
Table 2.
2.11. Centerline and streambank sampling
The data acquired in this study allow for the use of straightforward methods for determining WSE through sampling from photogrammetric DSM along the stream centerline and at the streambank [
15]. These readings will be used for baseline comparison with the proposed method. The polylines used for sampling were determined manually, without employing any algorithm. Sampling was performed with care, especially in the water-edge method, where attention was given to ensuring that samples were consistently taken from the water area, albeit possibly close to the streambank.
5. Conclusion
In this study, the feasibility of employing deep learning to extract the WSE of a small stream from photogrammetric DSM and orthophoto was investigated. The task proved to be non-trivial, as the most obvious solution of direct regression using an encoder proved to be ineffective. Only a properly adapted architecture, which involved predicting the mask of the weights and then using it for sampling the DSM, obtained a satisfactory result.
The principal steps of the proposed solution encompass the following: i) Acquiring a photogrammetric survey over the river area and incorporating it into a geographic reference system, for instance, by utilizing ground control points. ii) Extracting DSM and orthophoto raster samples that encompass the stream area and adjacent land. iii) Employing a trained model for prediction. Improving accuracy can be achieved by finetuning for a specific case.
The proposed solution is characterized by high flexibility, generalizability and explainability. It substantially amplifies the potential of utilizing UAV photogrammetry for WSE estimation as it outperforms current methods based on UAV photogrammetry. In some cases, it achieves results comparable to the radar-based method, which is regarded as the best remote sensing method for measuring water levels in small streams.
Author Contributions
Conceptualization, R.S.; methodology, R.S., P.C., P.W., M.Z., M.P.; software, R.S., M.P.; validation, R.S., P.C., P.W., M.Z., M.P.; formal analysis, M.Z., P.W., M.P.; investigation, R.S., M.Z., P.W., M.P., P.C; resources, P.W., M.Z., P.C., M.P.; data curation, R.S., M.Z.; writing—original draft preparation, R.S.; writing—review and editing, R.S., P.W., M.Z., M.P., P.C.; visualization, R.S.; supervision, M.Z., P.W., M.P.; project administration, P.W., M.Z.; funding acquisition, P.W., M.Z. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Orthophoto mosaics from Grodzisko (a) and Rybna (b) case studies from July 13, 2021.
Figure 1.
Orthophoto mosaics from Grodzisko (a) and Rybna (b) case studies from July 13, 2021.
Figure 2.
Part of the Kocinka stretch in Rybna (March 2022).
Figure 2.
Part of the Kocinka stretch in Rybna (March 2022).
Figure 3.
Schematic representation of the data set preparation workflow.
Figure 3.
Schematic representation of the data set preparation workflow.
Figure 5.
Direct regression approach – schematic representation. Numbers near arrows provide information about the dimensions of the flowing data.
Figure 5.
Direct regression approach – schematic representation. Numbers near arrows provide information about the dimensions of the flowing data.
Figure 6.
Mask averaging approach – schematic representation. Numbers near arrows provide information about the dimensions of the flowing data.
Figure 6.
Mask averaging approach – schematic representation. Numbers near arrows provide information about the dimensions of the flowing data.
Figure 7.
The selection of validation samples for each of the 5 folds in stratified resampling.
Figure 7.
The selection of validation samples for each of the 5 folds in stratified resampling.
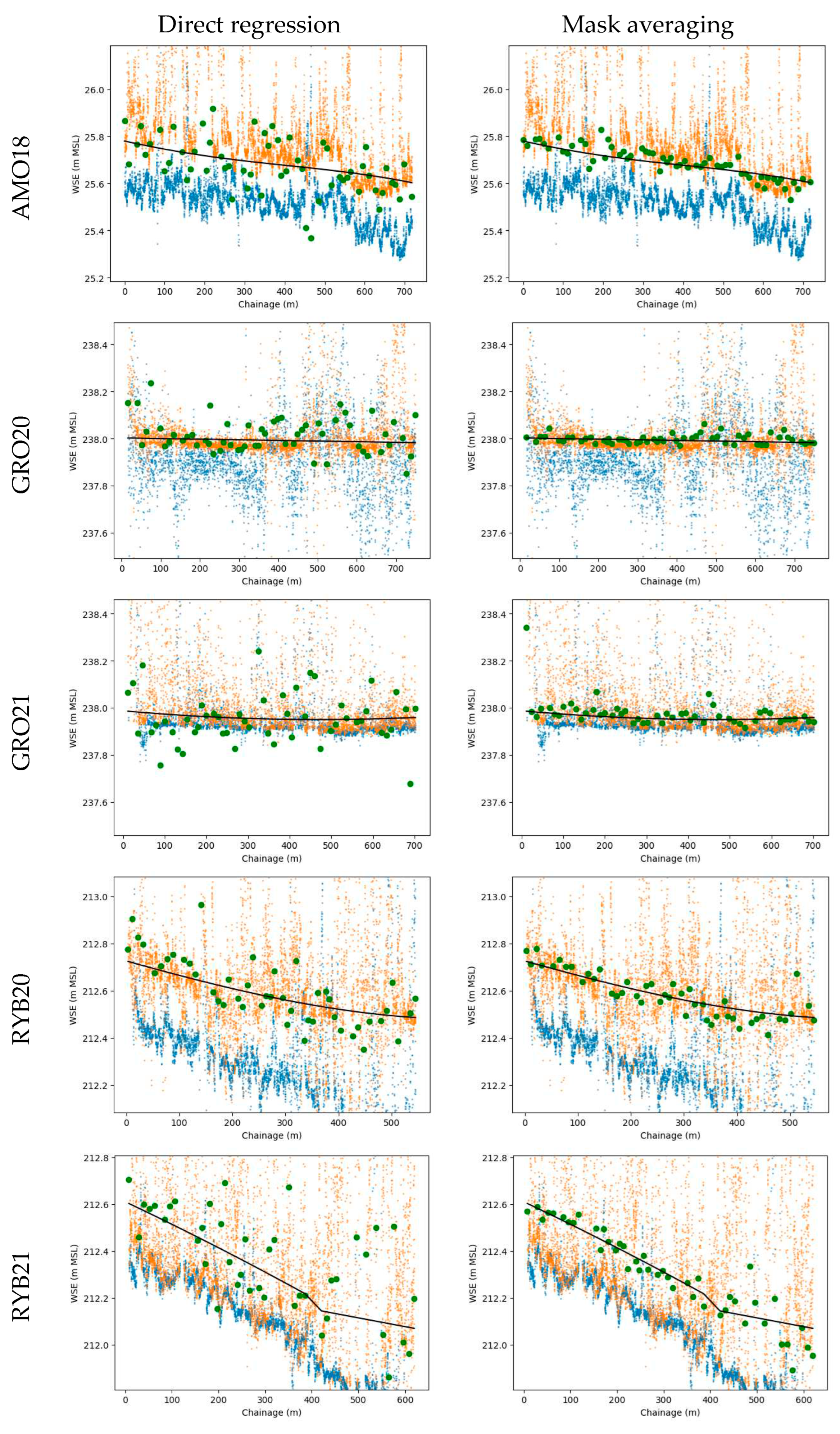
Figure 8.
Predictions on validation subsets from stratified cross-validation plotted against chainage (dark green points). Compared with ground-truth WSE (black line), DSM sampled near streambank (orange points), DSM sampled at stream centerline (blue points). Columns denote different approaches, and rows correspond to distinct case studies.
Figure 8.
Predictions on validation subsets from stratified cross-validation plotted against chainage (dark green points). Compared with ground-truth WSE (black line), DSM sampled near streambank (orange points), DSM sampled at stream centerline (blue points). Columns denote different approaches, and rows correspond to distinct case studies.
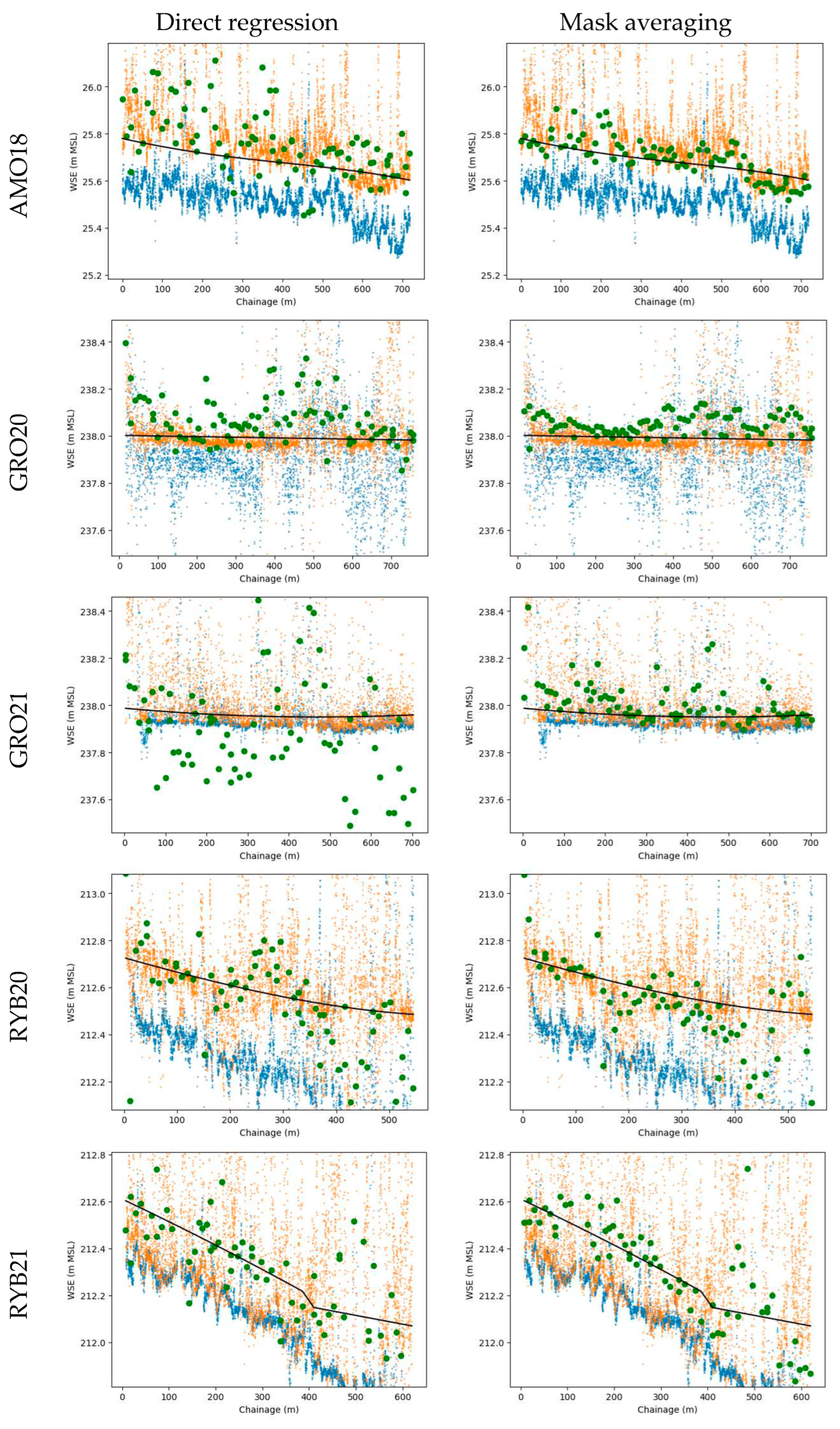
Figure 9.
Predictions on validation subsets from all-in-case-out cross-validation plotted against chainage (dark green points). Compared with ground-truth WSE (black line), DSM sampled near streambank (orange points), DSM sampled at stream centerline (blue points). Columns denote different approaches, and rows correspond to distinct case studies.
Figure 9.
Predictions on validation subsets from all-in-case-out cross-validation plotted against chainage (dark green points). Compared with ground-truth WSE (black line), DSM sampled near streambank (orange points), DSM sampled at stream centerline (blue points). Columns denote different approaches, and rows correspond to distinct case studies.
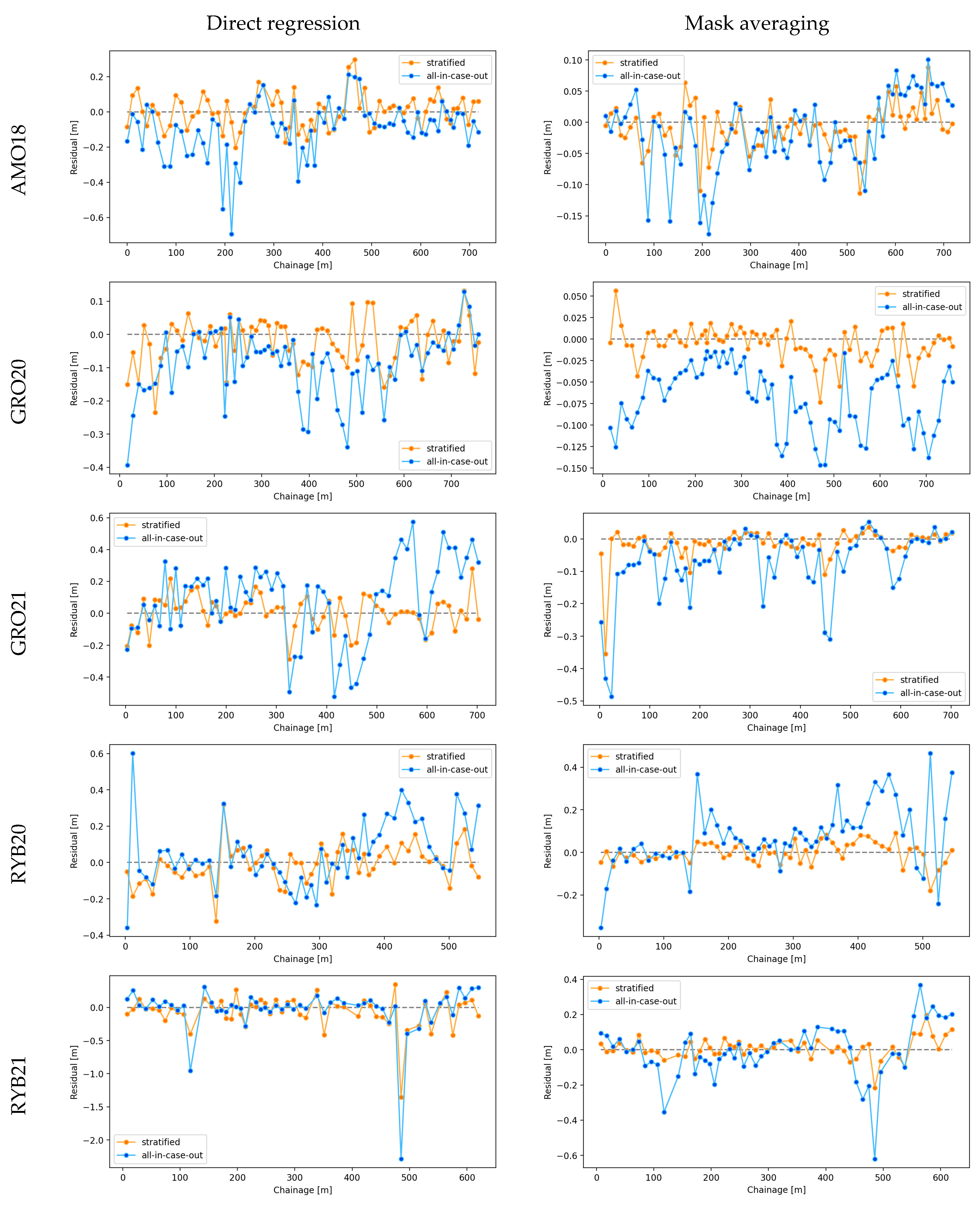
Figure 10.
Residuals (ground truth WSE minus predicted WSE) obtained during stratified and all-in-case-out cross-validations for each case study (rows) and method (columns) plotted against chainage.
Figure 10.
Residuals (ground truth WSE minus predicted WSE) obtained during stratified and all-in-case-out cross-validations for each case study (rows) and method (columns) plotted against chainage.
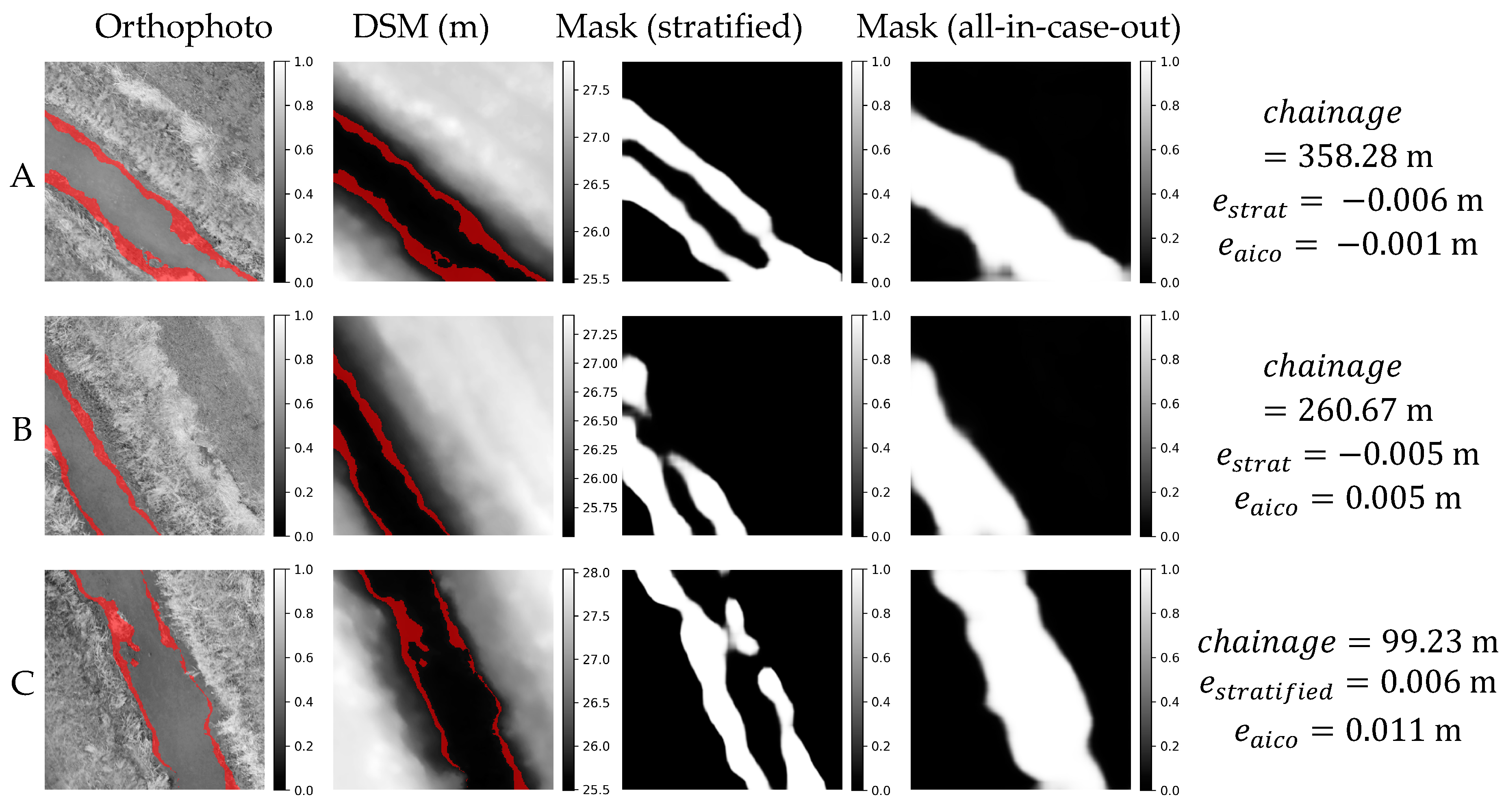
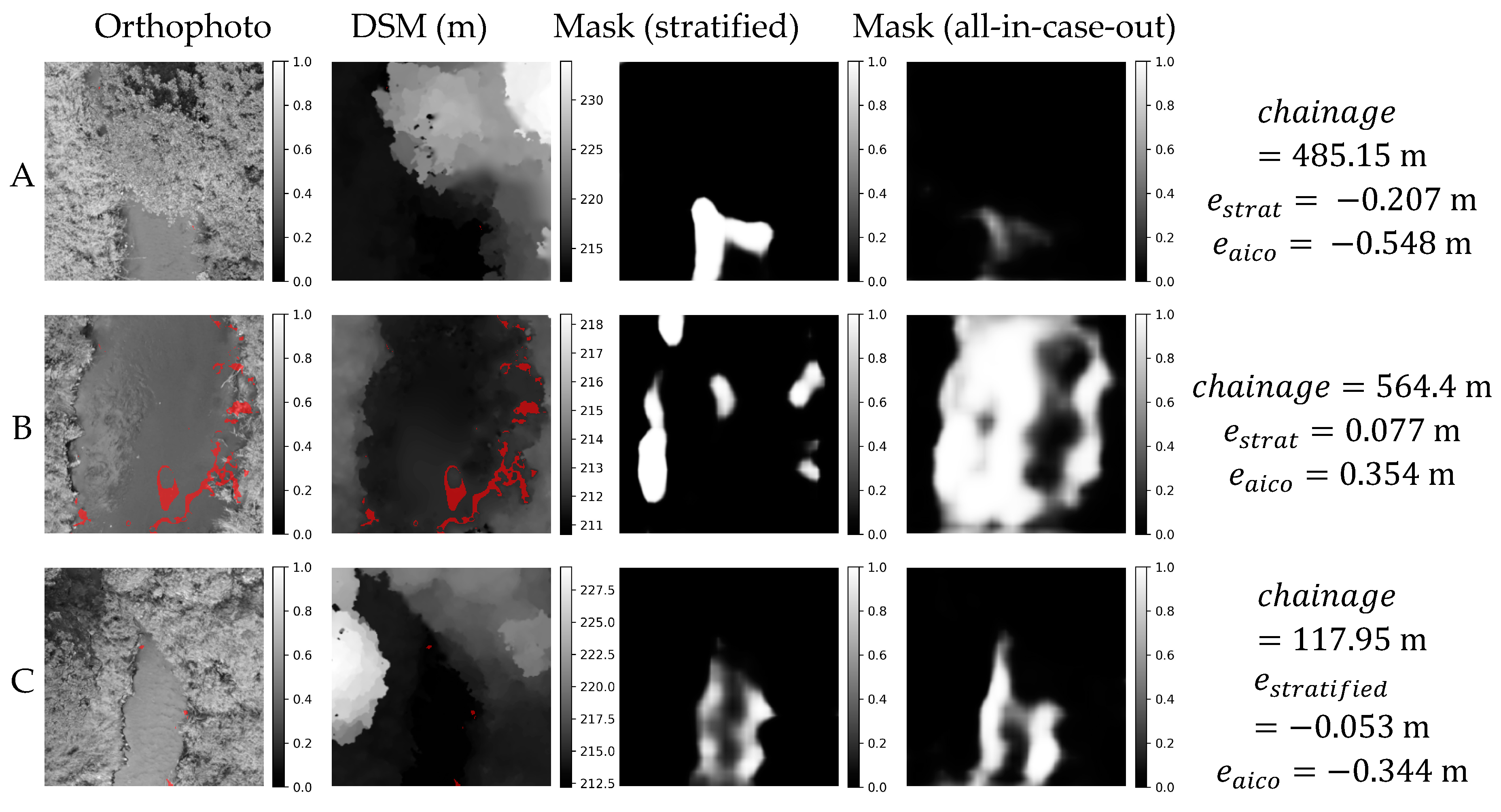
Figure 11.
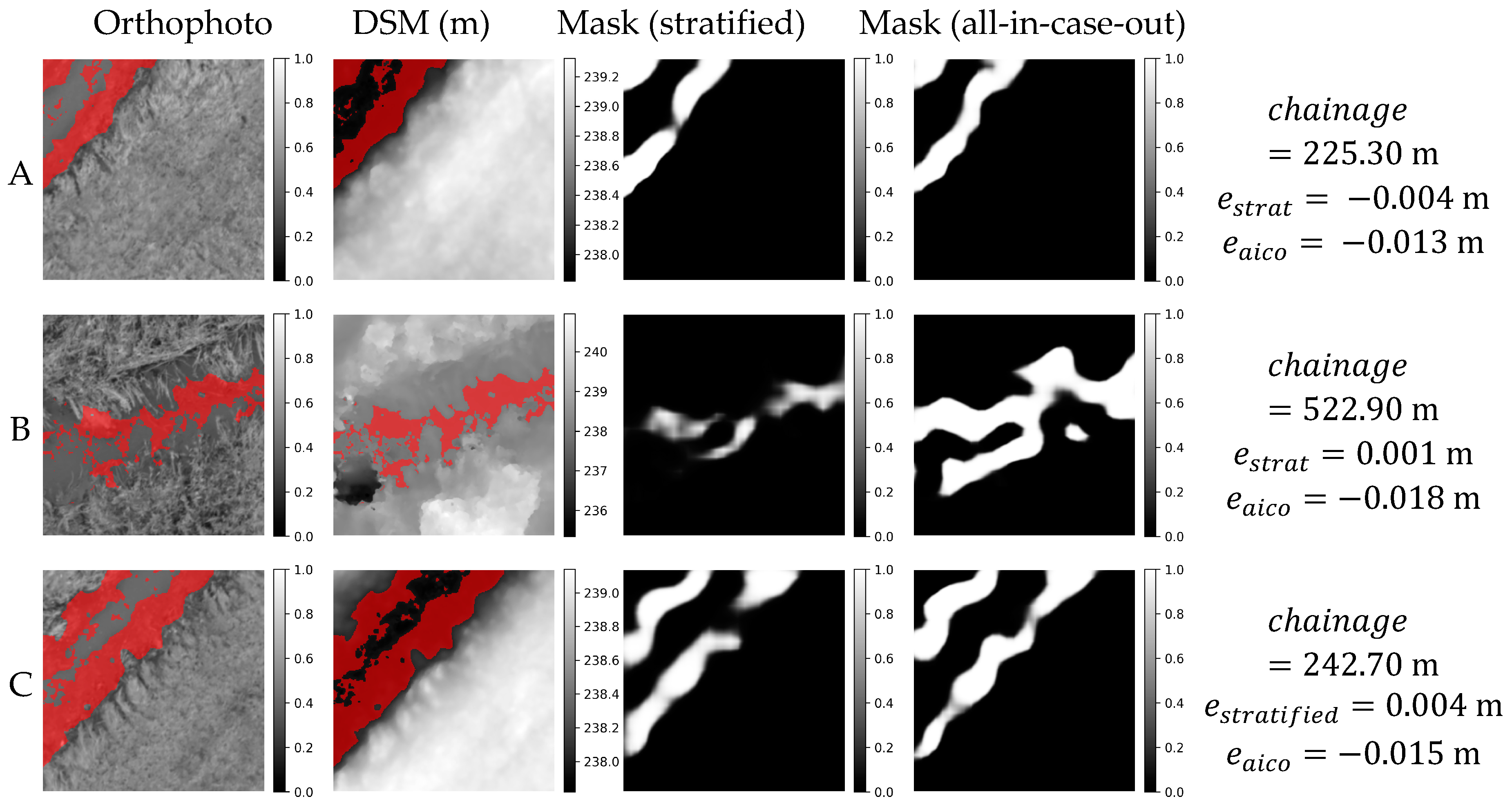
Orthophoto, DSM and weight masks obtained in stratified and all-in-case-out cross validations for three best performing samples from AMO18 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 11.
Orthophoto, DSM and weight masks obtained in stratified and all-in-case-out cross validations for three best performing samples from AMO18 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 12.
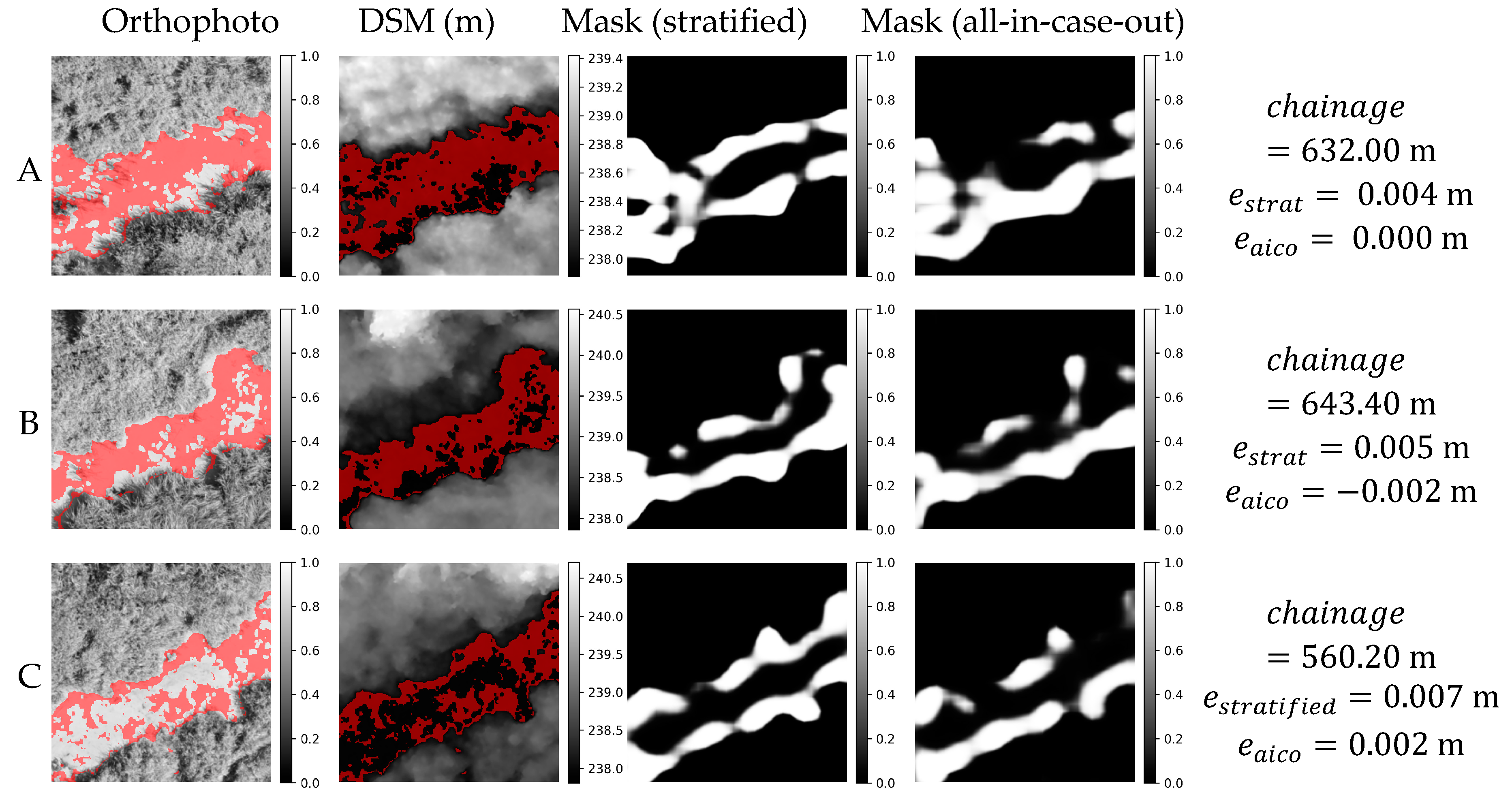
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from GRO20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 12.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from GRO20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 13.
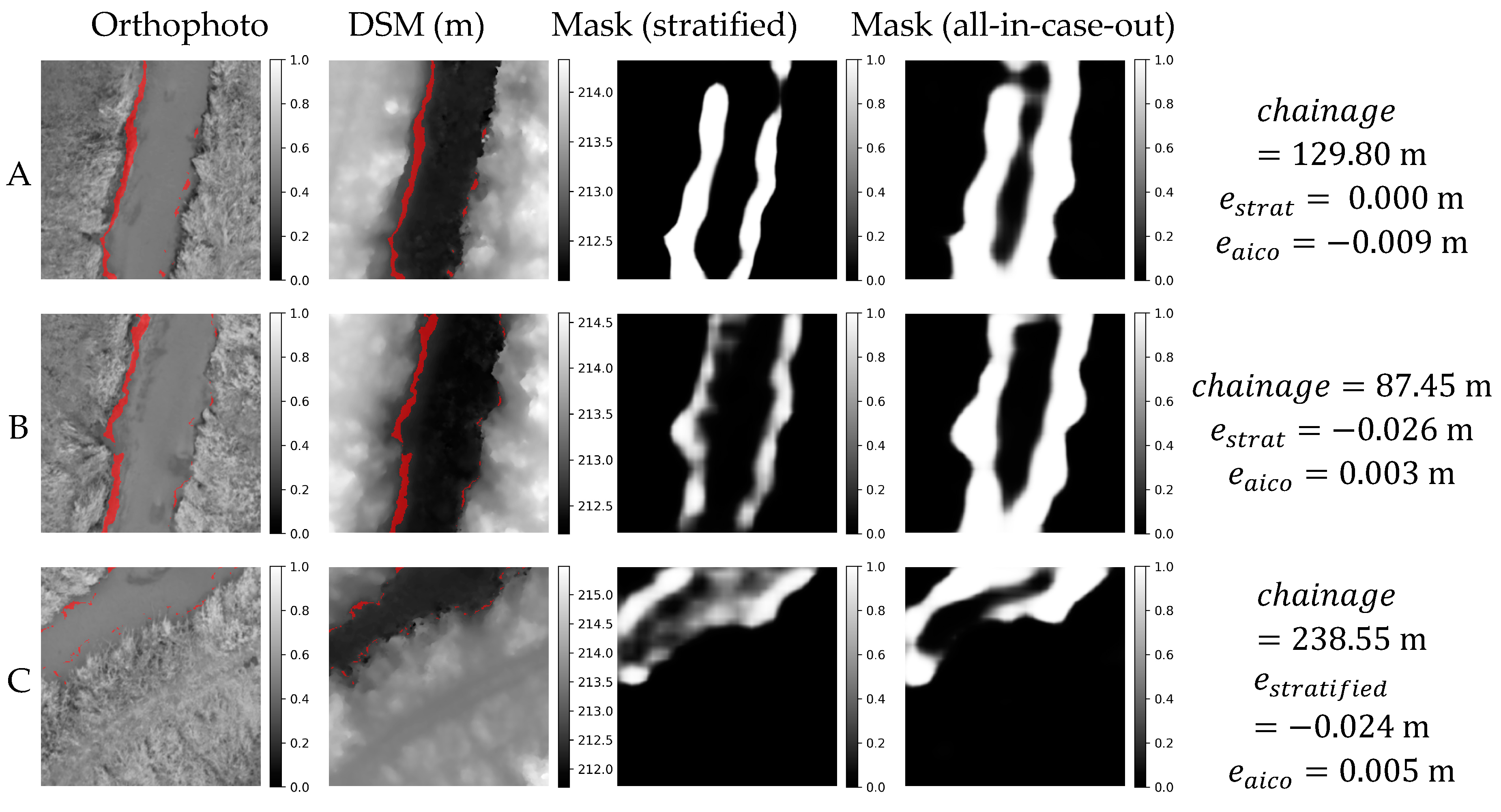
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from GRO21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 13.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from GRO21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 14.
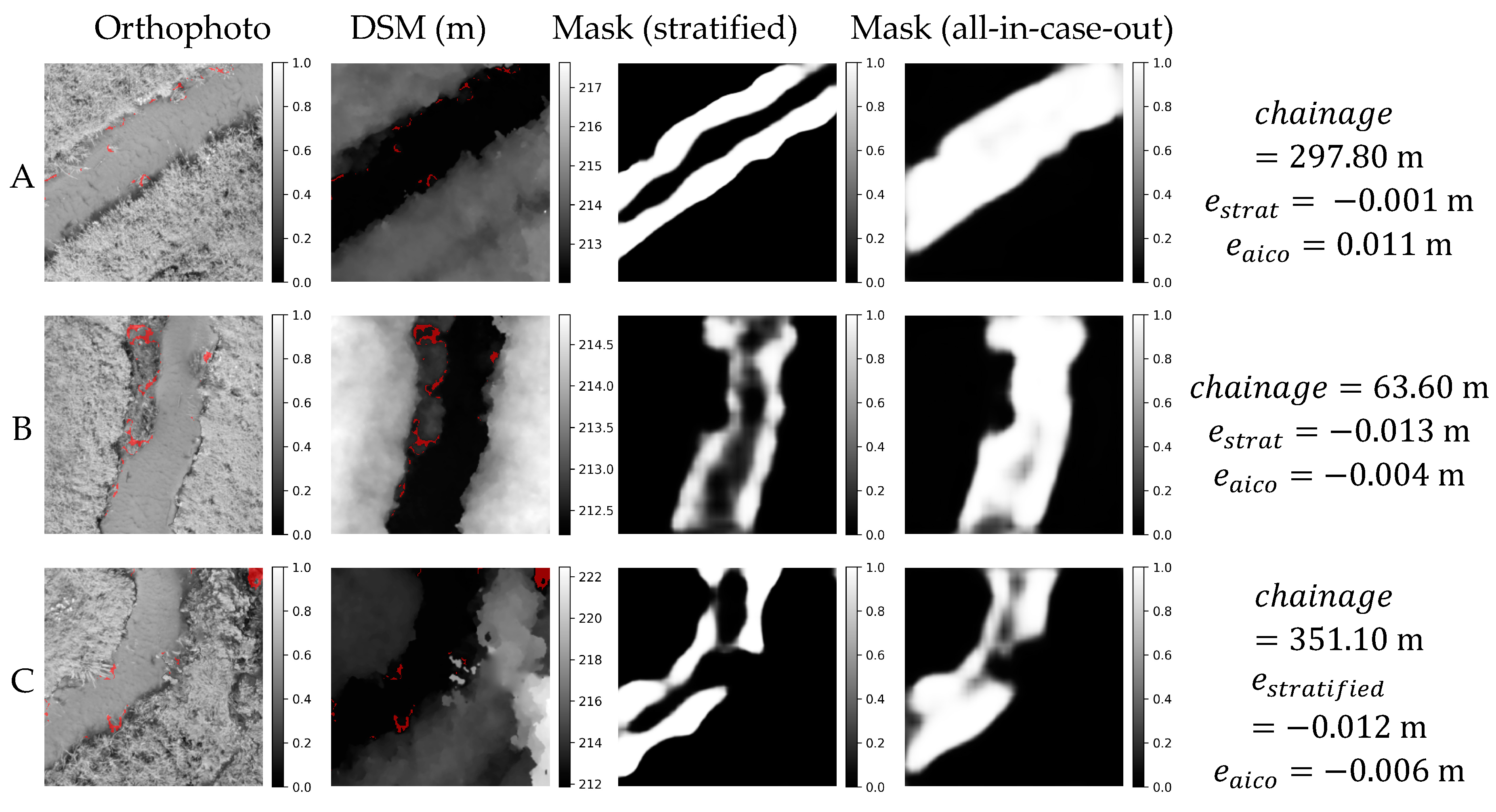
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from RYB20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 14.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from RYB20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 15.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from RYB21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 15.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three best performing samples from RYB21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
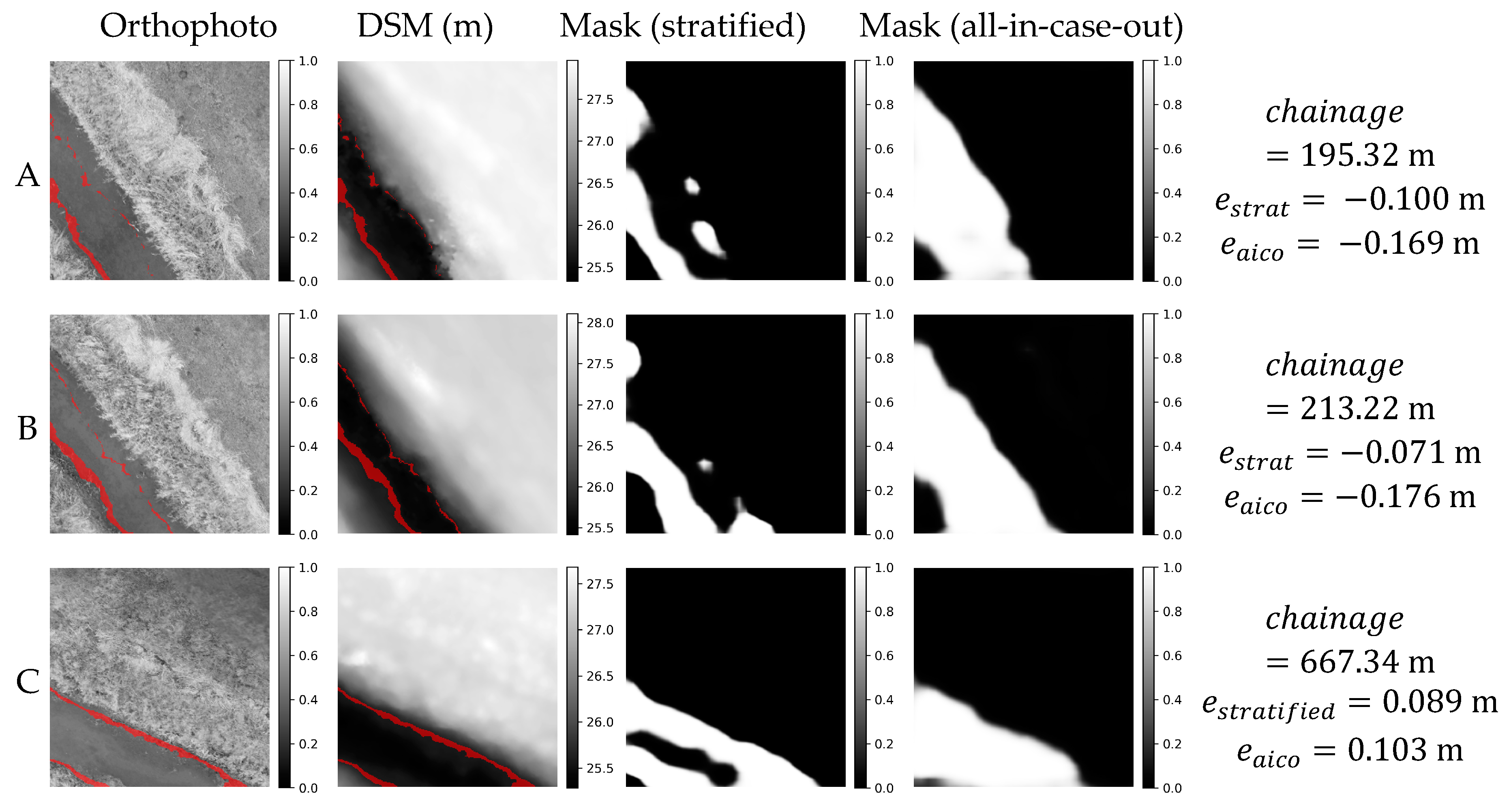
Figure 16.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from AMO18 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 16.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from AMO18 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
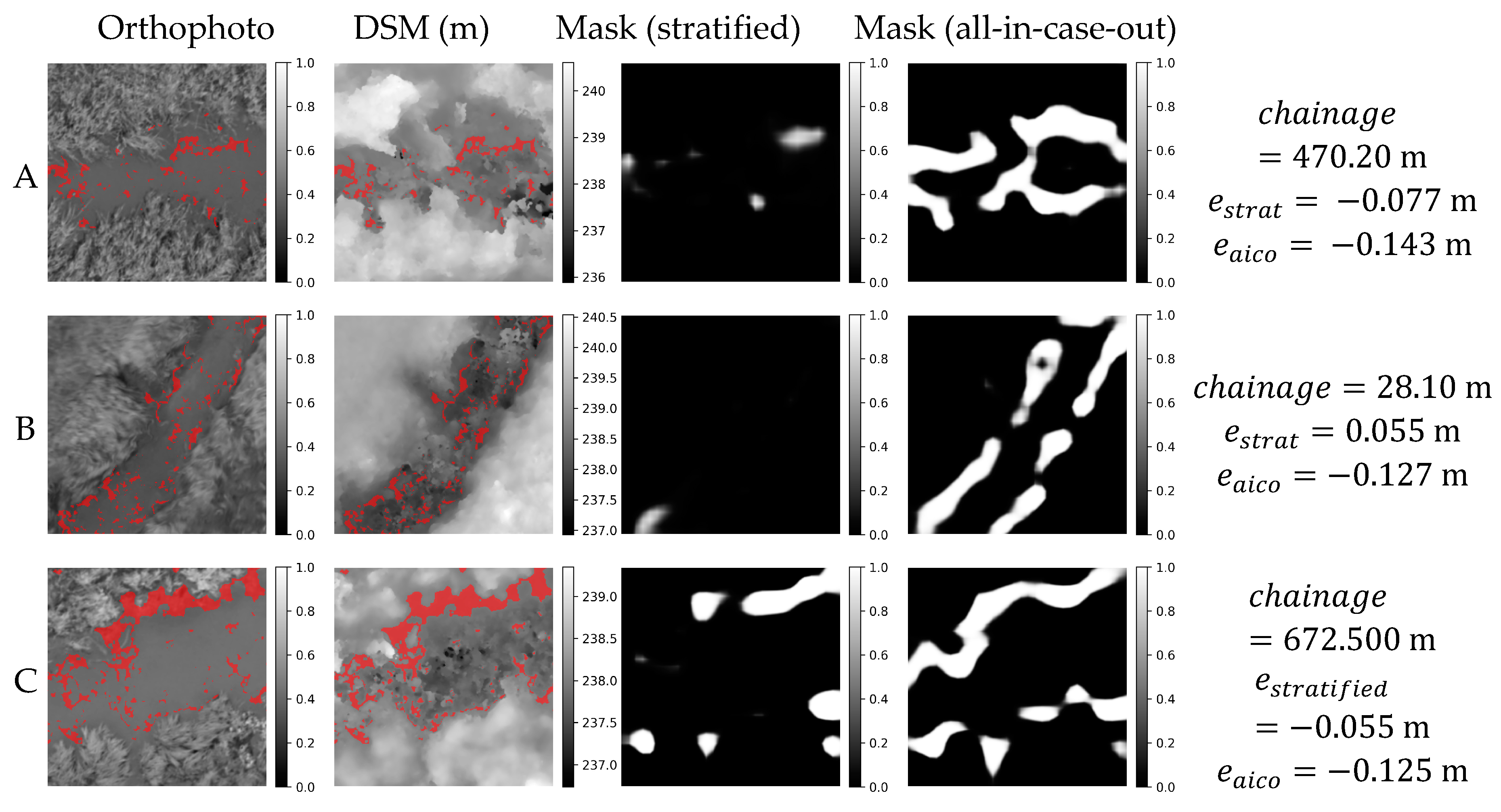
Figure 17.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from GRO20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 17.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from GRO20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
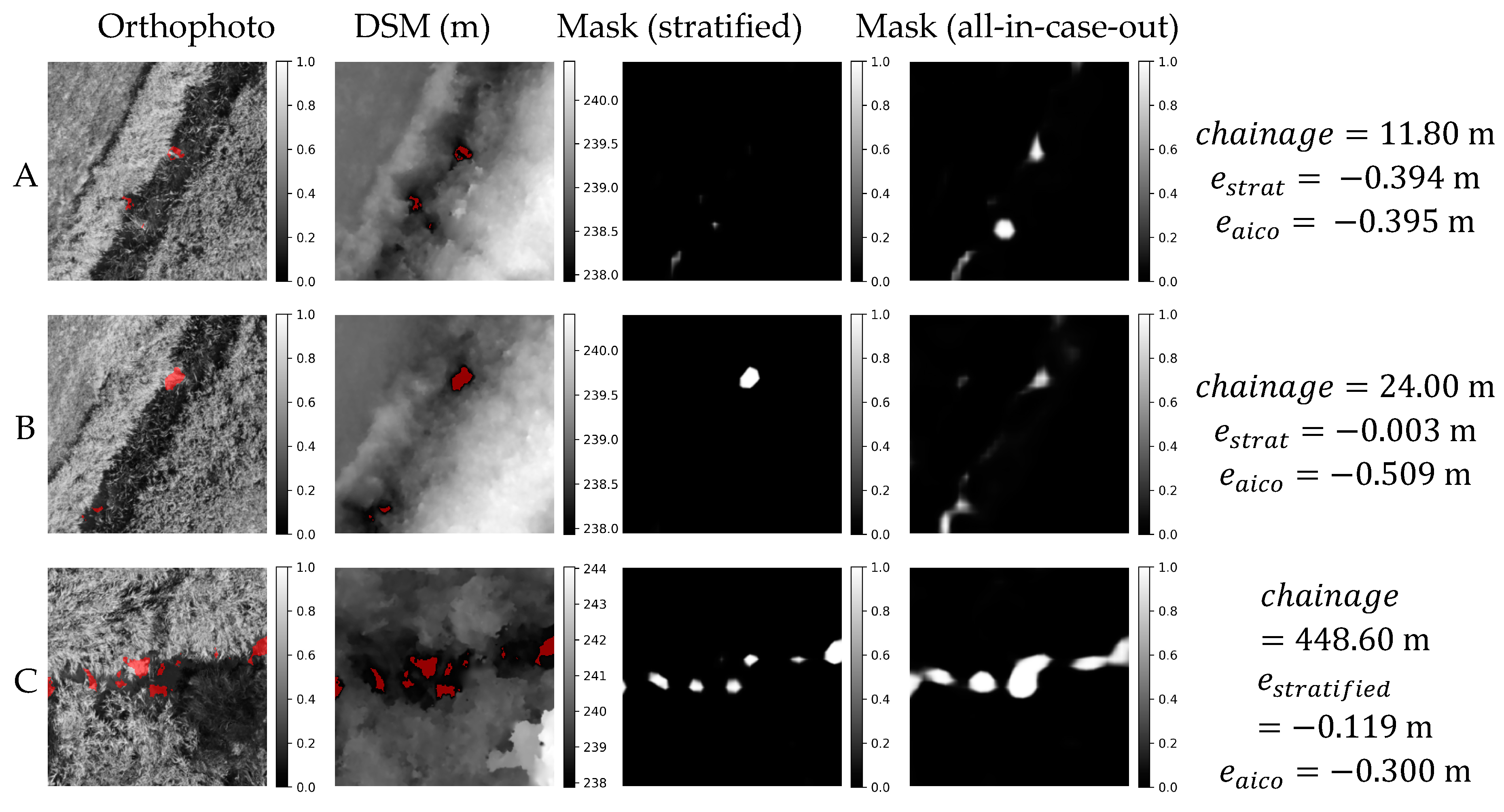
Figure 18.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from GRO21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 18.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from GRO21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
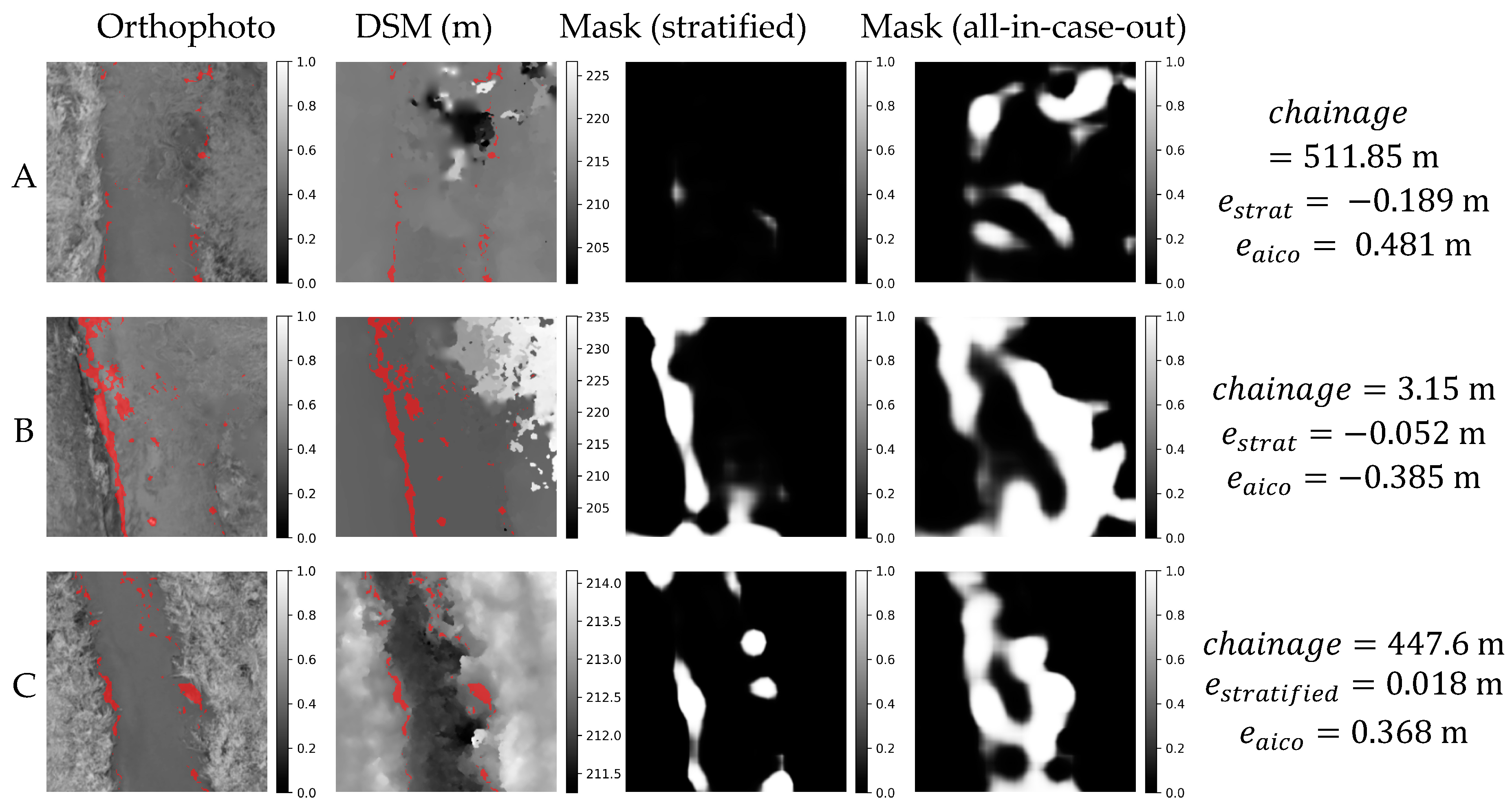
Figure 19.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from RYB20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 19.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from RYB20 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 20.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from RYB21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Figure 20.
Orthophoto, DSM and masks obtained in stratified and all-in-case-out cross validations for three worst performing samples from RYB21 case study. and correspond to residuals obtained using stratified and all-in-case-out cross-validation respectively. Areas where the DSM equals the actual WSE ±5 cm are marked with red color on orthophoto and DSM.
Table 1.
Number of WSE point measurements, standard error of estimate for ground truth WSE and number of extracted data set samples for each case study.
Table 1.
Number of WSE point measurements, standard error of estimate for ground truth WSE and number of extracted data set samples for each case study.
| Subset ID |
Number of WSE point measurements |
Standard Error of Estimate for ground-truth WSE (m) |
Number of extracted data set samples |
| GRO21 |
36 |
0.012 |
64 |
| RYB21 |
52 |
0.013 |
55 |
| GRO20 |
84 |
0.020 |
72 |
| RYB20 |
76 |
0.016 |
57 |
| AMO18 |
7235 |
0.020 |
74 |
Table 2.
Propositions of architecture, encoder, and batch size used in grid search.
Table 2.
Propositions of architecture, encoder, and batch size used in grid search.
| Approach |
Architectures |
Encoder |
Batch_size |
| Direct regression |
- |
ResNet18, ResNet50, VGG11, VGG13, VGG16, VGG19 |
1, 2, 4, 8, 16 |
| Mask averaging |
U-Net, MA-Net, PSP-Net |
VGG13, VGG16, VGG19 |
1, 2, 4, 8, 16 |
Table 3.
Best parameters configurations and achieved validation RMSEs averaged over all cross-validation folds.
Table 3.
Best parameters configurations and achieved validation RMSEs averaged over all cross-validation folds.
| |
direct |
mask |
| encoder |
VGG16 |
VGG19 |
| architecture |
- |
PSPNet |
| batch size |
1 |
4 |
| Mean RMSE |
0.170 |
0.077 |
Table 4.
RMSEs (m) achieved by proposed direct regression and mask averaging approaches and by straightforward sampling of DSM over centerline and near streambank. Both stratified and all-in-case-out cross-validation techniques results are given. Provided mean and sample standard deviation are calculated over all case studies.
Table 4.
RMSEs (m) achieved by proposed direct regression and mask averaging approaches and by straightforward sampling of DSM over centerline and near streambank. Both stratified and all-in-case-out cross-validation techniques results are given. Provided mean and sample standard deviation are calculated over all case studies.
| cross-validation |
stratified |
all-in-case-out |
- |
| method |
direct |
mask |
direct |
mask |
centerline |
wateredge |
| AMO18 |
0.099 |
0.035 |
0.170 |
0.059 |
0.219 |
0.308 |
| GRO20 |
0.076 |
0.021 |
0.124 |
0.072 |
0.185 |
0.228 |
| GRO21 |
0.107 |
0.058 |
0.243 |
0.117 |
0.27 |
0.277 |
| RYB20 |
0.095 |
0.048 |
0.176 |
0.156 |
0.449 |
0.259 |
| RYB21 |
0.274 |
0.063 |
0.337 |
0.142 |
0.282 |
0.404 |
| Mean |
0.130 |
0.045 |
0.210 |
0.109 |
0.281 |
0.295 |
| Sample St. Dev. |
0.081 |
0.017 |
0.083 |
0.043 |
0.102 |
0.067 |
Table 5.
MAEs (m) achieved by proposed direct regression and mask averaging approaches and by straightforward sampling of DSM over centerline and near streambank. Both stratified and all-in-case-out cross-validation techniques results are given. Provided mean and sample standard deviation are calculated over all case studies.
Table 5.
MAEs (m) achieved by proposed direct regression and mask averaging approaches and by straightforward sampling of DSM over centerline and near streambank. Both stratified and all-in-case-out cross-validation techniques results are given. Provided mean and sample standard deviation are calculated over all case studies.
| cross-validation |
stratified |
all-in-case-out |
- |
| method |
direct |
mask |
direct |
mask |
centerline |
wateredge |
| AMO18 |
0.078 |
0.026 |
0.121 |
0.045 |
0.179 |
0.176 |
| GRO20 |
0.059 |
0.015 |
0.091 |
0.06 |
0.139 |
0.104 |
| GRO21 |
0.082 |
0.028 |
0.195 |
0.072 |
0.117 |
0.138 |
| RYB20 |
0.074 |
0.037 |
0.127 |
0.111 |
0.373 |
0.157 |
| RYB21 |
0.169 |
0.045 |
0.154 |
0.096 |
0.249 |
0.276 |
| Mean |
0.092 |
0.030 |
0.138 |
0.077 |
0.211 |
0.17 |
| Sample St. Dev. |
0.044 |
0.011 |
0.039 |
0.027 |
0.103 |
0.065 |
Table 6.
MBEs (m) achieved by proposed direct regression and mask averaging approaches and by straightforward sampling of DSM over centerline and near streambank. Both stratified and all-in-case-out cross-validation techniques results are given. Provided mean and sample standard deviation are calculated over all case studies.
Table 6.
MBEs (m) achieved by proposed direct regression and mask averaging approaches and by straightforward sampling of DSM over centerline and near streambank. Both stratified and all-in-case-out cross-validation techniques results are given. Provided mean and sample standard deviation are calculated over all case studies.
| cross-validation |
stratified |
all-in-case-out |
- |
| method |
direct |
mask |
direct |
mask |
centerline |
wateredge |
| AMO18 |
0.008 |
-0.007 |
-0.084 |
-0.015 |
-0.149 |
0.161 |
| GRO20 |
-0.024 |
-0.007 |
-0.076 |
-0.057 |
-0.071 |
0.042 |
| GRO21 |
0.004 |
-0.018 |
0.069 |
-0.064 |
0.058 |
0.116 |
| RYB20 |
-0.013 |
0.002 |
0.042 |
0.059 |
-0.277 |
0.036 |
| RYB21 |
-0.076 |
0.006 |
-0.034 |
-0.004 |
-0.225 |
0.102 |
| Mean |
-0.020 |
-0.005 |
-0.017 |
-0.016 |
-0.133 |
0.091 |
| Sample St. Dev. |
0.034 |
0.009 |
0.069 |
0.049 |
0.132 |
0.053 |
Table 7.
RMSE, MAE, and MBE from this study (using mask averaging method with both stratified and all-in-case-out cross-validations) compared with Bandini et al. [
21] (using RADAR, photogrammetry, and LIDAR), arranged by RMSE. All data is from the same case study of stream Åmose Å in Denmark, collected on November 21, 2018.
Table 7.
RMSE, MAE, and MBE from this study (using mask averaging method with both stratified and all-in-case-out cross-validations) compared with Bandini et al. [
21] (using RADAR, photogrammetry, and LIDAR), arranged by RMSE. All data is from the same case study of stream Åmose Å in Denmark, collected on November 21, 2018.
| Method |
Source |
RMSE |
MAE |
MBE |
| UAV RADAR |
[21] |
0.030 |
0.033 |
0.033 |
| DL photogrammetry (stratified) |
This study |
0.035 |
0.026 |
-0.007 |
| DL photogrammetry (all-in-case-out) |
This study |
0.059 |
0.045 |
-0.015 |
| UAV photogrammetry DSM centerline |
[21] |
0.164 |
0.150 |
−0.151 |
| UAV photogrammetry point cloud |
[21] |
0.180 |
0.160 |
−0.160 |
| UAV LIDAR point cloud |
[21] |
0.222 |
0.159 |
0.033 |
| UAV LIDAR DSM centerline |
[21] |
0.358 |
0.238 |
0.076 |
| UAV photogrammetry DSM "water-edge" |
[21] |
0.450 |
0.385 |
0.385 |