1. Introduction
Soybean, as a kind of grain, and oil, feeding diversified economic crops [
1], is an important import and export material. With the upgrading of the consumption structure of the population, the demand for soybeans in China is also rising year by year. Nitrogen is one of the important factors affecting soybean yield [
2,
3], which cannot be absorbed directly by plants [
4]. The symbiosis with rhizobia can provide a stable nitrogen source [
5,
6], which improves the yield while reducing pollution and cost.
In recent years, rhizobial inoculation has become a hit in the process of growing soybeans [
7], which has been promoted in countries like Brazil, the United States, and Argentina to improve soybean crop yields, reduce chemical fertilizer inputs and the cost of soybean production with microbial nitrogen fixation properties [
8], making themselves the main soybean exports [
9]. In the 100-year history since the discovery of rhizobia, countries around the world have promoted the isolation of rhizobia and put studies on host compliance into practice. For example, the Encapsulation Method has been used to avoid harm to the environment and improve the efficiency of inoculation [
10]. Related studies have played a role in advancing the development, promotion, and utilization of rhizobacterial agents.
However, there is a great difference in the host compliance between the cultivars and rhizobia in different regions [
11], which leads to unstable nitrogen fixation efficiency and dim inoculation effect, which restricts the large-scale popularization of rhizobacterial agents. In order to improve the utilization of rhizobacterial agents, it is necessary to get strains with good host compliance, wide adaptation, and high nitrogen fixation efficiency according to local conditions. The combination of different strains of rhizobia with different advantages can be used. When inoculated with different rhizobia, the absorption of mineral elements (especially nitrogen and phosphorus) will be enhanced, significantly increasing the amount and dry weight of nodules, resulting in accelerated nutritional growth of soybean [
12] and crop yield [
13].
As the largest soybean growing area in China, Heilongjiang province has the largest grain production capacity [
14]and six cumulative temperature zones, whose active accumulative temperatures can differ by more than 800°C, where vary different cultivars. The complexity of the ecological environment reduces the efficiency of rhizobacterial agents. Collectively, this study provides theoretical resources for the production of rhizobacterial agents with compound traits such as good adaptability, high nitrogen fixation, and good resistance.
2. Materials and Methods
2.1. Isolation and Characterization of Rhizobia from Different Regions
Suinong 14 soybean, the cultivar in northeast China was sterilized by chlorine fumigation as the arrest host, which was sown in the soil isolated from Heilongjiang Province (43.26°~53.33°N, 121.11°~135.05°E), Shaanxi Province (31.95°~39.01°N, 105.29°~111.15°E), Yunnan Province (21.8°~29.15°N, 97.31°~106.11°E) and Guangdong Province (20.13°~25.31°N, 97.31°~106.11°E, 20.13° to 25.31° N and 109.39° to 117.19° E) and cultivated at 25 °C and a 16/8 h for 25-day incubation, then dug out when mature. The roots were rinsed with running water and then full and bulky rhizomes were picked with forceps and placed in Petri dishes. The fresh rhizobia were surface sterilized with sterile water and anhydrous ethanol under aseptic conditions and then cut using a sterilized scalpel. Its inner cut surface was affixed to the yeast juice agar (YMA) Congo red solid medium for delineation, then incubated in a constant temperature incubator at 28°C. According to the surface polysaccharide morphology of rhizobia, the milky white or white jam morphology was picked and continuously streaked and purified until monoclines appeared. The purified monoclonal colonies obtained were streaked on YMA Congo red solid medium and incubated at 28℃ for two days. Then the size and shape, edge, hardness, transparency, and other apparent properties of the single colonies were observed.
The isolated monoclonal strains were cultured in TY liquid medium on a shaker (28°C, 2000 r-min -1) until the OD
600 was 0.6 and then removed. Bacterial genome DNA extraction was performed with a bacterial genome DNA extraction kit (Tiangen Biochemical Technology Beijing Co., Ltd.) according to the manufacturer's instructions. A large fragment of the 16S rDNA gene of several bacterial fluids was amplified to 1500 bp with bacterial universal primers (F: AGAGTTTGATCCTGGCTCAG; R: AAGGAGGTGATCCAGCC) [
15], and then identified by 16S
rDNA. The identification results were compared with the reported gene sequences on NCBI by BLAST, and then MAGE was applied to construct a phylogenetic tree to determine their relative positions in the rhizobial system.
2.2. Re-inoculation with Arrest Host
The identified rhizobia were used to formulate the bacterial agent with an OD
600 of about 0.2. The double-layered mantle was assembled, the upper layer filled with vermiculite and the lower layer filled with nitrogen-deficient nutrient solution [
16], connected in the middle with skimmed cotton, and covered with a lid for autoclaving. The arrest host Suinong 14 soybean seeds were sterilized using the same chlorine fumigation method as before, then planted in double-layered pots under aseptic conditions and placed in the plant room at 25°C for constant incubation. When soybean seedlings grew close to the lid, the lid was opened and sterilized white stones were spread flatly over the vermiculite surface of the upper layer of the double-layer mantle. At 25 °C and a 16/8 h cultivation, the rhizobial agent inoculation was performed when the opposite euphylla of seedlings were fully expanded. Four rhizobial agents and a blank control group with 10 mmol/L MgSO
4 were inoculated separately, and three Suinong 14 soybean materials were inoculated with 2 mL of rhizobial agent for each rhizobium. After 28 days, the nodulation phenotypes were identified, including traits such as the amount of nodules, nodule size, nodule cut color, and nodule dry weight [
17].
2.3. Identification of Rhizobial Resistance
Several stress conditions of antibiotics, salinity, acid and alkali were selected to identify rhizobial resistance. The solid medium with three concentrations of different antibiotics, 20 mmol/L, 50 mmol/L, and 100 mmol/L, was configured for the cultivation of rhizobium. After 5-day cultivation, the growth status of the colonies on the medium with different concentrations of different antibiotics was observed.
TY solid medium with different concentrations of NaCl of 0 mmol/L, 50 mmol/L, 100 mmol/L and 150 mmol/L was configured. The monoclonal strains were picked and streaked on the TY solid medium with different salt contents, then cultivated at 28℃ for 5 days to observe the growth of colonies on the medium.
The pH of the medium was adjusted with HCl and NaOH to configure TY solid medium with pH 5-10. The monoclonal strains were picked and streaked on different pH values of TY solid medium. After the cultivation at 28°C for 5 days, the growth of colonies on the medium was observed.
2.4. Identification of Rhizobium Host Compliance with Cultivars
The seeds were pretreated with chlorine fumigation. Four rhizobial agents with OD600 of 0.2 and a control group of fast-growing rhizobium Sinorhizobium fredii HH103, were inoculated with eight cultivars selected from the six cumulative temperature zones in Heilongjiang Province, a land cultivar, a wild cultivar and the arrest host Suinong 14, respectively. To minimize the error, 10 replications were carried out for each variety, and each strain was inoculated with 2 mL of bacterial solution. At 25 °C and 16/8 h for 30-day incubation, the amount of nodules, nodule size, nitrogen-fixing enzyme activity, and dry weight of nodules were determined.
3. Results
3.1. Isolation and Identification of Rhizobia from Different Regions
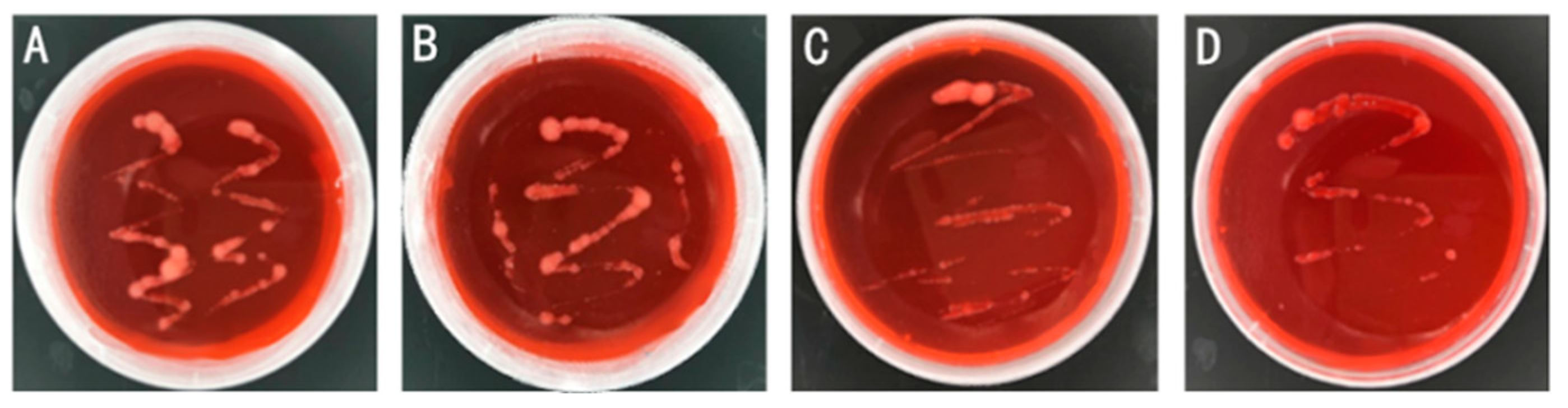
Obtained rhizobia from four regions, Heilongjiang Province, Shaanxi Province, Yunnan Province, and Guangdong Province were named according to the geographic characteristics of high latitude in Heilongjiang Province and Northeast Agricultural University: High Latitude Northeast Agricultural University 1 (HLNEAU1), HLNEAU2, HLNEAU3, HLNEAU4. Monoclonal strains with morphological characteristics of rhizobium were obtained and observed on YMA (Congo red) medium plates (
Figure 1).
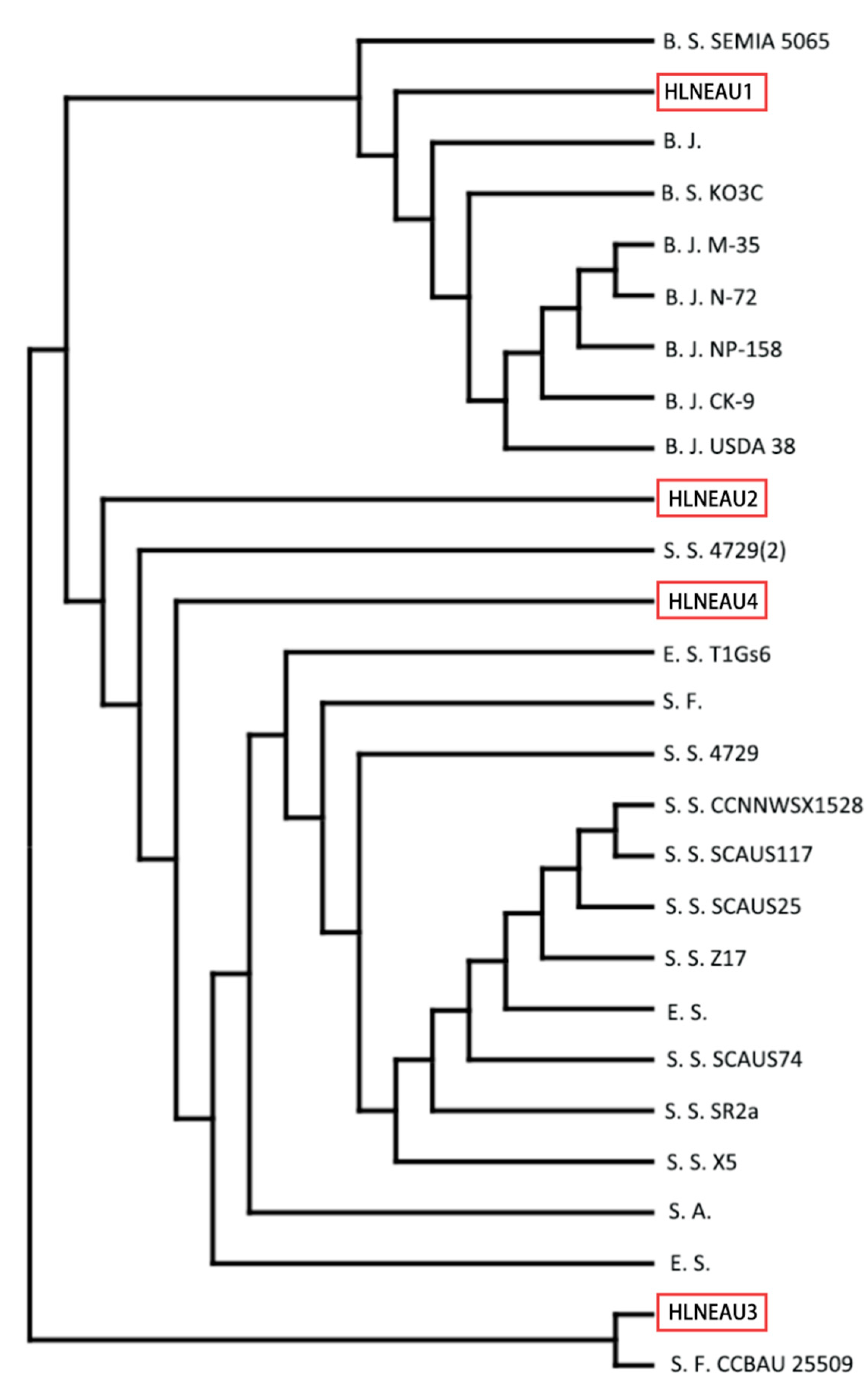
Based on the results of PCR amplification and sequencing of strain 16S rDNA (
Figure 2) and the results of plotting the phylogenetic tree with known sequences (
Figure 3), it was shown that the 16S rDNA of the conserved sequence of HLNEAU1 had 99% similarity to the slow-growing strain Bradyrhizobium japonicum, the 16S rDNA of HLNEAU2 had 96% similarity to the 16S rDNA of Sinorhizobium fredii and Sinorhizobium sp, HLNEAU3 conserved sequence 16S rDNA had 98% similarity to Sinorhizobium fredii, and HLNEAU4 had 97% similarity to Sinorhizobium fredii and Sinorhizobium sp.. The isolated strain HLNEAU1 in this study was a slow-growing rhizobium and the other three were fast-growing rhizobia.
3.2. Phenotypic Identification of the Re-Inoculation with SUINONG14
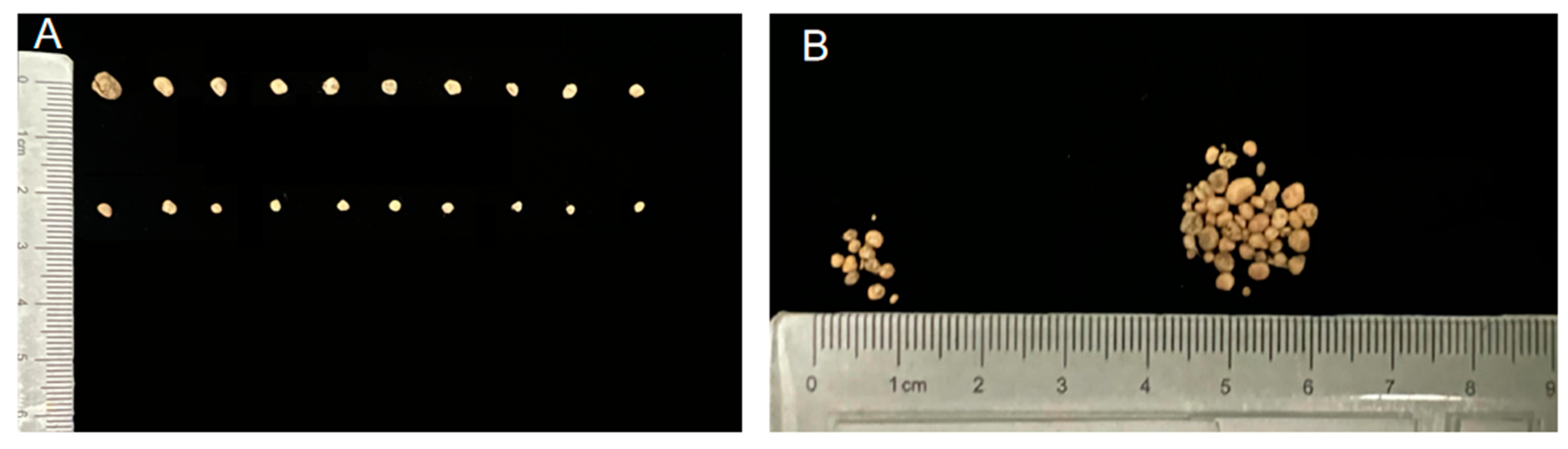
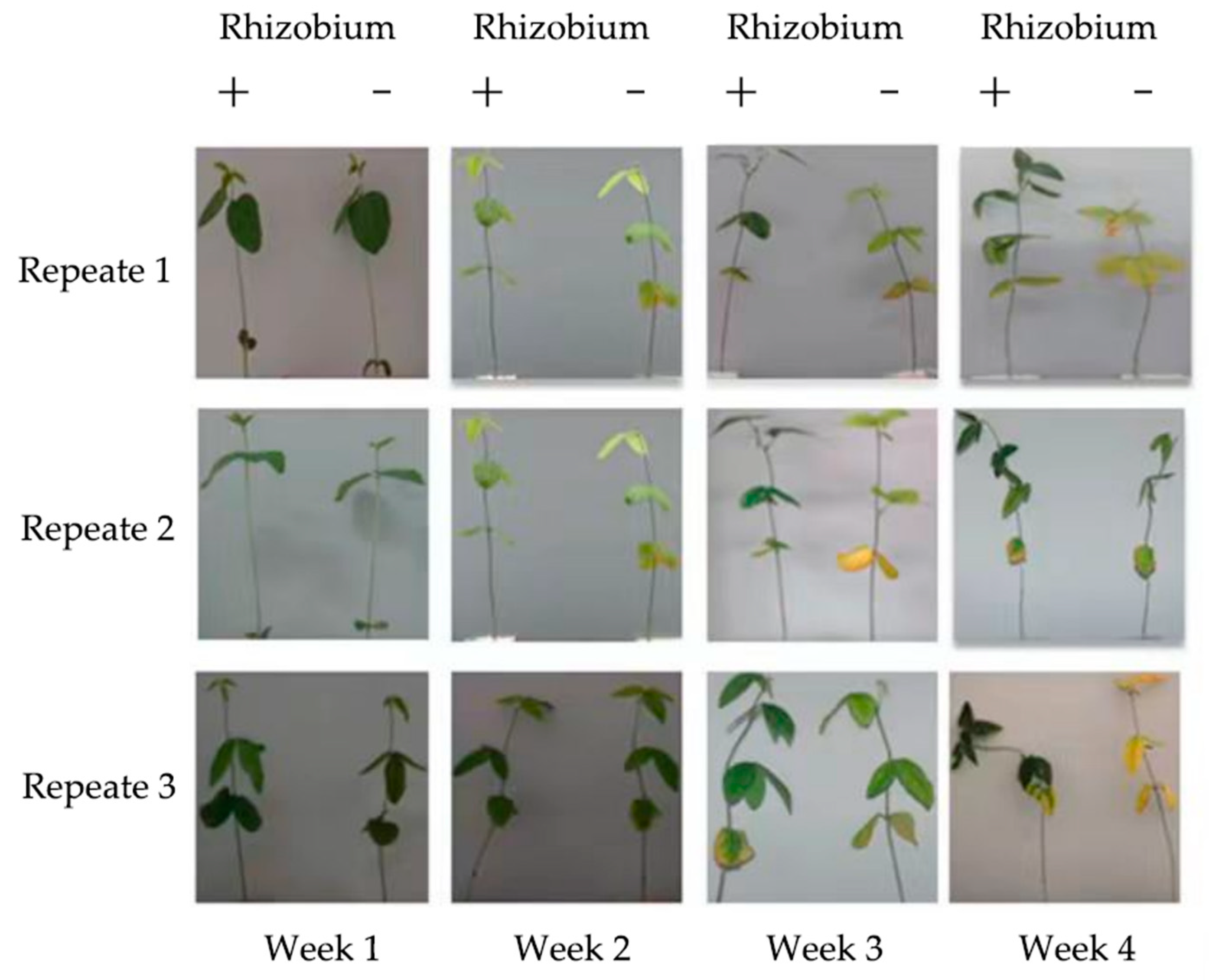
A nodulation test was conducted with Suinong 14 to further validate the nodulation ability of the isolated rhizobia. These strains and a blank control group were each inoculated with the arrest host Suinong 14, cultured with nitrogen-deficient nutrient solution for 28 days to get the nodule amount, size (
Figure 4) and plant height (
Figure 5). Then the size, amount, cut color, and dry weight of the obtained nodules from each rhizobial solution-inoculated plant were systematically counted (
Table 1). It was found that HLNEAU1, HLNEAU2, HLNEAU3 and HLNEAU4 were able to symbiotically colonize all soybeans, which proved that these rhizobial symbioses had a high degree of broad-spectrum, and the nodules were pink in color on the cut surface, indicating that they were effective nodules with normal nitrogen fixation capacity. In the aspect of the number of nodules, HLNEAU1 and HLNEAU4 had significantly more nodules than HLNEAU2 and HLNEAU3. While HLNEAU2 and HLNEAU3 had larger nodules. Due to the nitrogen-deficient nutrient solution, Suinong 14 can only absorb nitrogen source through symbiotic nitrogen fixation with rhizobia.
3.3. Identification of Rhizobial Antibresistance
3.3.1. Antibiotic Resistance Identification of Rhizobia
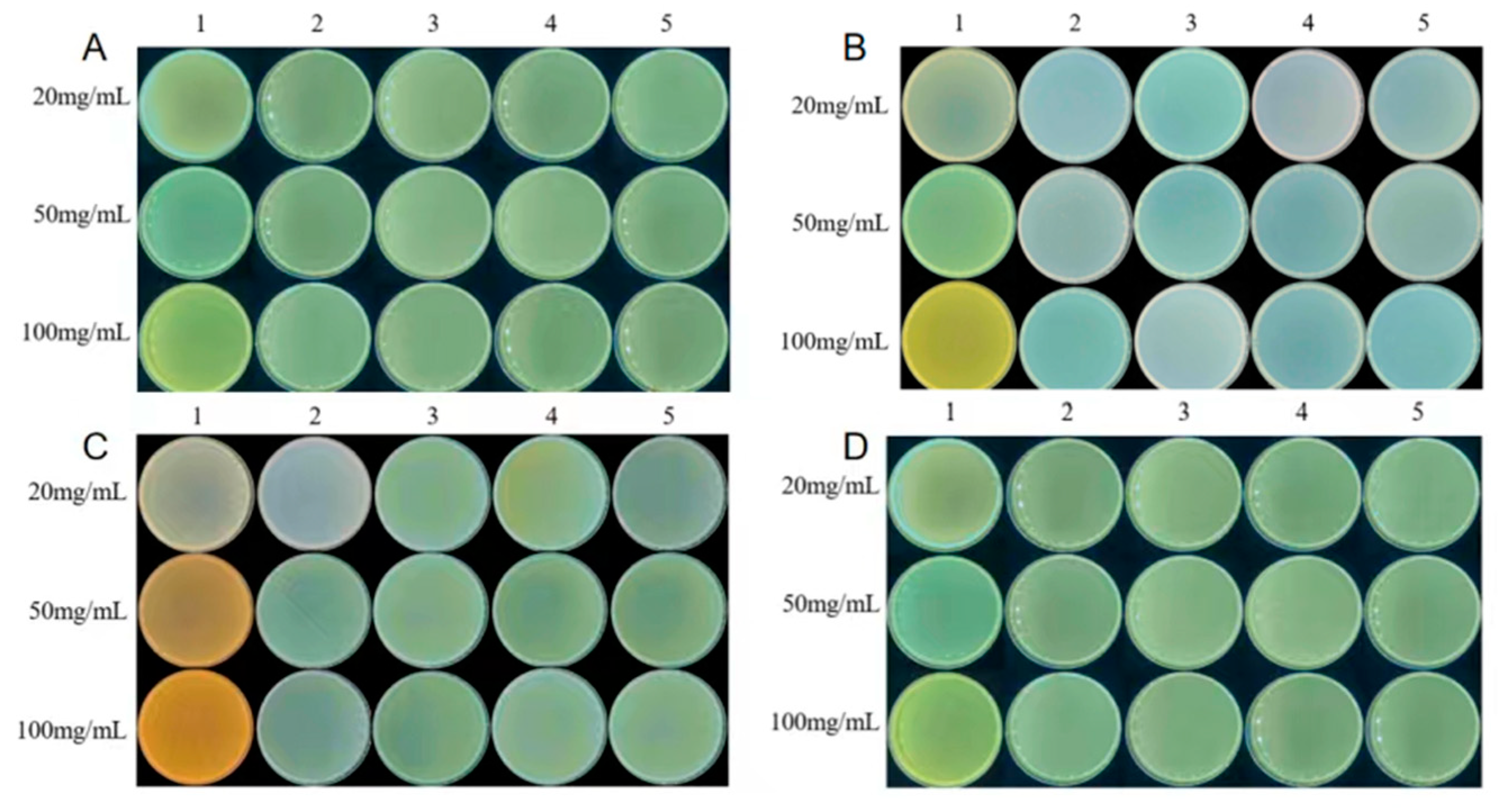
It can be found in the identification of antibiotic resistance in rhizobia (
Figure 6) that the four rhizobia were non-resistant to the common antibiotics Gentamicin (Gent), Kanamycin (Km), Tetracycline (Tet), Rifampicin (Rif), and Spectinomycin (Spe).
3.3.2. Salt-Resistance, Alkali-Resistance and Acid-Resistance Identification
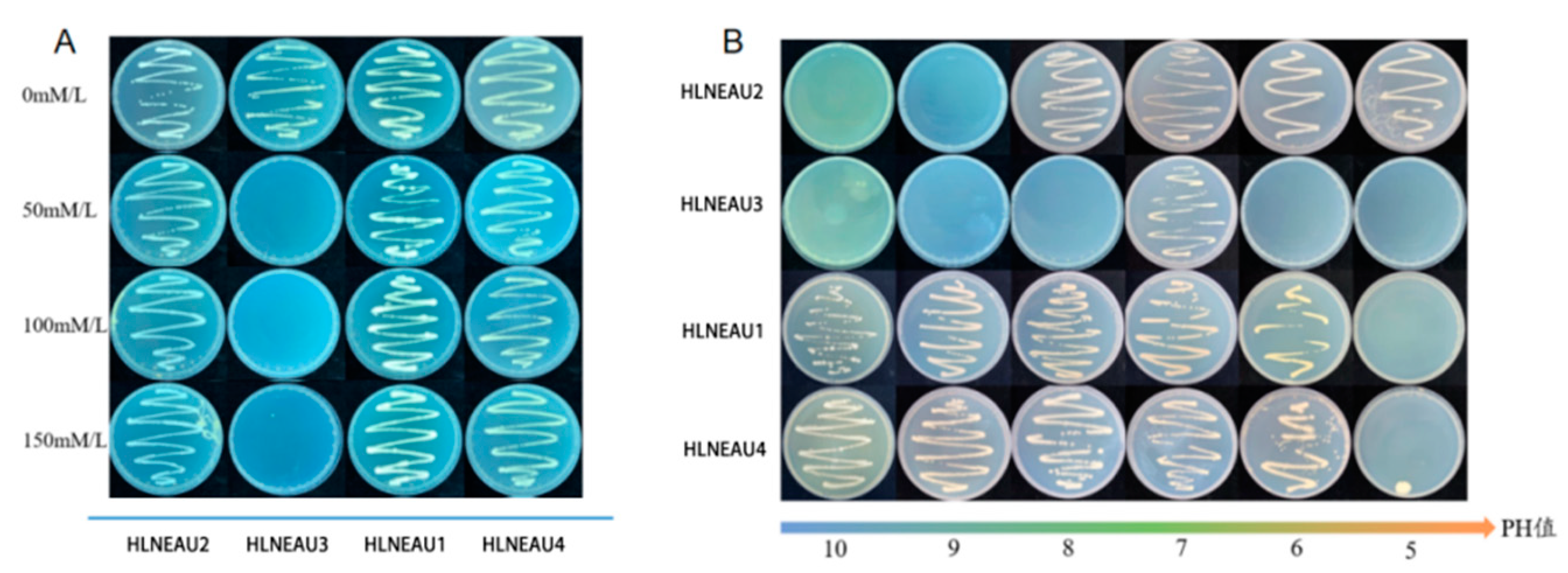
It was shown that HLNEAU4 could only grow on the medium with 0 mmol/L NaCl concentration while HLNEAU1, HLNEAU2, and HLNEAU3 could grow normally on the medium with 150 mmol/L NaCl concentration. HLNEAU4 could only grow on a neutral medium at pH 7, could not grow on any other pH medium, neither acid nor alkali-resistant and only surviving normally in neutral soil at pH 7. HLNEAU1 could grow on medium with pH 5-8, indicating that this strain was extremely resistant to acid and weakly resistant to alkali, and could grow normally in acidic soil. HLNEAU2 and HLNEAU3 could grow on medium with pH 6~10. These two rhizobia had strong alkali resistance, and weak acid resistance and could grow normally in alkaline soil, showing good salinity resistance (
Figure 7).
3.4. Identification of Rhizobium Host Compliance with Cultivars
3.4.1. Identification of Nodulation Capacity
Isolated rhizobia were inoculated with eight cultivars selected from the six cumulative temperature zones in Heilongjiang Province, a land cultivar, a wild cultivar and the arrest host Suinong 14, respectively, to determine the host compliance, with HH103 as a control group and examined in aspects of nodule amounts, nodule size (greater than 2 mm and less than 2 mm), nodule dry weight and so on after 28-day cultivation.
HLNEAU4 had the most nodules of 38.5-121.3. HLNEAU2 had the least nodules of 4.7-16.2. The nodule amount of HH103 was between 8.6 and 58.0 while which of HLNEAU1 between 25.7 and 56.3 and which of HLNEAU3 between 5.7 and 33.2 (
Table 2).
HLNEAU4 had the heaviest nodules of 2.3-9.3mg, while HLNEAU2 had the lightest nodules of 1.1-6.1mg. The nodule dry weight of HH103 was between 0.8-11.4mg, while which of HLNEAU1 was between 1.4 and 10.0mg, and which of HLNEAU3 between 0.9 and 4.9 (
Table 3).
Except the re-inoculation with Heihe 33, HLNEAU1 and HLNEAU4 had more nodules than the control group HH103. Combined with dry weight and size, it was found that Dongnongdou 252 and ZYD00006 were phenotypically superior to the control group and other rhizobia in all aspects when symbiotic with HLNEAU4, while Heinong 56, Heinong 35, and Suinong 14 were superior to the control group and other rhizobia in dry weight when symbiotic with HLNEAU1, even though they did not have the highest amount of nodules
1 (
Table 4).
3.4.2. Identification of Nitrogen-Fixation Capacity
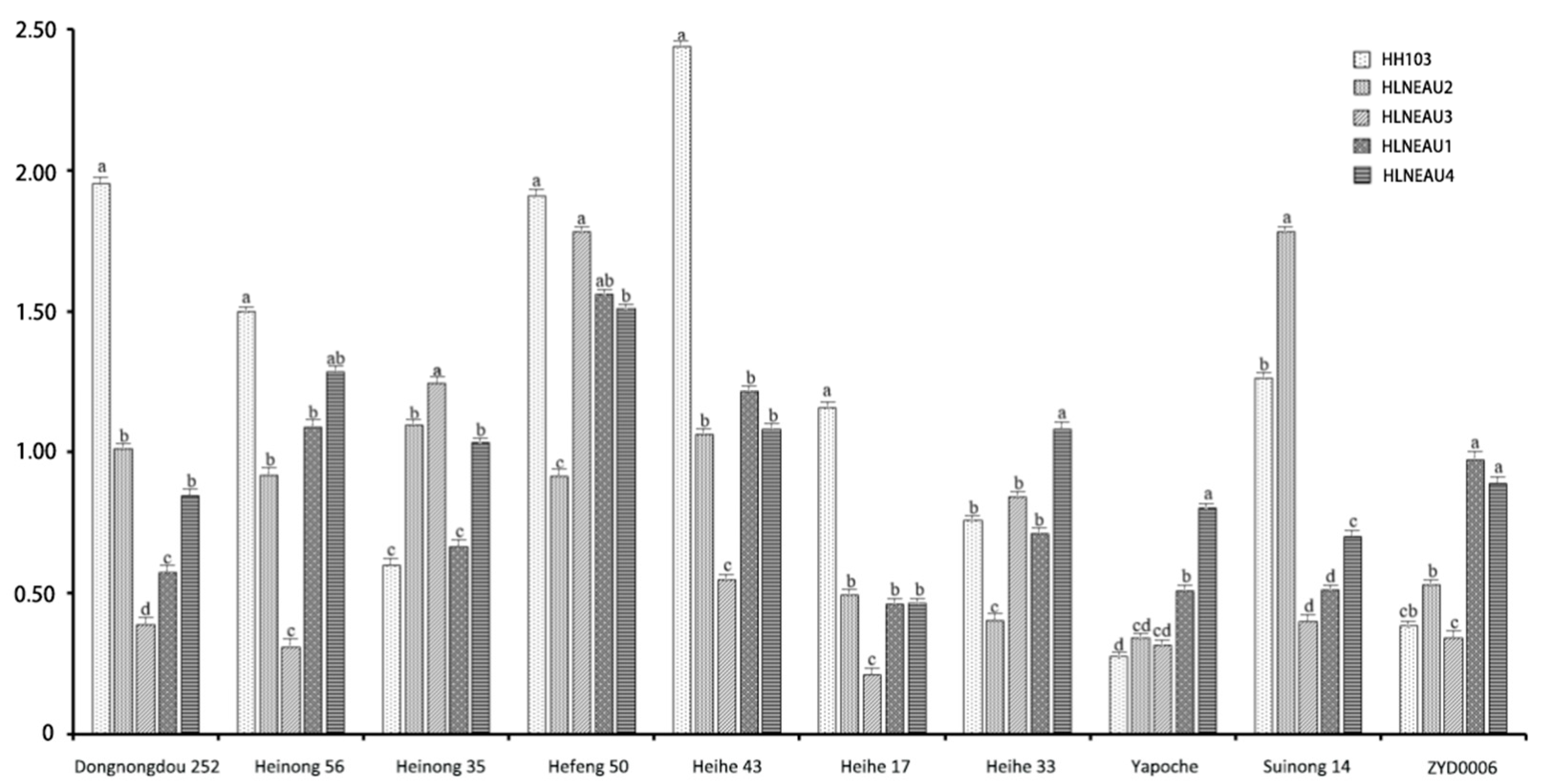
It is an important indicator to identify the nitrogenase activity by Acetylene Reduction Assay (ARA) for detecting the nitrogen-fixation and host compliance capacity of rhizobia (
Figure 8). All rhizobial nodules were effective to carry out normal nitrogen-fixation activities. Where there existed differences when the same culticar was inoculated with different rhizobia. Compared with the control group HH103, it was found that the nitrogenase activity of HLNEAU2 in symbiosis with Suinong 14 was significantly (p < 0.05) higher than that of other control group and other three rhizobia, presenting excellent symbiotic nitrogen-fixation effect. The nitrogenase activity of Heihe 33 were higher than those of other rhizobia in symbiosis with HLNEAU4, while which of HLNEAU1 and HLNEAU4 were significantly (p < 0.05) higher than those of other rhizobia when inoculated land landrace Yapoche and wild soybean ZYD00006, suggesting that these two strains may have better symbiotic host compliance with the original cultivar resources.
4. Discussion
4.1. Application Analysis of Rhizobia
As the greenest and most environmentally friendly way of nitrogen supply for plants [
18], rhizobium nitrogen fixation can not only provide stable nitrogen sources but also fertilize the soil [
19], which is the most economical and effective way for legume crops to obtain nitrogen sources [
20,
21]. Whether rhizobium inoculation can exert its maximum benefits involves a variety of external factors, such as the host compliance between cultivars and rhizobia, the adaptation of rhizobia to local soil pH, salt content, and heavy metal content, etc., the competitiveness of applied rhizobia against indigenous rhizobia and the effect of compound rhizobium [
22]. Heilongjiang is the main producer of soybeans in China, in which the climate of each accumulative temperate zone is quite different. The accumulated temperature of the first cumulative temperate zone can reach more than 2700℃, while the accumulated temperature of the sixth cumulative temperate zone is even below 1900℃, which leads to multiple cultivars, increasing the difficulty of the widespread application of rhizobia.
In this study, the comprehensive factors of isolated rhizobia from different areas such as acid-base adaptation, salt resistance, adaptation to cultivars in different accumulated temperate zones in Heilongjiang, and nitrogenase activity were determined to screen out the excellent rhizobium strains with high host compliance. Four strains of rhizobia were isolated from soils of different regions in China, three fast-growing strains and one slow-growing strain. The results of host compliance experiments with 10 Heilongjiang cultivars showed that both fast-growing and slow-growing rhizobia could nodulate with Heilongjiang cultivars, carrying out nitrogen fixation normally, which was consistent with the finding of Ji Zhaojun et al., that both Chinese rhizobia and slow rhizobia could achieve symbiosis and nitrogen fixation with Luhuang No. 1 soybean. There were great differences in nodulation number, nodule dry weight, and nitrogenase activity among different cultivars inoculated with the same kind of rhizobium, which was the same as the results of Wu Hui et al. [
23]. Nodulation ability varied significantly between different fast-growing rhizobia and cultivars. For example, HLNEAU3 and HLNEAU4 were both fast-growing rhizobia, among 70% of which, their nitrogenase activity varied by more than 50% as obvious differences, in accord with the research results of Ma Zhongyu et al. [
24].
It has been shown in numerous studies that the symbiotic host compliance of compound rhizobium with soybean is complex with multiple regulators acting together, which are critical for soybean, due to its reliance on biological processes of organic or eco-friendly agricultural practices instead of agrochemical inputs. Tripartite symbioses among plants, bacteria, and fungi occur naturally in soil ecosystems, which is facilitated by a variety of factors such as antibiotics, signaling molecules, synergistic metabolism and physical interactions. In practice, different rhizobia can be selected for compound application according to different soybean varieties, which may greatly improve the utilization efficiency of rhizobia. And this study may provide effective information for the use of rhizobia in these excellent cultivars.
4.2. Application Analysis of Rhizobia in Different Areas
As the demand for soybeans has been rising, so does the demand for planting soybeans. However, corn and rice have occupied most of the land resources, making it meaningful to develop some saline-alkali or acidic land for planting. The area of saline-alkali lands in China reaches about 99.13 million hectares, accounting for about 5% of the available land area [
25], whose utilization will greatly improve the supply of agricultural products. For example, about 23% of the land in Inner Mongolia is saline land, covering a comprehensive area of up to 7.63 million hectares. In recent years, many scientists have conducted a lot of researches on the negative effects of salt stress on crops. As a moderately salt-resistant crop, salt-resistant cultivars have also appeared in years of research and cultivation. Therefore, it is the screening of corresponding salt-resistant rhizobia that can also promote the cultivation of soybeans in saline-alkali lands.
In this study, three strains of rhizobia from different regions resistant to 150mM/L were found through the growth status on the media with different salt concentrations (0mM/L, 50mM/L, 100mM/L, 150mM/L), which can be used in salt-resistant soil of most salt-resistant cultivars. Investigating the salt resistance of 58 rhizobium strains, Chen Wenxin et al. found that some rhizobia could grow normally on 1% NaCl medium, but could not grow on 1.5% NaCl medium, and only a few rhizobia could grow on 4.5% NaCl medium. Qin Jie et al. found a bradyrhizobium strain from Jinda 53 soybean, which can resist 5% NaCl medium and has good salt resistance [
26]. Three strains of rhizobia found in this experiment could be normal in 150mM/L, having reached the application level in salt resistance with soybean with the undetermined limit for developing.
4.3. Effects of Rhizobia on the Environment
There are many kinds of rhizobia, among which S. F. and B. J. are two different groups. In early studies, it was found that S. F. was a class of fast-growing and acid-producing soybean rhizobium, while B. J. was a slow-growing and alkali-producing soybean rhizobium. Because of this physiological characteristic, the pH of the environment has a great impact on the survival of rhizobia. For example, the black soil from the cultivated land in northern China has more humus, dark color, loose and mostly acidic soil, where exists B. J.. However, The central and western regions of China are mostly alkaline soil, with the color of yellow or red, so most of the rhizobia are fast-growing rhizobia. For instance, the rhizobia isolated from soybean in some areas of Heilongjiang Province by Wang Jingyu et al. were found to be slow-growing soybean rhizobia. While Li Qinqin et al. isolated soybean rhizobia in Hebei and found that there were both fast-growing and slow-growing rhizobia, with the fast-growing rhizobium of S. F. as the dominant group [
27]. This is also consistent with the results of rhizobia isolated in this experiment, where slow-growing rhizobia were isolated in soil from Heilongjiang Province, and fast-growing rhizobia were isolated in Xi'an, Shaanxi Province, Guangzhou, Guangdong Province, and Yunnan Province. Furthermore, In the determination of pH resistance, Heilongjiang strain could grow on the medium with pH 5-8, showing acid resistance and slight alkali resistance, while The strains from Yunnan and Shaanxi Province could grow on media with pH 6 to 10, showing alkali and slightly acid resistant, which is consistent with the results of Yang Jiangke et al., showing the strong resistance of S. F. under alkaline conditions and that of B. J. under acidic conditions. However, perhaps due to the different inter-individual differences of rhizobia, the strains in Guangdong differ from this conclusion, neither resistant to acid nor alkali. For the physiological characteristics of acid-producing B. J. and alkali-producing S. F., rhizobia can also be applied in the bio-modification of acidified soils and saline-alkaline soils by planting legume plants with rhizobium characteristics of different rhizobia, which adjusts the soil pH, and leguminous crops belong to the land-raising crops can cultivate the land power to achieve the purpose of improving the soil.
5. Conclusions
Four rhizobia strains, HLNEAU1, HLNEAU2, HLNEAU3, and HLNEAU4, were not antibiotic resistant, and HLNEAU1 was more advantageous in acid resistance. While HLNEAU2 and HLNEAU3 were more resistant to salinity. In terms of host compliance, HLNEAU1 and HLNEAU4 were found to be better matches on the more primitive soybean cultivars, and HLNEAU2 has a better host compliance with the arrest host Suinong 14. In response to the saline soil characteristics of Northeast China, Rhizobium HLNEAU2 can better help the growth of common cultivars in Heilongjiang Province when planting soybean varieties, which presents a valuable theoretical basis and future practicable space for growing cultivars.
Author Contributions
Conceptualization, D.X. and Q.C.; methodology, T.Y., H.L., X.W. and G.Z.; software, W.T.; validation, Z.Z., X.W. and Y.G.; formal analysis, Y.S.; investigation, T.Y., J.L. and W.T.; resources, X.W.; data curation, X.Z.; writing—original draft preparation, T.Y. and H.L.; writing—review and editing, D.X.; visualization, Y.S. and T.Y.; supervision, D.X. and Q.C.; project administration, T.Y.; funding acquisition, D.X. and Q.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant numbers: 32070274, 31771882, 32072014 and U20A2027).
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Dawei Xin for the technical help.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, Z.; Tian, Z. Genomics progress will facilitate molecular breeding in soybean. Sci. China Life Sci. 2015, 58, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, K.; Yin, T.; Zhao, Y.; Liu, W.; Shen, Y.; Ding, Y.; Tang, S. Nitrogen Fertilizer Regulated Grain Storage Protein Synthesis and Reduced Chalkiness of Rice Under Actual Field Warming. Front. Plant Sci. 2021, 12, 715436. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef] [PubMed]
- Cesco, S.; Neumann, G.; Tomasi, N.; Pinton, R.; Weisskopf, L. Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 2010, 329, 1–25. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, Q.; Li, X.; Ai, W.; Liu, D.; Qi, W.; Zhang, M.; Yang, C.; Liao, H. Characterization of Genetic Basis on Synergistic Interactions between Root Architecture and Biological Nitrogen Fixation in Soybean. Front. Plant Sci. 2017, 8, 1466. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M.; Tegeder, M. Increasing Nitrogen Fixation and Seed Development in Soybean Requires Complex Adjustments of Nodule Nitrogen Metabolism and Partitioning Processes. Curr. Biol. 2016, 26, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Guan, D.; Li, L.; Jiang, X.; Ma, M.; Li, J. Screening and Characterization of Superior Nitrogen-Tolerance Soybean Rhizobia. Soybean Science 2014, 33, 861–864. [Google Scholar]
- Ni, H.; Tian, S.; Zhang, G.; Huo, J.; Tian, H.; Peng, Y.; Yu, K.; Chen, Q.; Wang, J.; Xin, D.; et al. QTL Mapping and Functional Identification of Candidate Genes Regulated by Sinorhizobium fredii HH103 and Associated with Nodulation Traits in Soybean. AGRONOMY-BASEL 2023, 13. [Google Scholar] [CrossRef]
- Atieno, M.; Lesueur, D. Opportunities for improved legume inoculants: enhanced stress tolerance of rhizobia and benefits to agroecosystems. Symbiosis 2019, 77, 191–205. [Google Scholar] [CrossRef]
- D, K.; B, L.; A, P.; S, J.; M, V.; S, K. The Storage Efficiency of Immobilized Bradyrhizobium japonicum Strain Using Encapsulation Method[J]. Contemporary Agriculture 2019, 68. [Google Scholar]
- Innosensia, N.L.P.C.; Suputra, I.P.W.; Wirya, G.N.A.S.; Narisawa, K. First Report of Tripartite Symbiosis Potential among Soybean, Bradyrhizobium japonicum, and Dark Septate Endophytes. Agronomy 2023, 13, 1788. [Google Scholar] [CrossRef]
- Ni, H.; Peng, Y.; Wang, J.; Wang, J.; Yuan, Y.; Fu, T.; Zhu, Z.; Zhang, J.; Pan, X.; Cui, Z.; et al. Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT. AGRONOMY-BASEL 2022, 12. [Google Scholar] [CrossRef]
- Nadarajah, K.; Abdul Rahman, N.S.N. The Microbial Connection to Sustainable Agriculture. Plants 2023, 12, 2307. [Google Scholar] [CrossRef]
- Sun, J.; Wei, D.; Ma, X.; Liu, D.; Guo, W.; Liu, X.; Lu, W. Establishing Fertilization Recommendation Index of Soybean in Black Soil Region of Heilongjiang Province. Soybean Science 2013, 32, 512–516. [Google Scholar]
- Feng Mei, Wen Kong, Yang Li, Tingting Ma, Ting Pi, Donglan He, Guojun Cheng, Tao Liu, Xiaohua Li. Isolation and gene characterization of a nicotine-degradation strain Agrobacterium tumefacience SCUEC1[J]. Acta Microbiologica Sinica, 2017, 57, 701–709.
- Kiers, E. T.; Rousseau, R. A.; West, S. A.; Denison, R. F. Host sanctions and the legume-rhizobium mutualism. NATURE 2003, 425, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhu, Z.; Deng, X.; Zou, J.; Ma, C.; Li, C.; Yin, T.; Liu, C.; Wang, J.; Chen, Q.; et al. GmPBS1,a Hub Gene Interacting with Rhizobial Type-III Effectors NopT and NopP, Regulates Soybean Nodulation. AGRONOMY-BASEL 2023, 13. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Liu, C.; Ma, C.; Li, C.; Zhang, Y.; Qi, Z.; Zhu, R.; Shi, Y.; Zou, J.; et al. Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP. Int. J. Mol. Sci. 2018, 19, 3438. [Google Scholar] [CrossRef] [PubMed]
- Esseling, J.J.; Lhuissier, F.G.; Emons, A.M.C. Nod factor-induced root hair curling: Continuous polar growth towards the point of nod factor application. Plant Physiol. 2003, 132, 1982–1988. [Google Scholar] [CrossRef]
- Huo, H.; Wang, X.; Liu, Y.; Chen, J.; Wei, G. A Nod factor- and type III secretion system-dependent manner for Robinia pseudoacacia to establish symbiosis with Mesorhizobium amorphae CCNWGS0123. Tree Physiol. 2021, 41, 817–835. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, M.; Sun, Y.; Zhao, P.; Liu, C.; Qing, K.; Hu, X.; Zhong, Z.; Cheng, J.; Wang, H.; et al. Glycine max NNL1 restricts symbiotic compatibility with widely distributed bradyrhizobia via root hair infection. Nat. Plants. 2021, 7, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Xiuhong, X. U. Effects of rhizobium, soybean variety, soil type on nitrogenase activity. Journal of Northeast Agricultural University 2008, 39, 36–39. [Google Scholar]
- Wu, H.; Zhong, Z.; Wang, X.; Chen, D.; Li, Y. Screening and identification of elite rhizobia matched with main cultivars of soybean in Heilongjiang Province. Chinese Journal of Applied and Environmental Biology 2018, 24, 39–46. [Google Scholar]
- Zhongyu, M. A.; Jun, L. I.; Yongfang, Z.; Hui, F. A. N.; Li, L. I. Symbiotic Matching between Soybean Rhizobium and Soybean Cultivars. Soybean Science 2008, 27, 221–227. [Google Scholar]
- Jiali, W.; Xianjin, H.; Taiyang, Z.; Zhigang, C. Review on Sustainable Utilization of Salt-affected Land. Acta Geographica Sinica 2011, 66, 673–684. [Google Scholar]
- Qin, J.; Gao, Z.; Yue, A.; Zhang, Y.; Gao, C.; Zhao, J.; Wang, M.; Du, W. Isolation,Identification and Stress Resistance Analysis of A Mesorhizobium Isolated from Soybean Variety Jinda 53. Soybean Science 2020, 39, 898–905. [Google Scholar]
- Tian, C. F.; Zhou, Y. J.; Zhang, Y. M.; Li, Q. Q.; Zhang, Y. Z.; Li, D. F.; Wang, S.; Wang, J.; Gilbert, L. B.; Li, Y. R.; et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proceedings of the National Academy of Sciences of the United States of America 2012, 109, 8629–8634. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).