1. Introduction
The Bcr-Abl oncoprotein, alternatively referred to as the Philadelphia chromosome or Philadelphia fusion protein, emerges as a fusion protein due to a genetic anomaly detected in certain leukemia types, notably chronic myeloid leukemia (CML) and a subgroup of acute lymphoblastic leukemia (ALL). This anomaly stems from a chromosomal translocation involving chromosome 9 and chromosome 22, recognized as the Philadelphia chromosome[
1,
2,
3].
Leukemia, characterized by abnormal production of white blood cells, is a cancer affecting the blood and bone marrow. It encompasses four main types: acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML), and chronic lymphocytic leukemia (CLL)[
4,
5,
6,
7]. Its etiology is multifactorial, involving factors like exposure to ionizing radiation, certain chemicals, genetic abnormalities, and hereditary influences. Symptoms of leukemia vary by type and stage but commonly include fatigue, weakness, pallor, fever, excessive sweating, weight loss, enlarged lymph nodes, easy bruising or bleeding, and frequent infections[
4,
5,
6,
7]. Diagnosis typically includes blood tests, bone marrow biopsy, and imaging studies.Treatment options for leukemia are diverse and may encompass chemotherapy, radiation therapy, bone marrow transplantation, and targeted therapies like tyrosine kinase inhibitors [
4,
5,
6,
7].
In summary,Tyrosine kinase inhibitors (TKIs) are a vital therapeutic approach in leukemia management, offering targeted and efficient treatment while reducing the adverse effects linked with traditional chemotherapy.
Current research efforts are focused on developing novel TKIs and combination therapies to enhance outcomes for patients with leukemia [
8,
9,
10]. They constitute a class of targeted therapy medications that have demonstrated substantial efficacy against leukemia, notably in treating chronic myeloid leukemia (CML) and certain types of acute lymphoblastic leukemia (ALL). These drugs function by impeding the activity of specific tyrosine kinases, pivotal enzymes involved in cellular signaling pathways crucial for cancer growth and advancement.
In the case of CML, the Bcr-Abl tyrosine kinase inhibitor marked the inception of TKI utilization in clinical practice. Agents such as imatinib, dasatinib, and nilotinib are designed to target the Bcr-Abl fusion protein, a hallmark feature of CML. By effectively inhibiting its activity, these medications suppress the growth and proliferation of leukemic cells [
11,
12]. The current study is centered on Molecular Docking simulations [
13,
14] utilizing Autodock Vina in conjunction with the Pyrx program ]16]. The primary objective is to explore the potential of drugs and natural compounds in targeting the Bcr-Abl oncoprotein. By employing computational docking techniques, this research aims to identify promising candidates capable of interacting with and potentially inhibiting the activity of the Bcr-Abl oncoprotein, thereby offering insights into novel therapeutic strategies for diseases associated with its aberrant function.
3. Results and Discussion
The focus of this study involves Molecular Docking simulations [
13,
14], utilizing Autodock Vina along with the Pyrx program [
16]. The main aim is to investigate the effectiveness of drugs and natural compounds in targeting the Bcr-Abl oncoprotein. Through computational docking techniques, this research seeks to identify potential candidates capable of interacting with and potentially inhibiting the activity of the Bcr-Abl oncoprotein. The findings aim to provide insights into novel therapeutic strategies for diseases associated with the aberrant function of this protein.
According to the docking results assessed using the Pyrx program, compounds such as Amentoflavone, Gingetin, Hesperidin, and Diosmin exhibited excellent binding energy scores of approximately -10/-11 kcal/mol when complexed with the Bcr-Abl oncoprotein. Similarly, Bafetinib, Bemcentinib, Candesartan_cilexetil, Eltrombopag, Lapatinib, Nilotinib, and Mocetinostat demonstrated outstanding binding values with the oncoprotein. These findings suggest that these compounds have strong potential for interacting with and potentially inhibiting the activity of the Bcr-Abl oncoprotein, highlighting their promising role as candidates for further investigation in therapeutic strategies targeting diseases associated with this protein's aberrant function.
The selection of 11 potential compounds against the Bcr-Abl oncoprotein using Autodock Vina out of a total of 200 initial compounds via a virtual screening tool represented only the initial phase of the process. Further investigations were conducted to further reduce the number of potential compounds, with particular emphasis on predicting the toxicity of these 11 compounds using the pKCSM server [
17].
From the results of these investigations, it was observed that Diosmin and Hesperidin exhibited a relatively high maximum tolerated dose (MTD) in humans of approximately 0.5 logmg/kg/day and low toxicity, as indicated by the parameters of acute oral toxicity in rats (LD50) of approximately 2,500 mol/kg and oral chronic toxicity in rats (LOAEL) of approximately 3000 log mg/kg_bw/day.
As for the selected drugs, only Eltrombopag showed excellent parameters, with low toxicity similar to the two natural substances mentioned earlier and a high value of human MTD of approximately logmg/kg/day. However, it is important to note that Eltrombopag may pose the risk of liver damage if its dosage is high, unlike the natural substances.
In summary, while Diosmin and Hesperidin demonstrated good toxicity properties and human MTD, Eltrombopag showed similar potential but with the additional risk of liver damage at high dosages. These results highlight the importance of carefully evaluating both the efficacy and safety of selected compounds for further development as potential treatments against the Bcr-Abl oncoprotein.
Table 1.
Comparison best binding energies scores (kcal/mol) of natural compounds in complex with Structure of the Bcr-Abl Oncoprotein, evaluated by Blind Docking method with Pyrx program.
Table 1.
Comparison best binding energies scores (kcal/mol) of natural compounds in complex with Structure of the Bcr-Abl Oncoprotein, evaluated by Blind Docking method with Pyrx program.
| Ligand |
Binding Energy (kcal/mol) |
| Amentoflavone |
-10.7 |
| Ginkgetin |
-10.3 |
| Hesperidin |
-10.8 |
| Diosmin |
-10 |
| Bafetinib |
-10.3 |
| Bemcentinib |
-11.2 |
| Candesartan_cilexetil |
-9.7 |
| Eltrombopag |
-9.6 |
| Lapatinib |
-9.8 |
| Nilotinib |
-10.3 |
| Mocetinostat |
-9.7 |
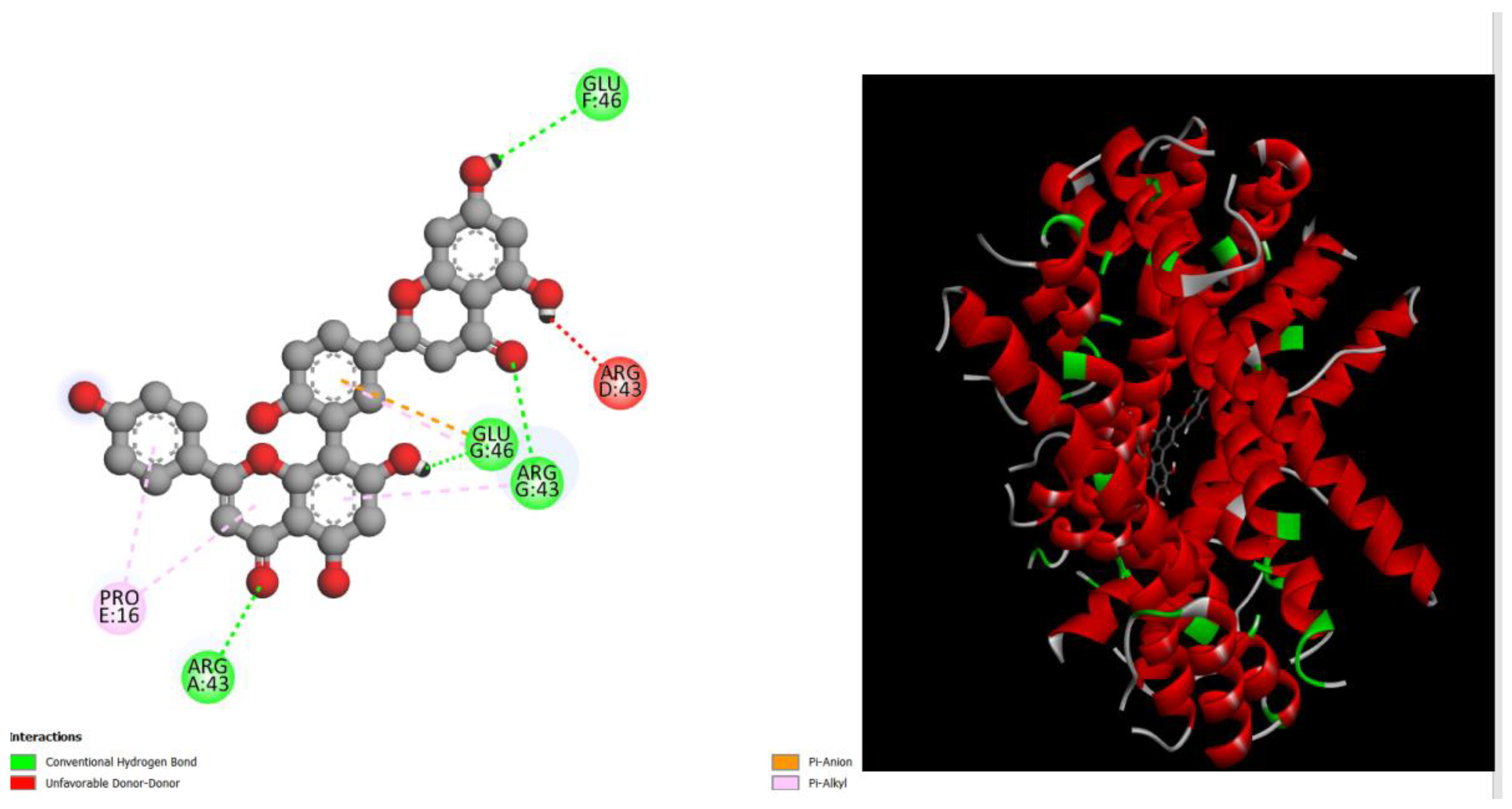
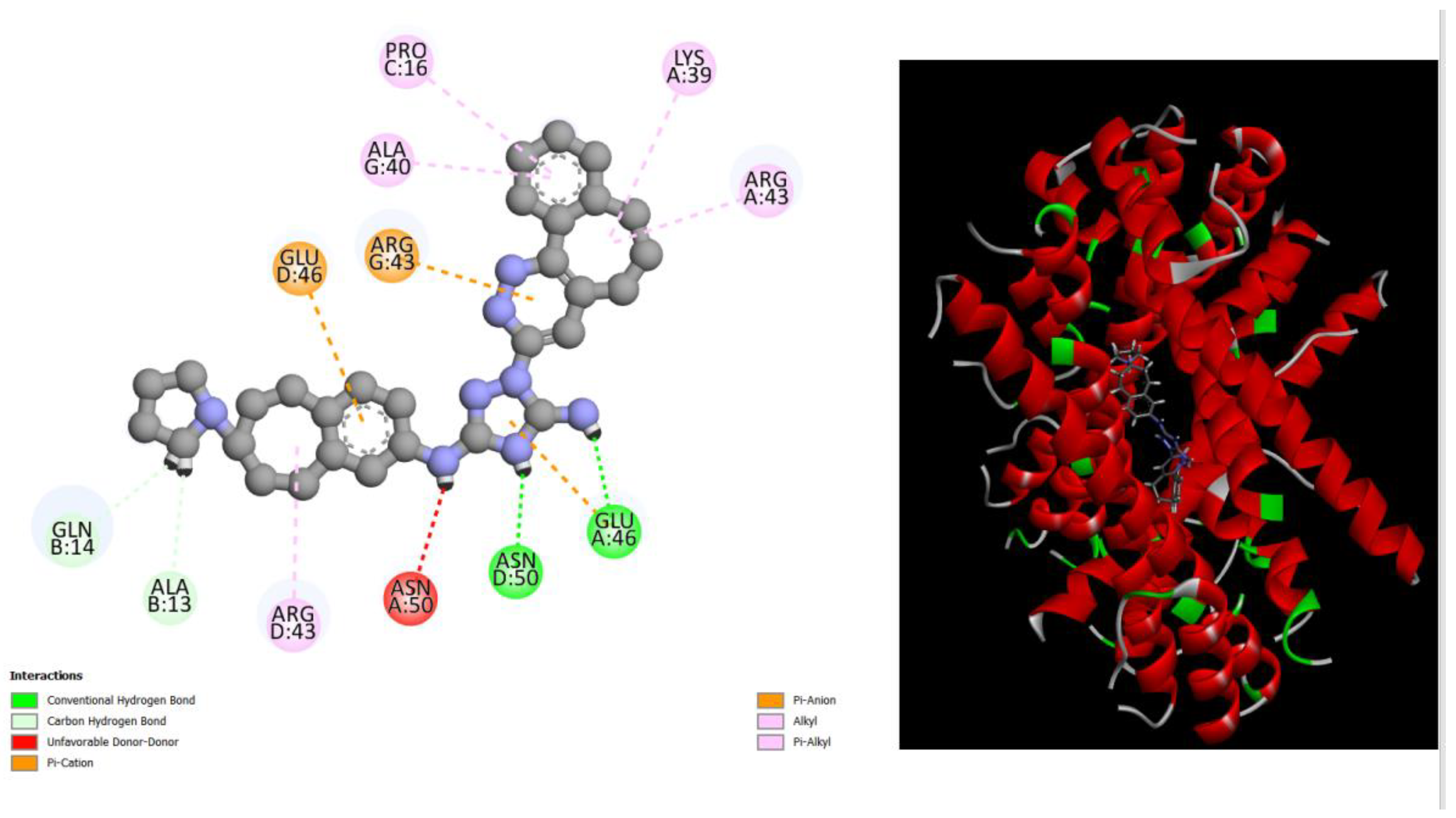
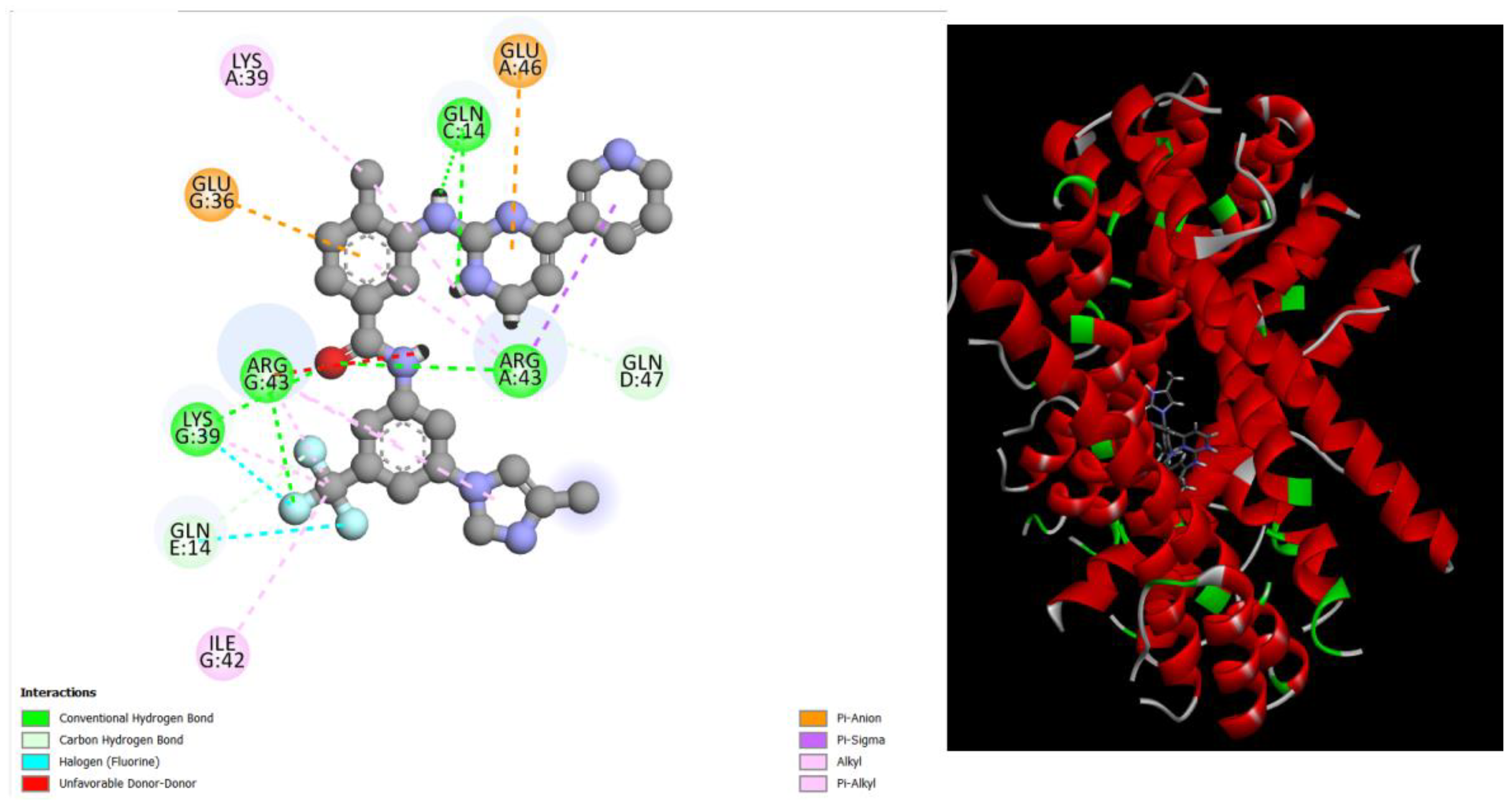
Figure 1.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked amentoflavone -11.7 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and amentoflavone. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of amentoflavone.
Figure 1.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked amentoflavone -11.7 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and amentoflavone. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of amentoflavone.
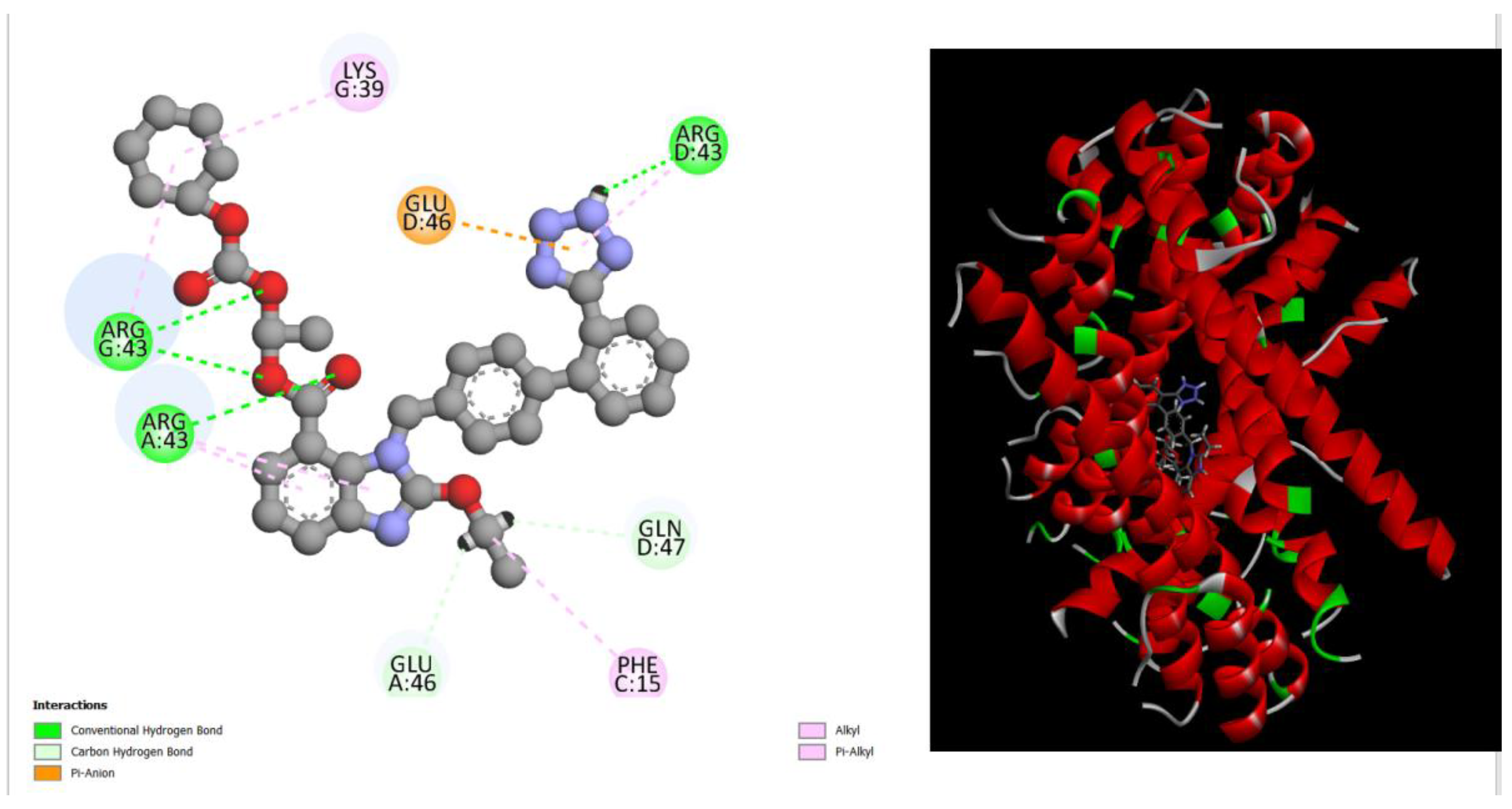
Figure 2.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Ginkgetin -10.3 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Ginkgetin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Ginkgetin.
Figure 2.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Ginkgetin -10.3 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Ginkgetin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Ginkgetin.
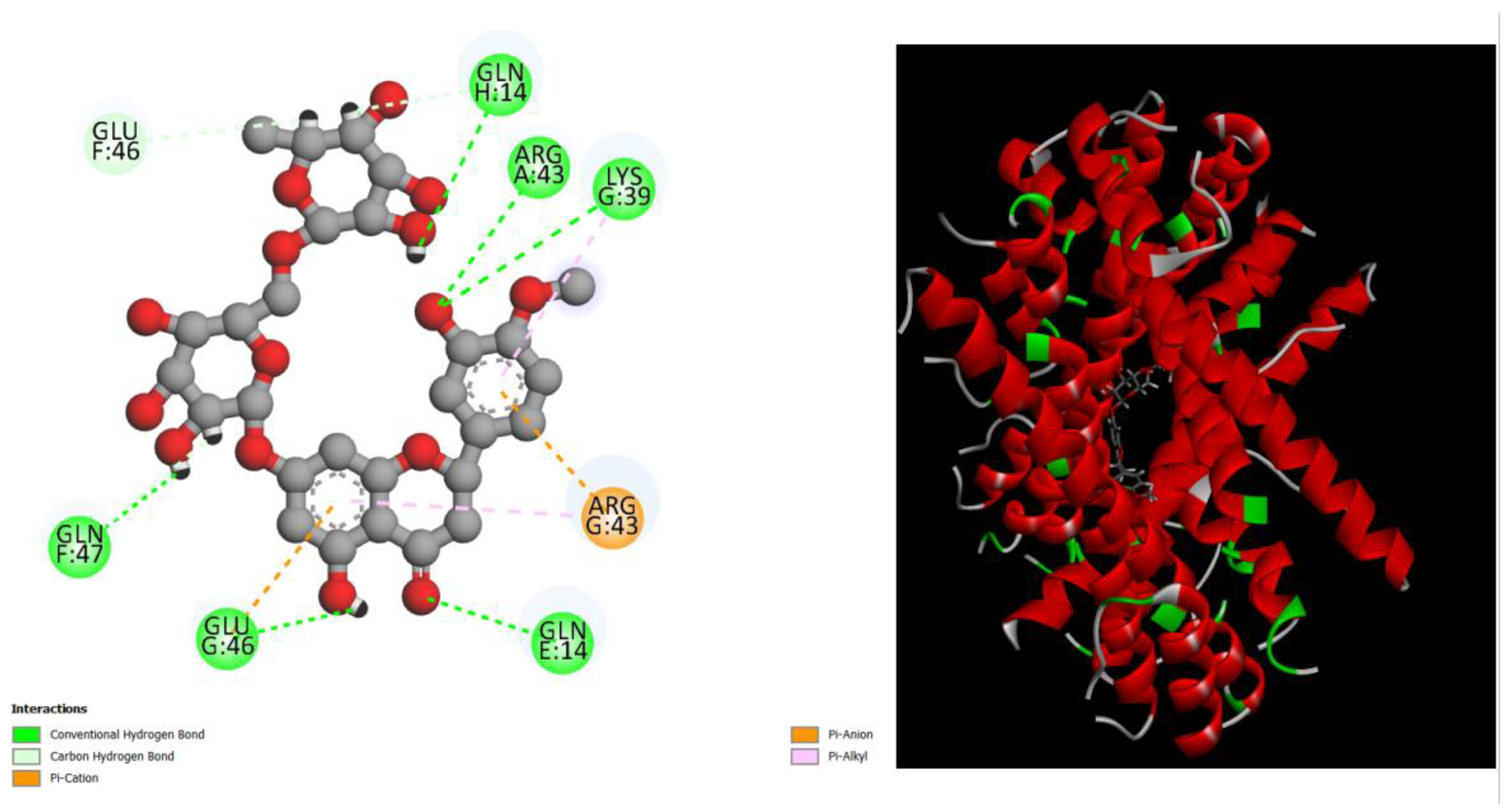
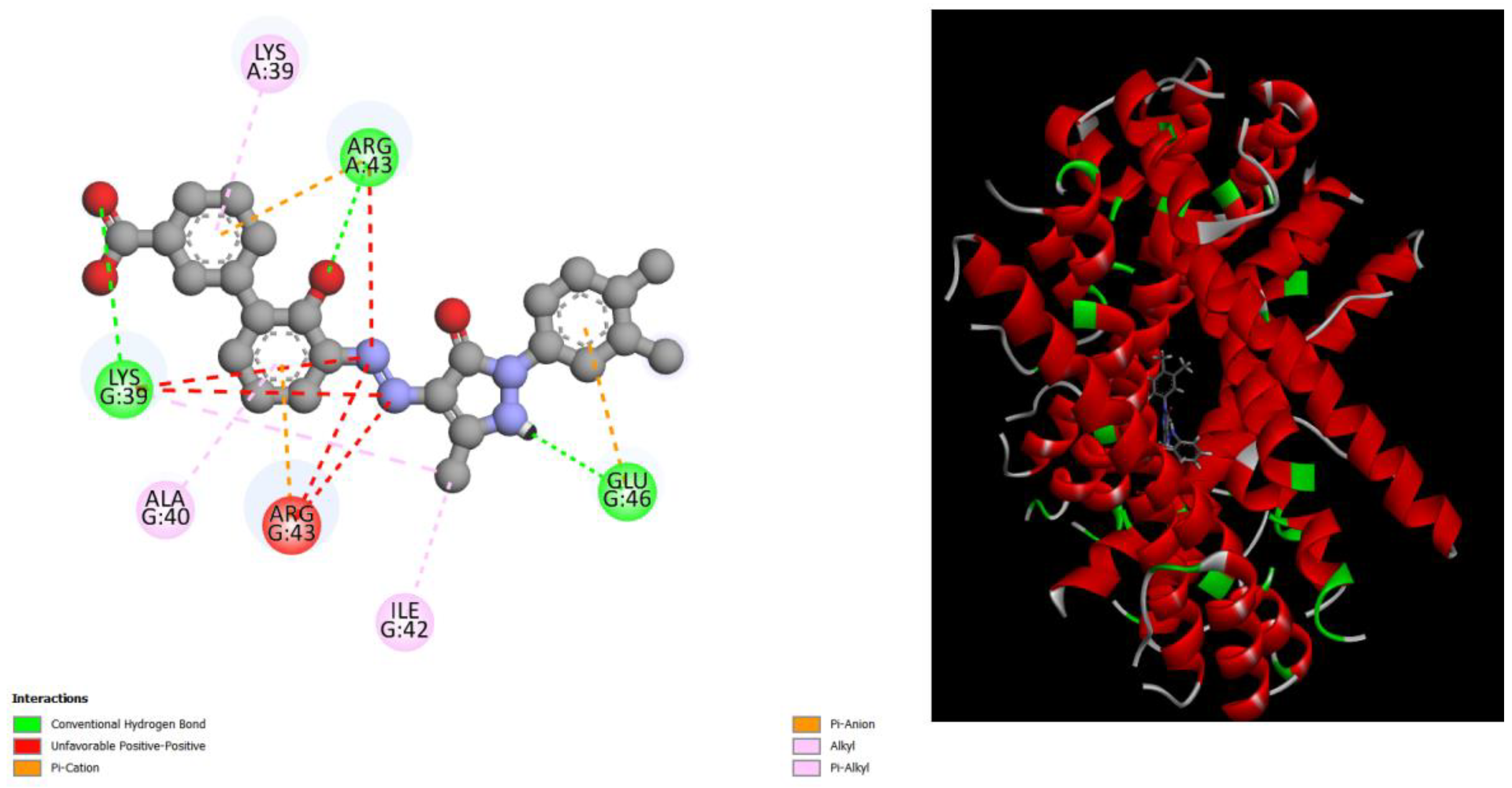
Figure 3.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Hesperidin -10.8kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hesperidin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hesperidin.
Figure 3.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Hesperidin -10.8kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hesperidin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hesperidin.
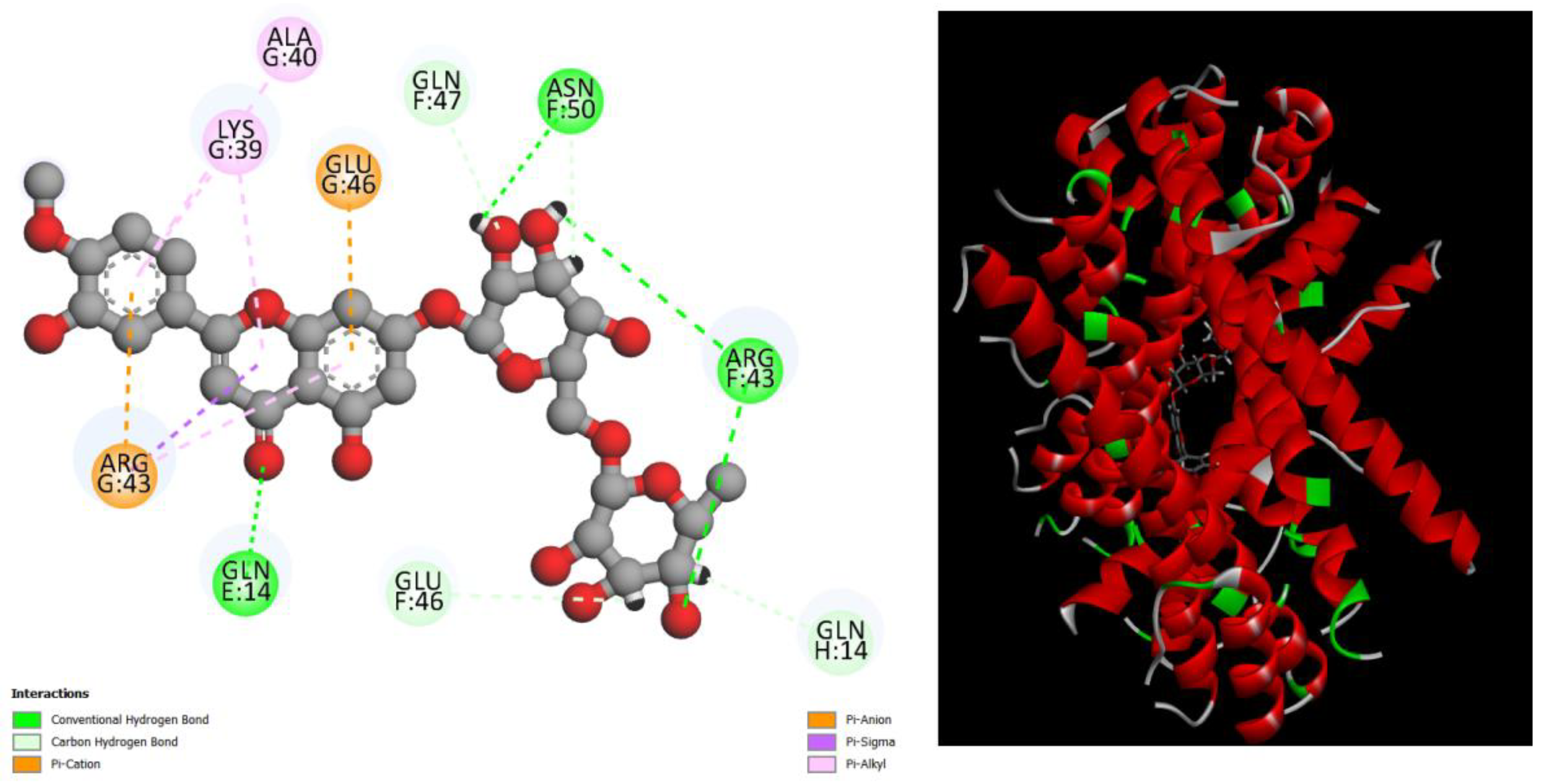
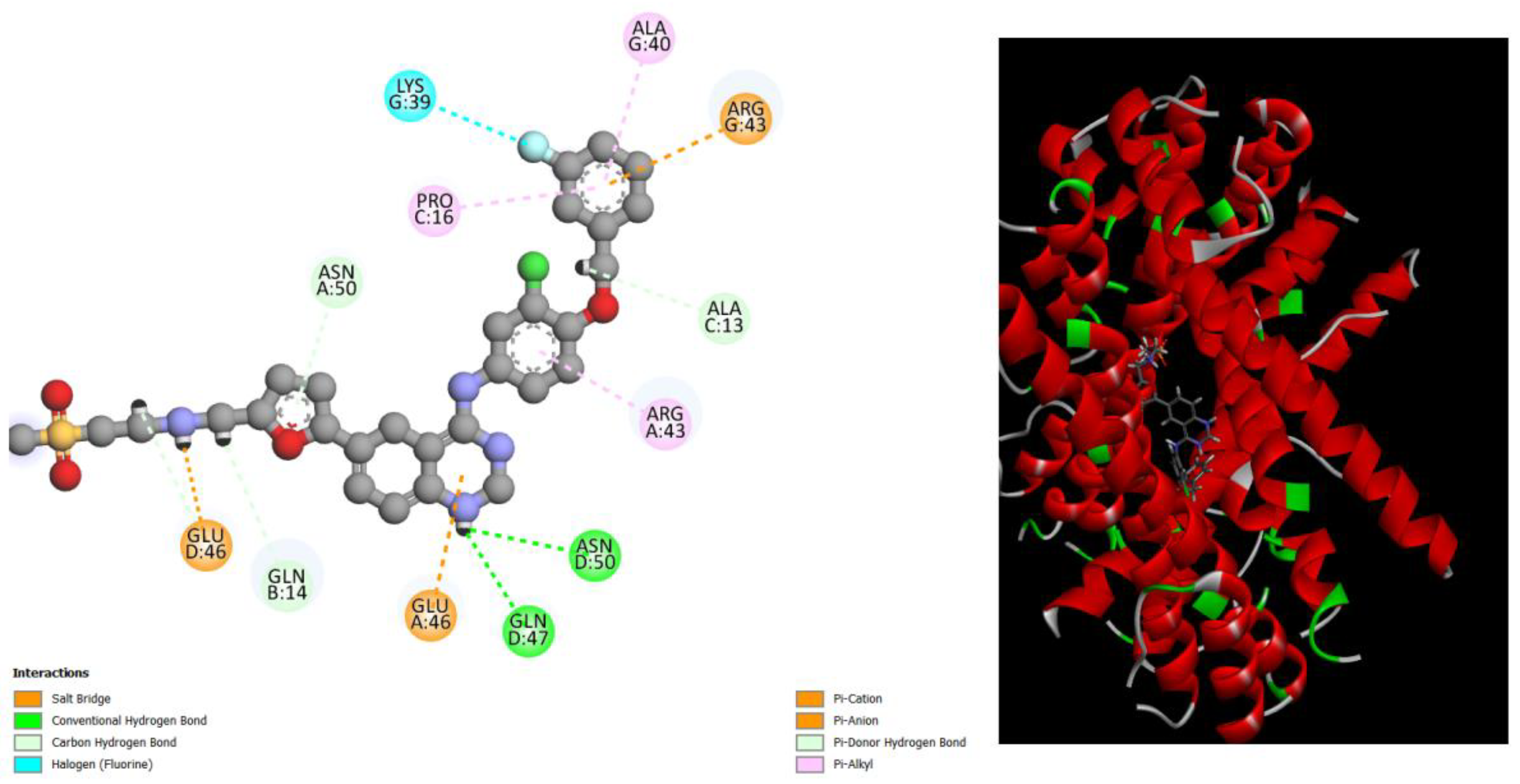
Figure 4.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Diosmin -10 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Diosmin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Diosmin.
Figure 4.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Diosmin -10 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Diosmin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Diosmin.
Figure 5.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Bafetinib -10.3 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Bafetinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bafetinib.
Figure 5.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Bafetinib -10.3 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Bafetinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bafetinib.
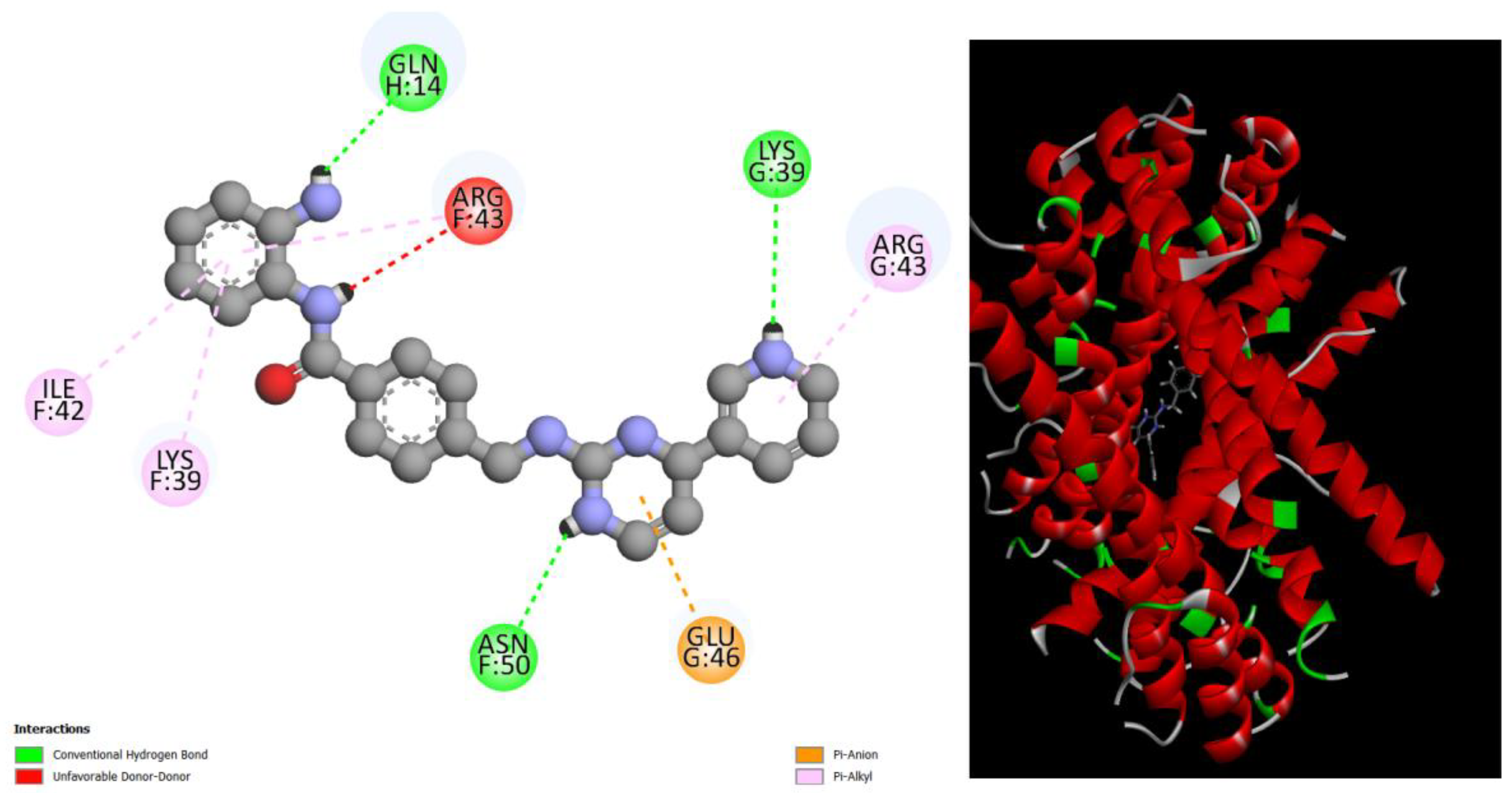
Figure 6.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Bemcentinib -11.2 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Bemcentinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bemcentinib.
Figure 6.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Bemcentinib -11.2 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Bemcentinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bemcentinib.
Figure 7.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Candesartan_cilexetil -9.7 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Candesartan_cilexetil. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Candesartan_cilexetil.
Figure 7.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Candesartan_cilexetil -9.7 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Candesartan_cilexetil. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Candesartan_cilexetil.
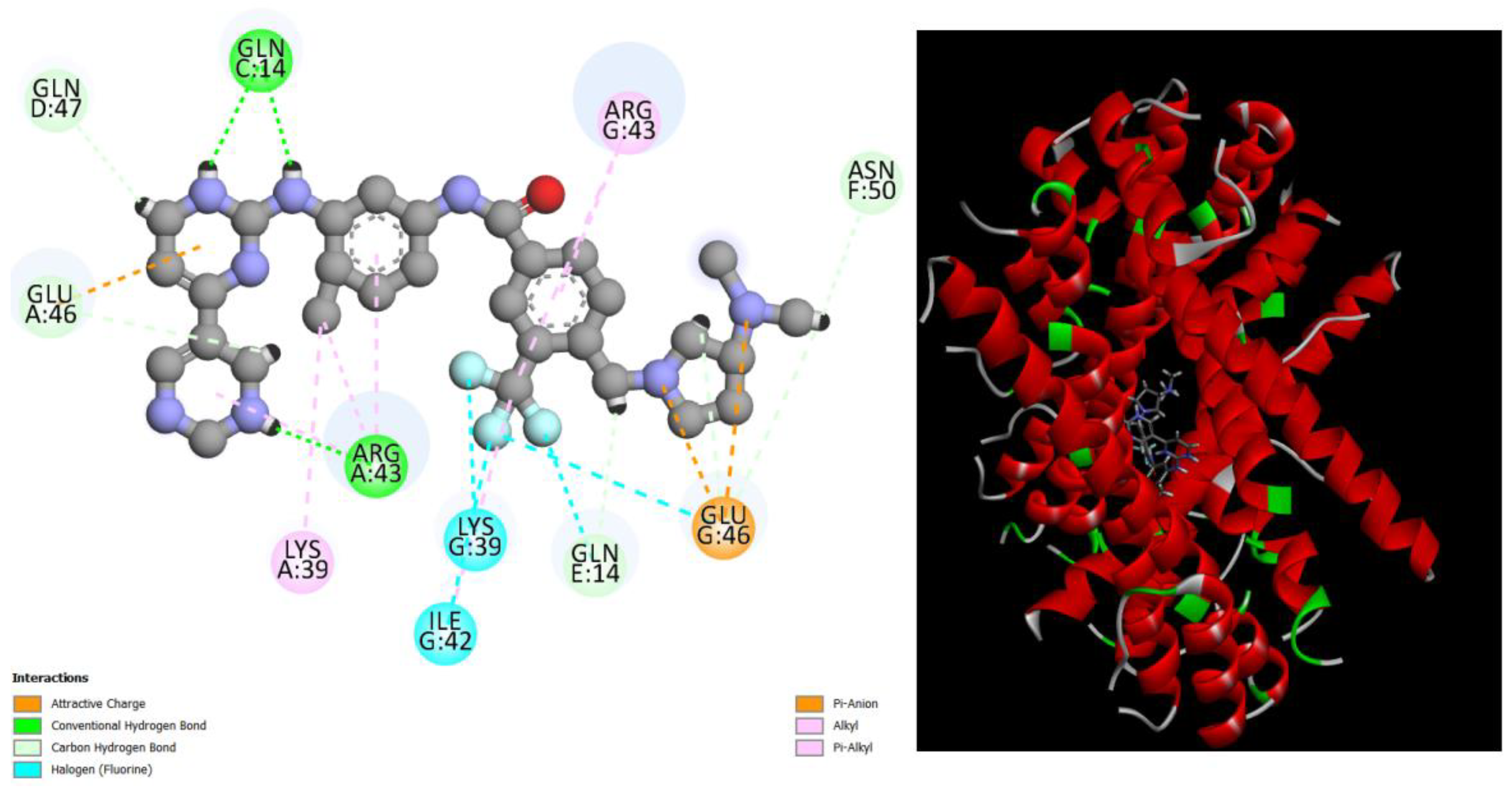
Figure 8.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Eltrombopag -9.6 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Eltrombopag. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Eltrombopag.
Figure 8.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Eltrombopag -9.6 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Eltrombopag. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Eltrombopag.
Figure 9.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Lapatinib -9.8 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Lapatinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Lapatinib.
Figure 9.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Lapatinib -9.8 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Lapatinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Lapatinib.
Figure 10.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Nilotinib -10.3 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Nilotinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Nilotinib.
Figure 10.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Nilotinib -10.3 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Nilotinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Nilotinib.
Figure 11.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Mocetinostat -9.7 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Mocetinostat. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Mocetinostat.
Figure 11.
displays the docking outcomes of Structure of Crystal Human Bcr-Abl Oncoprotein in conjunction with docked Mocetinostat -9.7 kcal mol , within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Mocetinostat. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Mocetinostat.
Table 1.
shows the comparison of predicted toxicity parameters by pKCSM Server with best compounds evaluated by Autodock Vina in complex with Structure of the Bcr-Abl Oncoprotein.
Table 1.
shows the comparison of predicted toxicity parameters by pKCSM Server with best compounds evaluated by Autodock Vina in complex with Structure of the Bcr-Abl Oncoprotein.
| Compounds |
AMES
toxicity |
|
Max.
tolerated dose(human) (logmg/kg/day) |
hERG I inhibitor |
hERG II inhibitor |
Oral Rat Acute Toxicity (LD50) (mol/kg) |
Oral Rat Chronic Toxicity (LOAEL)
(log mg/kg_bw/day) |
Hepatotoxicity |
| Amentoflavone |
no |
|
0.438 |
no |
yes |
2.527 |
3.572 |
no |
| Ginkgetin |
no |
|
0.427 |
no |
yes |
2.733 |
2.475 |
no |
| Hesperidin |
no |
|
0.525 |
no |
yes |
2.506 |
3.167 |
no |
| Diosmin |
no |
|
0.565 |
no |
yes |
2.512 |
3.343 |
no |
| Bafetinib |
no |
|
0.383 |
no |
yes |
2.85 |
1.321 |
yes |
| Bemcentinib |
no |
|
0.174 |
no |
yes |
2.86 |
1.736 |
yes |
| Candesartan_cilexetil |
Yes |
|
0.315 |
no |
yes |
2.517 |
2.736 |
yes |
| Eltrombopag |
no |
|
0.503 |
no |
no |
2.424 |
2.263 |
yes |
| Lapatinib |
Yes |
|
0.436 |
no |
yes |
3.171 |
0.064 |
yes |
| Nilotinib |
no |
|
0.199 |
no |
yes |
2.489 |
0.923 |
yes |
| Mocetinostat |
no |
|
0.249 |
no |
yes |
2.473 |
0.268 |
yes |