Submitted:
01 February 2024
Posted:
05 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
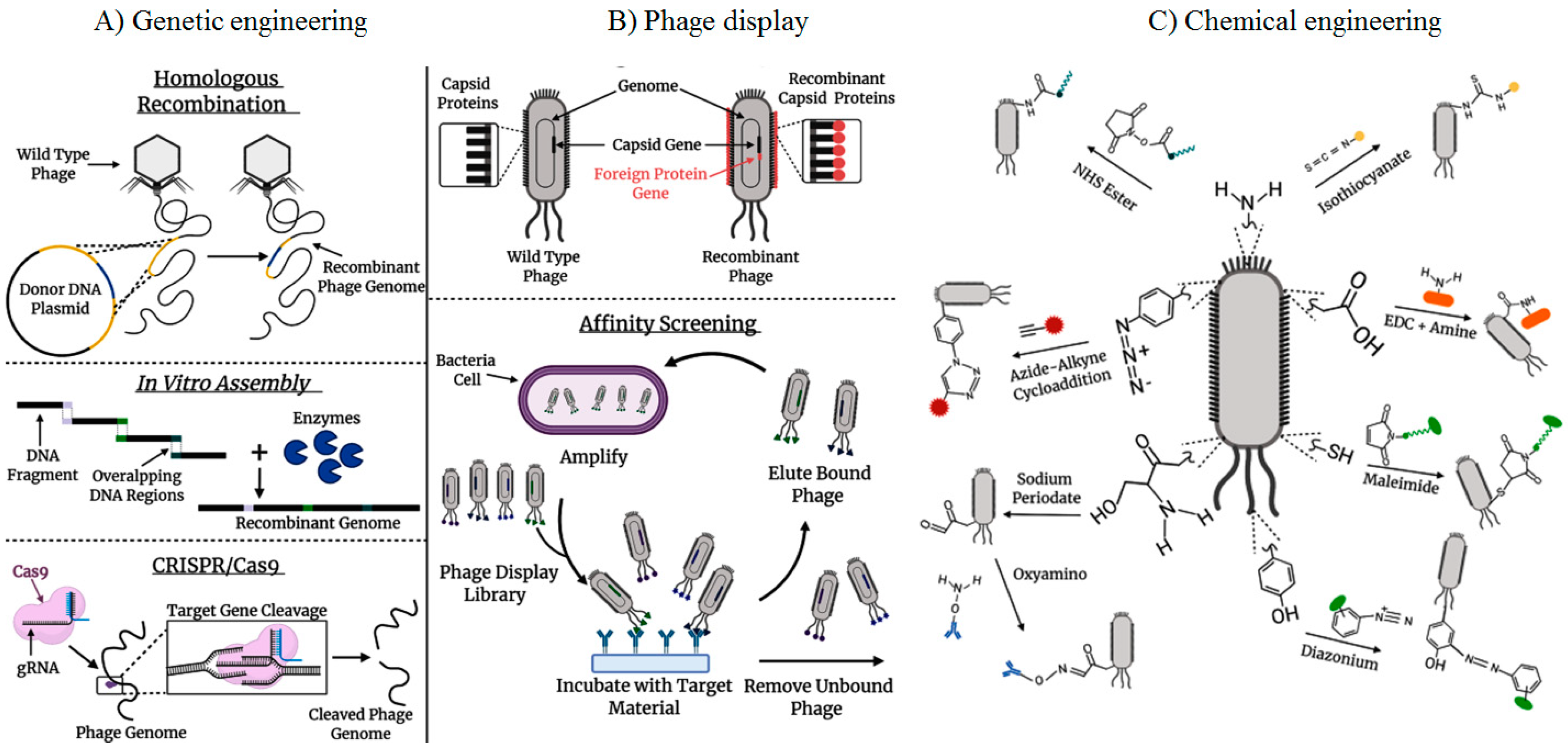
2. The technology of phage display
3. Phage display biopanning strategy
4. Infection therapy using bacteriophages
5. Infections of the soft tissues and the skin
6. Oral infections
7. Infections of the gastrointestinal system
8. Infections of the respiratory system
9. Infections of the urinary tract
10. Infection of the eyes
11. Infections of the ears
12. Nasal infections
13. Complications due to bacteremia / sepsis
14. SARS-CoV-2 pneumonia
15. Liver disease
16. Orthopaedic infections
17. Bovine mastitis
18. Production of mAbs using phage display technology
19. Phage display technology in diagnostics and therapy of neurodegenerative diseases
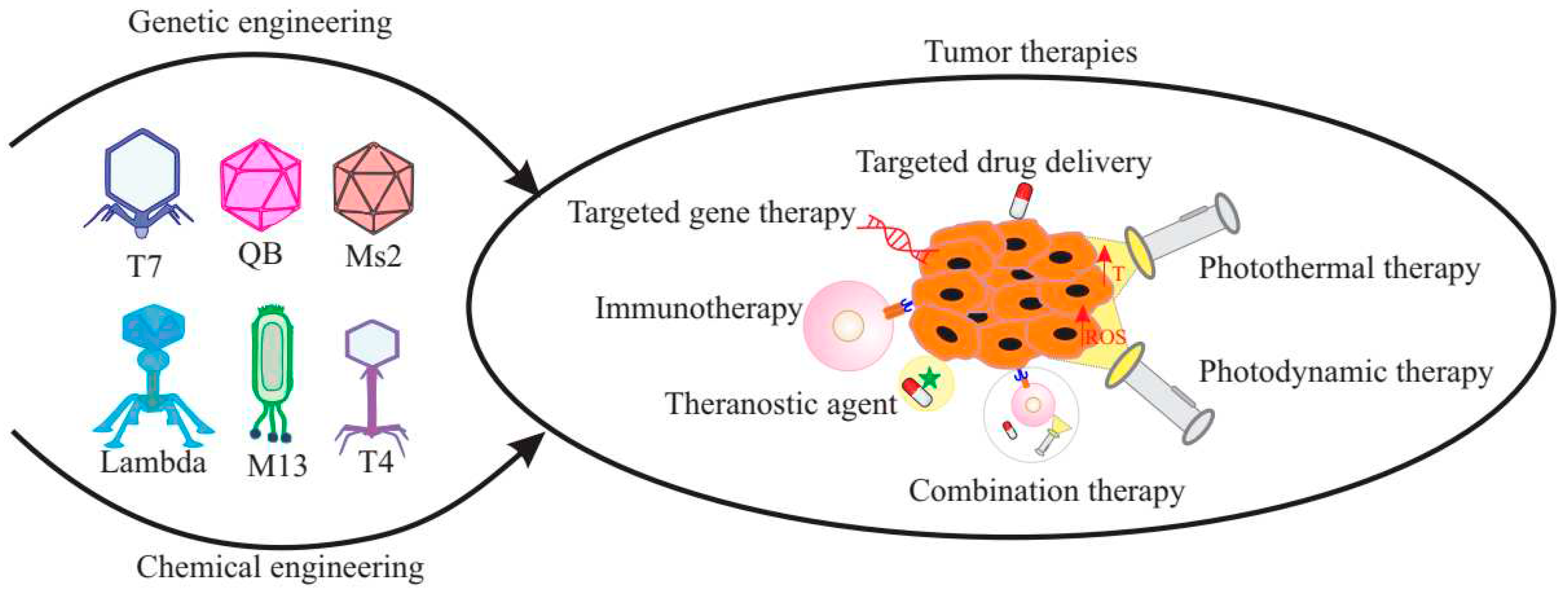
20. Multifunctional Bacteriophages
21. Bacteriophages as anticancer therapeutic agents
22. Bacteriophages in targeted drug delivery
23. Bacteriophages in targeted gene therapy
24. Bacteriophages in photothermal and photodynamic therapy
25. Bacteriophages in cancer immunotherapy
26. Bacteriophages in combination therapy
27. Bacteriophages as bioimaging agents
28. Bacteriophages as theranostic agents
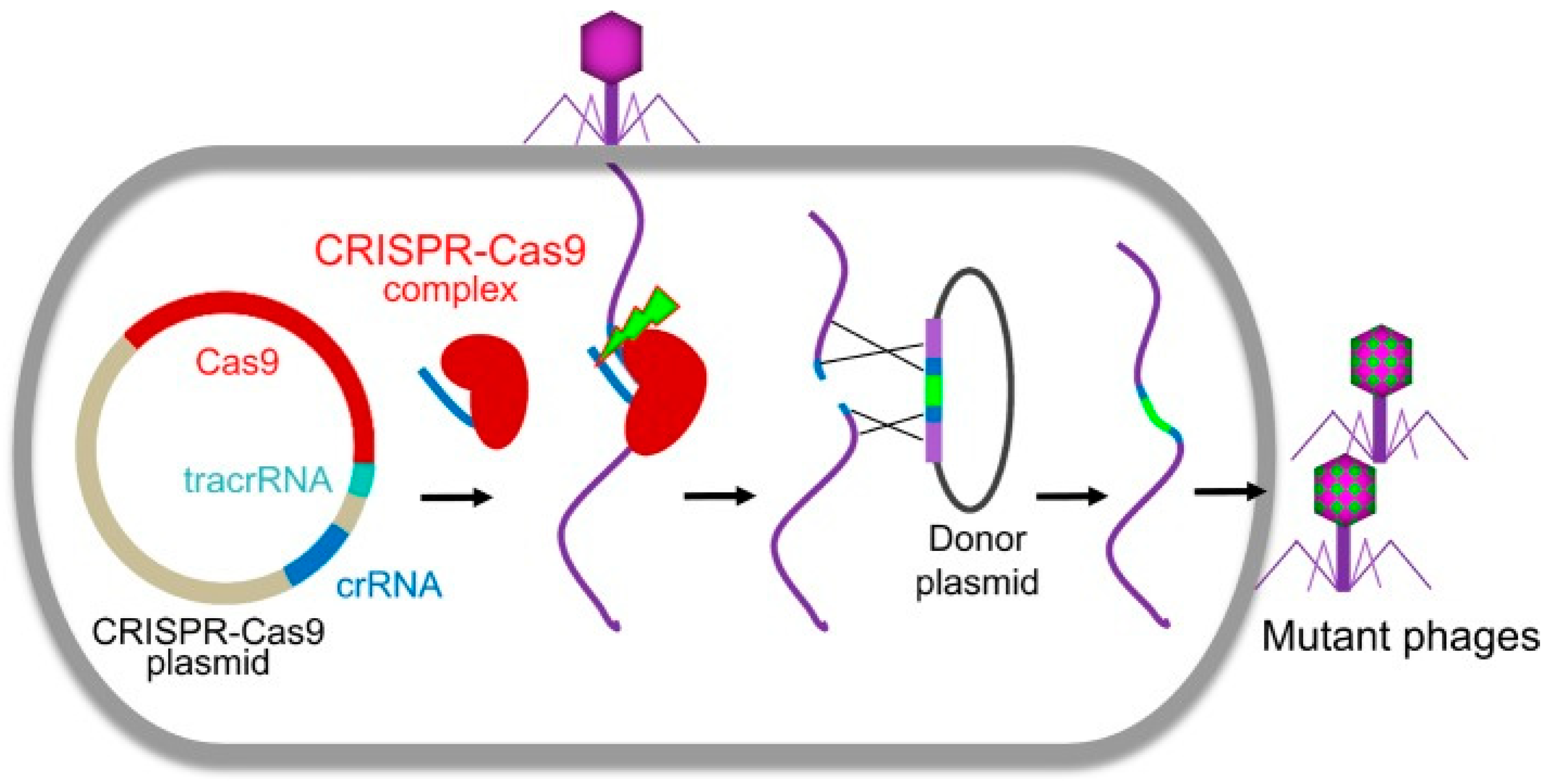
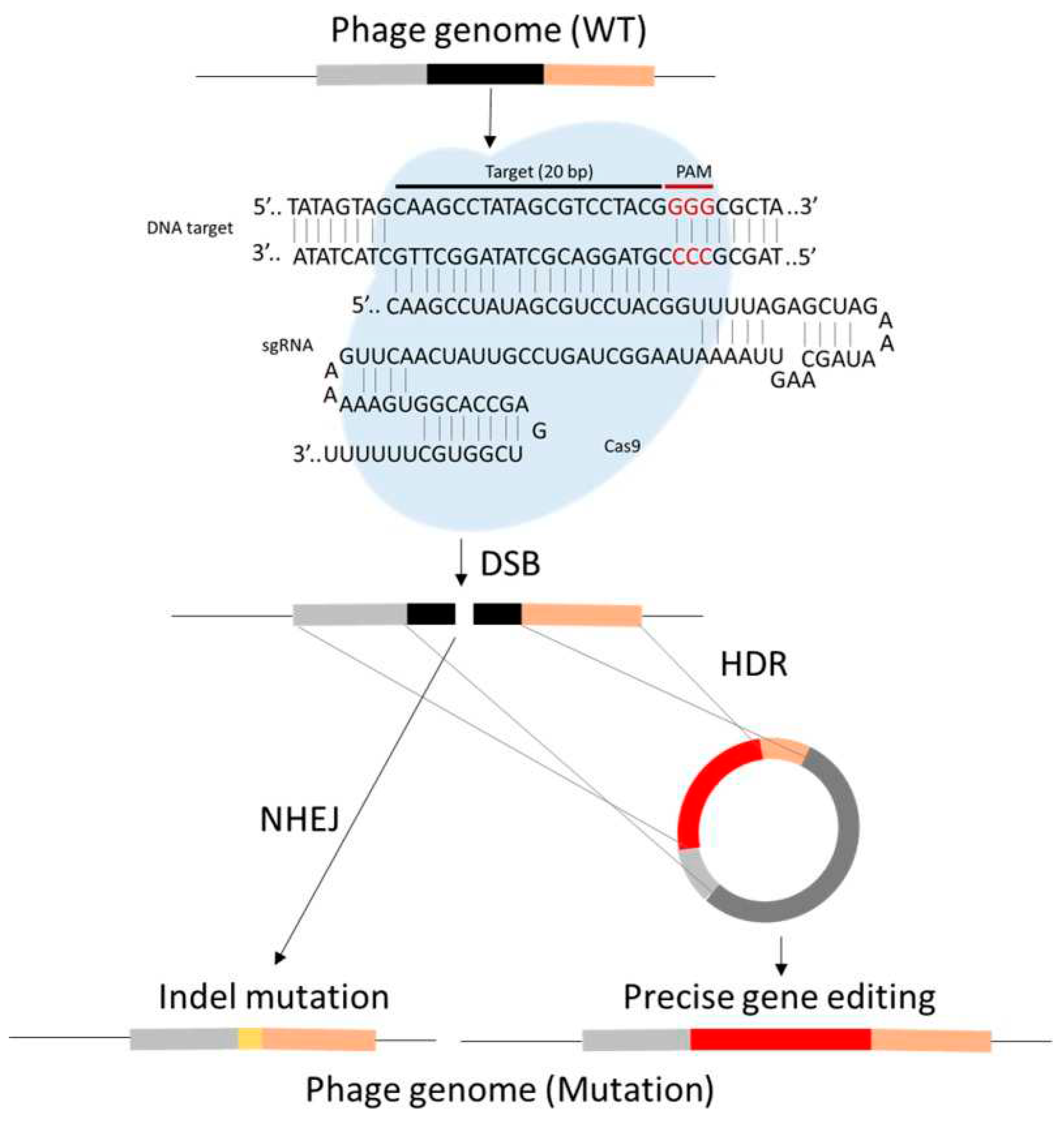
29. Phage therapy and CRISPR Cas9
30. Advantages of phage therapy
31. Disadvantages of phage therapy
32. Regulatory issues associated with phage therapy
33. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wommack, K.E.; Colwell, R.R. Virioplankton: viruses in aquatic ecosystems. Microbiology and molecular biology reviews 2000, 64, (1), 69–114. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS microbiology letters 2016, 363, (4), fnw002. [Google Scholar] [CrossRef] [PubMed]
- Doss, J.; Culbertson, K.; Hahn, D.; Camacho, J.; Barekzi, N. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses 2017, 9, (3), 50. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, (4705), 1315–1317. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chemical reviews 1997, 97, (2), 391–410. [Google Scholar] [CrossRef]
- Xu, P.; Ghosh, S.; Gul, A.R.; Bhamore, J.R.; Park, J.P.; Park, T.J. Screening of specific binding peptides using phage-display techniques and their biosensing applications. TrAC Trends in Analytical Chemistry 2021, 137, 116229. [Google Scholar] [CrossRef]
- Nixon, A.E.; Sexton, D.J.; Ladner, R.C. In Drugs derived from phage display: from candidate identification to clinical practice, MAbs, 2014; Taylor & Francis: 2014; pp 73-85. [CrossRef]
- Winter, G.; Griffiths, A.D.; Hawkins, R.E.; Hoogenboom, H.R. Making antibodies by phage display technology. Annual review of immunology 1994, 12, (1), 433–455. [Google Scholar] [CrossRef]
- Gibb, B.P.; Hadjiargyrou, M. Bacteriophage therapy for bone and joint infections: an instructional review. The bone & joint journal 2021, 103, (2), 234–244. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Gao, H.; Qing, G. Phage display derived peptides for alzheimer's disease therapy and diagnosis. Theranostics 2022, 12, (5), 2041. [Google Scholar] [CrossRef]
- Anand, T.; Virmani, N.; Bera, B.C.; Vaid, R.K.; Vashisth, M.; Bardajatya, P.; Kumar, A.; Tripathi, B.N. Phage display technique as a tool for diagnosis and antibody selection for coronaviruses. Current Microbiology 2021, 78, (4), 1124–1134. [Google Scholar] [CrossRef]
- Love, K.R.; Swoboda, J.G.; Noren, C.J.; Walker, S. Enabling glycosyltransferase evolution: a facile substrate-attachment strategy for phage-display enzyme evolution. ChemBioChem 2006, 7, (5), 753–756. [Google Scholar] [CrossRef]
- Yang, S.H.; Chung, W.J.; McFarland, S.; Lee, S.W. Assembly of bacteriophage into functional materials. The Chemical Record 2013, 13, (1), 43–59. [Google Scholar] [CrossRef]
- Zhou, X.; Cao, P.; Zhu, Y.; Lu, W.; Gu, N.; Mao, C. Phage-mediated counting by the naked eye of miRNA molecules at attomolar concentrations in a Petri dish. Nature materials 2015, 14, (10), 1058–1064. [Google Scholar] [CrossRef]
- Gray, B.P.; Brown, K.C. Combinatorial peptide libraries: mining for cell-binding peptides. Chemical reviews 2014, 114, (2), 1020–1081. [Google Scholar] [CrossRef]
- Piggott, A.M.; Karuso, P. Identifying the cellular targets of natural products using T7 phage display. Natural product reports 2016, 33, (5), 626–636. [Google Scholar] [CrossRef]
- Yang, M.; Sunderland, K.; Mao, C. Virus-derived peptides for clinical applications. Chemical reviews 2017, 117, (15), 10377–10402. [Google Scholar] [CrossRef]
- Hussein, A.H.; Davis, E.M.; Halperin, S.A.; Lee, S.F. Construction and characterization of single-chain variable fragment antibodies directed against the Bordetella pertussis surface adhesins filamentous hemagglutinin and pertactin. Infection and immunity 2007, 75, (11), 5476–5482. [Google Scholar] [CrossRef]
- Braganza, A.; Wallace, K.; Pell, L.; Parrish, C.R.; Siegel, D.L.; Mason, N.J. Generation and validation of canine single chain variable fragment phage display libraries. Veterinary immunology and immunopathology 2011, 139, (1), 27–40. [Google Scholar] [CrossRef]
- Galán, A.; Comor, L.; Horvatić, A.; Kuleš, J.; Guillemin, N.; Mrljak, V.; Bhide, M. Library-based display technologies: where do we stand? Molecular BioSystems 2016, 12, (8), 2342–2358. [Google Scholar] [CrossRef]
- Zhang, Y. In Evolution of phage display libraries for therapeutic antibody discovery, Mabs, 2023; Taylor & Francis: 2023; p 2213793. [CrossRef]
- Lee, T.-Y.; Lin, C.-T.; Kuo, S.-Y.; Chang, D.-K.; Wu, H.-C. Peptide-mediated targeting to tumor blood vessels of lung cancer for drug delivery. Cancer research 2007, 67, (22), 10958–10965. [Google Scholar] [CrossRef]
- Li, J.; Feng, L.; Fan, L.; Zha, Y.; Guo, L.; Zhang, Q.; Chen, J.; Pang, Z.; Wang, Y.; Jiang, X. Targeting the brain with PEG–PLGA nanoparticles modified with phage-displayed peptides. Biomaterials 2011, 32, (21), 4943–4950. [Google Scholar] [CrossRef]
- Akanda, Z.Z.; Taha, M.; Abdelbary, H. Current review—the rise of bacteriophage as a unique therapeutic platform in treating peri-prosthetic joint infections. Journal of Orthopaedic Research® 2018, 36, (4), 1051–1060. [Google Scholar] [CrossRef]
- Weber-Dąbrowska, B.; Jończyk-Matysiak, E.; Żaczek, M.; Łobocka, M.; Łusiak-Szelachowska, M.; Górski, A. Bacteriophage procurement for therapeutic purposes. Frontiers in microbiology 2016, 7, 1177. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Frontiers in microbiology 2014, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, L.D. Designing phage therapeutics. Current pharmaceutical biotechnology 2010, 11, (1), 15–27. [Google Scholar] [CrossRef]
- Romero-Calle, D.; Guimarães Benevides, R.; Góes-Neto, A.; Billington, C. Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics 2019, 8, (3), 138. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, S.; Hauwa-Suleiman, B.; Ali Abbagana, B.; Alhaji-Mustafa, I.; Abbas-Musa, I. Novel uses of bacteriophages in the treatment of human infections and antibiotic resistance. Am. J. Biosci 2016, 4, (3), 34. [Google Scholar] [CrossRef]
- Abedon, S.T.; Thomas-Abedon, C. Phage therapy pharmacology. Current pharmaceutical biotechnology 2010, 11, (1), 28–47. [Google Scholar] [CrossRef]
- Sharun, K.; Dhama, K.; Tiwari, R.; Gugjoo, M.B.; Iqbal Yatoo, M.; Patel, S.K.; Pathak, M.; Karthik, K.; Khurana, S.K.; Singh, R. Advances in therapeutic and managemental approaches of bovine mastitis: a comprehensive review. Veterinary Quarterly 2021, 41, (1), 107–136. [Google Scholar] [CrossRef]
- Haq, I.U.; Chaudhry, W.N.; Akhtar, M.N.; Andleeb, S.; Qadri, I. Bacteriophages and their implications on future biotechnology: a review. Virology journal 2012, 9, (1), 1–8. [Google Scholar] [CrossRef]
- Tiwari, R.; Dhama, K.; Kumar, A.; Rahal, A.; Kapoor, S. Bacteriophage therapy for safeguarding animal and human health: a review. Pakistan journal of biological sciences: PJBS 2014, 17, (3), 301–315. [Google Scholar] [CrossRef]
- Carson, L.; Gorman, S.P.; Gilmore, B.F. The use of lytic bacteriophages in the prevention and eradication of biofilms of Proteus mirabilis and Escherichia coli. FEMS Immunology & Medical Microbiology 2010, 59, (3), 447–455. [Google Scholar] [CrossRef]
- Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Leitner, L.; Mehnert, U.; Chkhotua, A.; Kessler, T.M.; Sybesma, W. Adapted bacteriophages for treating urinary tract infections. Frontiers in microbiology 2018, 9, 1832. [Google Scholar] [CrossRef] [PubMed]
- Khawaldeh, A.; Morales, S.; Dillon, B.; Alavidze, Z.; Ginn, A.; Thomas, L.; Chapman, S.; Dublanchet, A.; Smithyman, A.; Iredell, J. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. Journal of medical microbiology 2011, 60, (11), 1697–1700. [Google Scholar] [CrossRef]
- Marčuk, L.; Nikiforov, V.; Ščerbak, J.F.; Levitov, T.; Kotljarova, R.; Naumšina, M.; Davydov, S.; Monsur, K.; Rahman, M.; Latif, M. Clinical studies of the use of bacteriophage in the treatment of cholera. Bulletin of the World Health Organization 1971, 45, (1), 77. [Google Scholar] [PubMed]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, M.; Hill, C.; Ross, R.P. Exploiting gut bacteriophages for human health. Trends in microbiology 2014, 22, (7), 399–405. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrobial agents and chemotherapy 2017, 61, (10), 10.1128/aac. 00954–17. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature medicine 2019, 25, (5), 730–733. [Google Scholar] [CrossRef]
- Fish, R.; Kutter, E.; Wheat, G.; Blasdel, B.; Kutateladze, M.; Kuhl, S. Compassionate use of bacteriophage therapy for foot ulcer treatment as an effective step for moving toward clinical trials. Bacteriophage therapy: from lab to clinical practice 2018, 159-170. [CrossRef]
- Weber-Dąbrowska, B.; Mulczyk, M.; Górski, A. Bacteriophage therapy of bacterial infections: an update of our institute’s experience. Inflammation 2001, 201–209. [Google Scholar] [CrossRef]
- Markoishvili, K.; Tsitlanadze, G.; Katsarava, R.; Glenn, J.; Morris Jr, M.; Sulakvelidze, A. A novel sustained-release matrix based on biodegradable poly (ester amide) s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. International journal of dermatology 2002, 41, (7), 453–458. [Google Scholar] [CrossRef]
- Morozova, V.V.; Kozlova, Y.N.; Ganichev, D.A.; Tikunova, N.V. Bacteriophage treatment of infected diabetic foot ulcers. In Bacteriophage therapy: from lab to clinical practice, Springer: 2017; pp 151-158. [CrossRef]
- Kifelew, L.G.; Warner, M.S.; Morales, S.; Vaughan, L.; Woodman, R.; Fitridge, R.; Mitchell, J.G.; Speck, P. Efficacy of phage cocktail AB-SA01 therapy in diabetic mouse wound infections caused by multidrug-resistant Staphylococcus aureus. BMC microbiology 2020, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Harjai, K.; Chhibber, S. Bacteriophage versus antimicrobial agents for the treatment of murine burn wound infection caused by Klebsiella pneumoniae B5055. Journal of medical microbiology 2011, 60, (2), 205–210. [Google Scholar] [CrossRef]
- Yin, S.; Huang, G.; Zhang, Y.; Jiang, B.; Yang, Z.; Dong, Z.; You, B.; Yuan, Z.; Hu, F.; Zhao, Y. Phage Abp1 rescues human cells and mice from infection by pan-drug resistant Acinetobacter baumannii. Cellular Physiology and Biochemistry 2018, 44, (6), 2337–2345. [Google Scholar] [CrossRef]
- Totté, J.E.; van Doorn, M.B.; Pasmans, S.G. Successful treatment of chronic Staphylococcus aureus-related dermatoses with the topical endolysin Staphefekt SA. 100: a report of 3 cases. Case reports in dermatology 2017, 9, (2), 19–25. [Google Scholar] [CrossRef]
- Castillo-Ruiz, M.; Vinés, E.D.; Montt, C.; Fernández, J.; Delgado, J.M.; Hormazábal, J.C.; Bittner, M. Isolation of a novel Aggregatibacter actinomycetemcomitans serotype b bacteriophage capable of lysing bacteria within a biofilm. Applied and environmental microbiology 2011, 77, (9), 3157–3159. [Google Scholar] [CrossRef]
- Guo, L.; McLean, J.S.; Yang, Y.; Eckert, R.; Kaplan, C.W.; Kyme, P.; Sheikh, O.; Varnum, B.; Lux, R.; Shi, W. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proceedings of the National Academy of Sciences 2015, 112, (24), 7569–7574. [Google Scholar] [CrossRef]
- Tinoco, J.M.; Liss, N.; Zhang, H.; Nissan, R.; Gordon, W.; Tinoco, E.; Sassone, L.; Stevens, R. Antibacterial effect of genetically-engineered bacteriophage ϕEf11/ϕFL1C (Δ36) PnisA on dentin infected with antibiotic-resistant Enterococcus faecalis. Archives of Oral Biology 2017, 82, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, H.; Bi, Y.; Li, W.; Wei, H.; Li, Y. Activity of the chimeric lysin ClyR against common Gram-positive oral microbes and its anticaries efficacy in rat models. Viruses 2018, 10, (7), 380. [Google Scholar] [CrossRef]
- Li, W.; Yang, H.; Gong, Y.; Wang, S.; Li, Y.; Wei, H. Effects of a chimeric lysin against planktonic and sessile Enterococcus faecalis hint at potential application in endodontic therapy. Viruses 2018, 10, (6), 290. [Google Scholar] [CrossRef]
- Ott, S.J.; Waetzig, G.H.; Rehman, A.; Moltzau-Anderson, J.; Bharti, R.; Grasis, J.A.; Cassidy, L.; Tholey, A.; Fickenscher, H.; Seegert, D. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 2017, 152, (4), 799–811.e7. [Google Scholar] [CrossRef]
- Vahedi, A.; Soltan Dallal, M.M.; Douraghi, M.; Nikkhahi, F.; Rajabi, Z.; Yousefi, M.; Mousavi, M. Isolation and identification of specific bacteriophage against enteropathogenic Escherichia coli (EPEC) and in vitro and in vivo characterization of bacteriophage. FEMS microbiology letters 2018, 365, (16), fny136. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.; Koley, H.; Ghosh, A.; Palit, A.; Sarkar, B. Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes and Infection 2013, 15, (2), 152–156. [Google Scholar] [CrossRef]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepiński, P.; Douce, G.R.; Clokie, M.R. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrobial agents and chemotherapy 2016, 60, (2), 968–981. [Google Scholar] [CrossRef]
- Galtier, M.; De Sordi, L.; Maura, D.; Arachchi, H.; Volant, S.; Dillies, M.A.; Debarbieux, L. Bacteriophages to reduce gut carriage of antibiotic resistant uropathogens with low impact on microbiota composition. Environmental microbiology 2016, 18, (7), 2237–2245. [Google Scholar] [CrossRef]
- Cao, F.; Wang, X.; Wang, L.; Li, Z.; Che, J.; Wang, L.; Li, X.; Cao, Z.; Zhang, J.; Jin, L. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. BioMed Research International 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Alemayehu, D.; Casey, P.G.; McAuliffe, O.; Guinane, C.M.; Martin, J.G.; Shanahan, F.; Coffey, A.; Ross, R.P.; Hill, C. Bacteriophages ϕMR299-2 and ϕNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. MBio 2012, 3, (2), 10.1128/mbio. 00029–12. [Google Scholar] [CrossRef]
- Oduor, J.M.O.; Onkoba, N.; Maloba, F.; Nyachieo, A. Experimental phage therapy against haematogenous multi-drug resistant Staphylococcus aureus pneumonia in mice. African Journal of Laboratory Medicine 2016, 5, (1), 1–7. [Google Scholar] [CrossRef]
- Waters, E.M.; Neill, D.R.; Kaman, B.; Sahota, J.S.; Clokie, M.R.; Winstanley, C.; Kadioglu, A. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 2017, thoraxjnl-2016-209265. [CrossRef]
- Bao, J.; Wu, N.; Zeng, Y.; Chen, L.; Li, L.; Yang, L.; Zhang, Y.; Guo, M.; Li, L.; Li, J. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerging microbes & infections 2020, 9, (1), 771–774. [Google Scholar] [CrossRef]
- Leitner, L.; Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Chkonia, I.; Rigvava, S.; Chkhotua, A.; Changashvili, G.; McCallin, S.; Schneider, M.P. Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: a randomised, placebo-controlled, double-blind clinical trial. The Lancet Infectious Diseases 2021, 21, (3), 427–436. [Google Scholar] [CrossRef]
- Kuipers, S.; Ruth, M.M.; Mientjes, M.; de Sévaux, R.G.; van Ingen, J. A Dutch case report of successful treatment of chronic relapsing urinary tract infection with bacteriophages in a renal transplant patient. Antimicrobial agents and Chemotherapy 2019, 64, (1), 10.1128/aac. 01281–19. [Google Scholar] [CrossRef]
- Rostkowska, O.M.; Międzybrodzki, R.; Miszewska-Szyszkowska, D.; Górski, A.; Durlik, M. Treatment of recurrent urinary tract infections in a 60-year-old kidney transplant recipient. The use of phage therapy. Transplant Infectious Disease 2021, 23, (1), e13391. [Google Scholar] [CrossRef]
- Fukuda, K.; Ishida, W.; Uchiyama, J.; Rashel, M.; Kato, S.-i.; Morita, T.; Muraoka, A.; Sumi, T.; Matsuzaki, S.; Daibata, M. Pseudomonas aeruginosa keratitis in mice: effects of topical bacteriophage KPP12 administration. 2012. [CrossRef]
- Furusawa, T.; Iwano, H.; Hiyashimizu, Y.; Matsubara, K.; Higuchi, H.; Nagahata, H.; Niwa, H.; Katayama, Y.; Kinoshita, Y.; Hagiwara, K. Phage therapy is effective in a mouse model of bacterial equine keratitis. Applied and Environmental Microbiology 2016, 82, (17), 5332–5339. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.; Änggård, E.; Harper, D. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clinical otolaryngology 2009, 34, (4), 349–357. [Google Scholar] [CrossRef]
- Hawkins, C.; Harper, D.; Burch, D.; Änggård, E.; Soothill, J. Topical treatment of Pseudomonas aeruginosa otitis of dogs with a bacteriophage mixture: a before/after clinical trial. Veterinary microbiology 2010, 146, (3-4), 309–313. [Google Scholar] [CrossRef] [PubMed]
- Ooi, M.L.; Drilling, A.J.; Morales, S.; Fong, S.; Moraitis, S.; Macias-Valle, L.; Vreugde, S.; Psaltis, A.J.; Wormald, P.-J. Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to Staphylococcus aureus. JAMA Otolaryngology–Head & Neck Surgery 2019, 145, (8), 723–729. [Google Scholar] [CrossRef]
- Schneider, G.; Szentes, N.; Horváth, M.; Dorn, Á.; Cox, A.; Nagy, G.; Doffkay, Z.; Maróti, G.; Rákhely, G.; Kovács, T. Kinetics of targeted phage rescue in a mouse model of systemic Escherichia coli K1. BioMed Research International 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Pouillot, F.; Chomton, M.; Blois, H.; Courroux, C.; Noelig, J.; Bidet, P.; Bingen, E.; Bonacorsi, S. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b: H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrobial Agents and Chemotherapy 2012, 56, (7), 3568–3575. [Google Scholar] [CrossRef]
- Li, W.; Schäfer, A.; Kulkarni, S.S.; Liu, X.; Martinez, D.R.; Chen, C.; Sun, Z.; Leist, S.R.; Drelich, A.; Zhang, L. High potency of a bivalent human VH domain in SARS-CoV-2 animal models. Cell 2020, 183, (2), 429–441.e16. [Google Scholar] [CrossRef]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. The lancet 2012, 380, (9859), 2095–2128. [Google Scholar] [CrossRef]
- Rehm, J.; Dawson, D.; Frick, U.; Gmel, G.; Roerecke, M.; Shield, K.D.; Grant, B. Burden of disease associated with alcohol use disorders in the United States. Alcoholism: Clinical and Experimental Research 2014, 38, (4), 1068–1077. [Google Scholar] [CrossRef]
- Duan, Y.; Llorente, C.; Lang, S.; Brandl, K.; Chu, H.; Jiang, L.; White, R.C.; Clarke, T.H.; Nguyen, K.; Torralba, M. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019, 575, (7783), 505–511. [Google Scholar] [CrossRef]
- Barros, J.; Melo, L.D.; Poeta, P.; Igrejas, G.; Ferraz, M.P.; Azeredo, J.; Monteiro, F.J. Lytic bacteriophages against multidrug-resistant Staphylococcus aureus, Enterococcus faecalis and Escherichia coli isolates from orthopaedic implant-associated infections. International journal of antimicrobial agents 2019, 54, (3), 329–337. [Google Scholar] [CrossRef]
- Dias, R.; Eller, M.; Duarte, V.; Pereira, A.; Silva, C.; Mantovani, H.; Oliveira, L.; de A. M Silva, E.; De Paula, S., Use of phages against antibiotic‐resistant Staphylococcus aureus isolated from bovine mastitis. Journal of animal science 2013, 91, (8), 3930‐3939. [CrossRef]
- Porter, J.; Anderson, J.; Carter, L.; Donjacour, E.; Paros, M. In vitro evaluation of a novel bacteriophage cocktail as a preventative for bovine coliform mastitis. Journal of Dairy Science 2016, 99, (3), 2053–2062. [Google Scholar] [CrossRef]
- Amiri Fahliyani, S.; Beheshti-Maal, K.; Ghandehari, F. Novel lytic bacteriophages of Klebsiella oxytoca ABG-IAUF-1 as the potential agents for mastitis phage therapy. FEMS microbiology letters 2018, 365, (20), fny223. [Google Scholar] [CrossRef] [PubMed]
- Varela-Ortiz, D.F.; Barboza-Corona, J.E.; González-Marrero, J.; León-Galván, M.F.; Valencia-Posadas, M.; Lechuga-Arana, A.A.; Sánchez-Felipe, C.G.; Ledezma-García, F.; Gutiérrez-Chávez, A.J. Antibiotic susceptibility of Staphylococcus aureus isolated from subclinical bovine mastitis cases and in vitro efficacy of bacteriophage. Veterinary research communications 2018, 42, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Zou, W.; Zhang, M.; Xu, L.; Liu, F.; Li, X.; Wang, L.; Xu, Y. Evaluation of phage therapy in the treatment of Staphylococcus aureus-induced mastitis in mice. Folia microbiologica 2020, 65, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Keary, R.; Mcauliffe, O.; O'Mahony, J.; Coffey, A. Bacteriophage-Derived Peptidase CHAPK Eliminates and Prevents Staphylococcal Biofilms. 2013. [CrossRef]
- Canter, R.G.; Penney, J.; Tsai, L.-H. The road to restoring neural circuits for the treatment of Alzheimer's disease. Nature 2016, 539, (7628), 187–196. [Google Scholar] [CrossRef]
- De Strooper, B.; Karran, E. The cellular phase of Alzheimer’s disease. Cell 2016, 164, (4), 603–615. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, X.-L.; Yang, Q.-G.; Xu, W.-H.; Wang, F.; Chen, Y.-P.; Chen, G.-H. A peptide that binds specifically to the β-amyloid of Alzheimer's disease: selection and assessment of anti-β-amyloid neurotoxic effects. PLoS One 2011, 6, (11), e27649. [Google Scholar] [CrossRef] [PubMed]
- Van Groen, T.; Wiesehan, K.; Funke, S.A.; Kadish, I.; Nagel-Steger, L.; Willbold, D. Reduction of Alzheimer’s Disease Amyloid Plaque Load in Transgenic Mice by D3, ad-Enantiomeric Peptide Identified by Mirror Image Phage Display. ChemMedChem: Chemistry Enabling Drug Discovery 2008, 3, (12), 1848–1852. [Google Scholar] [CrossRef]
- Funke, S.A.; van Groen, T.; Kadish, I.; Bartnik, D.; Nagel-Steger, L.; Brener, O.; Sehl, T.; Batra-Safferling, R.; Moriscot, C.; Schoehn, G. Oral treatment with the D-enantiomeric peptide D3 improves the pathology and behavior of Alzheimer’s disease transgenic mice. ACS chemical neuroscience 2010, 1, (9), 639–648. [Google Scholar] [CrossRef]
- van Groen, T.; Kadish, I.; Funke, S.A.; Bartnik, D.; Willbold, D. Treatment with D3 removes amyloid deposits, reduces inflammation, and improves cognition in aged AβPP/PS1 double transgenic mice. Journal of Alzheimer's Disease 2013, 34, (3), 609–620. [Google Scholar] [CrossRef]
- Jiang, N.; Leithold, L.H.; Post, J.; Ziehm, T.; Mauler, J.; Gremer, L.; Cremer, M.; Schartmann, E.; Shah, N.J.; Kutzsche, J. Preclinical pharmacokinetic studies of the tritium labelled D-enantiomeric peptide D3 developed for the treatment of Alzheimer s disease. PloS one 2015, 10, (6), e0128553. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Ito, S.; Masuda, T.; Couraud, P.-O.; Ohtsuki, S. Novel cyclic peptides facilitating transcellular blood-brain barrier transport of macromolecules in vitro and in vivo. Journal of Controlled Release 2020, 321, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Li, Y.; Zhong, M.; Zhao, P.; Guo, C.; Xu, H.; Wang, T.; Gao, H. Brain targeting and Aβ binding bifunctional nanoparticles inhibit amyloid protein aggregation in APP/PS1 transgenic mice. ACS Chemical Neuroscience 2021, 12, (12), 2110–2121. [Google Scholar] [CrossRef]
- Zhang, C.; Wan, X.; Zheng, X.; Shao, X.; Liu, Q.; Zhang, Q.; Qian, Y. Dual-functional nanoparticles targeting amyloid plaques in the brains of Alzheimer's disease mice. Biomaterials 2014, 35, (1), 456–465. [Google Scholar] [CrossRef]
- Janelidze, S.; Stomrud, E.; Smith, R.; Palmqvist, S.; Mattsson, N.; Airey, D.C.; Proctor, N.K.; Chai, X.; Shcherbinin, S.; Sims, J.R. Cerebrospinal fluid p-tau217 performs better than p-tau181 as a biomarker of Alzheimer’s disease. Nature communications 2020, 11, (1), 1683. [Google Scholar] [CrossRef]
- Janelidze, S.; Mattsson, N.; Palmqvist, S.; Smith, R.; Beach, T.G.; Serrano, G.E.; Chai, X.; Proctor, N.K.; Eichenlaub, U.; Zetterberg, H. Plasma P-tau181 in Alzheimer’s disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nature medicine 2020, 26, (3), 379–386. [Google Scholar] [CrossRef]
- Carmody, C.M.; Goddard, J.M.; Nugen, S.R. Bacteriophage capsid modification by genetic and chemical methods. Bioconjugate chemistry 2021, 32, (3), 466–481. [Google Scholar] [CrossRef]
- Veeranarayanan, S.; Azam, A.H.; Kiga, K.; Watanabe, S.; Cui, L. Bacteriophages as solid tumor theragnostic agents. International Journal of Molecular Sciences 2021, 23, (1), 402. [Google Scholar] [CrossRef]
- Marino, S.M.; Gladyshev, V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. Journal of molecular biology 2010, 404, (5), 902–916. [Google Scholar] [CrossRef]
- Liyanagedera, S.B.; Williams, J.; Wheatley, J.P.; Biketova, A.Y.; Hasan, M.; Sagona, A.P.; Purdy, K.J.; Puxty, R.J.; Feher, T.; Kulkarni, V. SpyPhage: a cell-free TXTL platform for rapid engineering of targeted phage therapies. ACS Synthetic Biology 2022, 11, (10), 3330–3342. [Google Scholar] [CrossRef]
- Sanmukh, S.G.; Felisbino, S.L. Bacteriophages in cancer biology and therapies. Clin Oncol 2017, 2, 1295. [Google Scholar]
- Abbineni, G.; Modali, S.; Safiejko-Mroczka, B.; Petrenko, V.A.; Mao, C. Evolutionary selection of new breast cancer cell-targeting peptides and phages with the cell-targeting peptides fully displayed on the major coat and their effects on actin dynamics during cell internalization. Molecular pharmaceutics 2010, 7, (5), 1629–1642. [Google Scholar] [CrossRef]
- Sunderland, K.S.; Yang, M.; Mao, C. Phage-enabled nanomedicine: from probes to therapeutics in precision medicine. Angewandte Chemie International Edition 2017, 56, (8), 1964–1992. [Google Scholar] [CrossRef]
- Sanmukh, S.G.; Dos Santos, N.J.; Barquilha, C.N.; De Carvalho, M.; Dos Reis, P.P.; Delella, F.K.; Carvalho, H.F.; Latek, D.; Fehér, T.; Felisbino, S.L. Bacterial RNA virus MS2 exposure increases the expression of cancer progression genes in the LNCaP prostate cancer cell line. Oncology Letters 2023, 25, (2), 1–14. [Google Scholar] [CrossRef]
- Gibb, B.; Hyman, P.; Schneider, C.L. The many applications of engineered bacteriophages—An overview. Pharmaceuticals 2021, 14, (7), 634. [Google Scholar] [CrossRef]
- Manivannan, A.C.; Dhandapani, R.; Velmurugan, P.; Thangavelu, S.; Paramasivam, R.; Ragunathan, L.; Saravanan, M. Phage in cancer treatment–Biology of therapeutic phage and screening of tumor targeting peptide. Expert Opinion on Drug Delivery 2022, 19, (7), 873–882. [Google Scholar] [CrossRef]
- Choi, D.S.; Jin, H.-E.; Yoo, S.Y.; Lee, S.-W. Cyclic RGD peptide incorporation on phage major coat proteins for improved internalization by HeLa cells. Bioconjugate chemistry 2014, 25, (2), 216–223. [Google Scholar] [CrossRef]
- Moradi, M.; Ghaleh, H.E.G.; Bolandian, M.; Dorostkar, R. New role of bacteriophages in medical oncology. Biotechnology and Applied Biochemistry 2023. [CrossRef] [PubMed]
- Azizi, M.; Shahgolzari, M.; Fathi-Karkan, S.; Ghasemi, M.; Samadian, H. Multifunctional plant virus nanoparticles: An emerging strategy for therapy of cancer. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology 2022, e1872. [CrossRef]
- Phumyen, A.; Jantasorn, S.; Jumnainsong, A.; Leelayuwat, C. Doxorubicin-conjugated bacteriophages carrying anti-MHC class I chain-related A for targeted cancer therapy in vitro. OncoTargets and therapy 2014, 2183–2195. [Google Scholar] [CrossRef]
- Ashley, C.E.; Carnes, E.C.; Phillips, G.K.; Durfee, P.N.; Buley, M.D.; Lino, C.A.; Padilla, D.P.; Phillips, B.; Carter, M.B.; Willman, C.L. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS nano 2011, 5, (7), 5729–5745. [Google Scholar] [CrossRef]
- Bar, H.; Yacoby, I.; Benhar, I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC biotechnology 2008, 8, (1), 1–14. [Google Scholar] [CrossRef]
- Du, B.; Han, H.; Wang, Z.; Kuang, L.; Wang, L.; Yu, L.; Wu, M.; Zhou, Z.; Qian, M. Targeted drug delivery to hepatocarcinoma in vivo by phage-displayed specific binding peptide. Molecular Cancer Research 2010, 8, (2), 135–144. [Google Scholar] [CrossRef]
- Suthiwangcharoen, N.; Li, T.; Li, K.; Thompson, P.; You, S.; Wang, Q. M13 bacteriophage-polymer nanoassemblies as drug delivery vehicles. Nano Research 2011, 4, 483–493. [Google Scholar] [CrossRef]
- Kim, H.; Choi, H.; Bae, Y.; Kang, S. Development of target-tunable P22 VLP-based delivery nanoplatforms using bacterial superglue. Biotechnology and bioengineering 2019, 116, (11), 2843–2851. [Google Scholar] [CrossRef]
- Botstein, D.; Waddell, C.H.; King, J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22: I. Genes, proteins, structures and DNA maturation. Journal of molecular biology 1973, 80, (4), 669–695. [Google Scholar] [CrossRef]
- Kim, K.R.; Lee, A.S.; Kim, S.M.; Heo, H.R.; Kim, C.S. Virus-like nanoparticles as a theranostic platform for cancer. Frontiers in Bioengineering and Biotechnology 2023, 10, 1106767. [Google Scholar] [CrossRef] [PubMed]
- Kolesanova, E.; Melnikova, M.; Bolshakova, T.; Rybalkina, E.Y.; Sivov, I. Bacteriophage MS2 as a tool for targeted delivery in solid tumor chemotherapy. Acta Naturae (англoязычная версия) 2019, 11, (2 (41)), 98–101. [Google Scholar] [CrossRef]
- Catala, A.; Dzieciatkowska, M.; Wang, G.; Gutierrez-Hartmann, A.; Simberg, D.; Hansen, K.C.; D’Alessandro, A.; Catalano, C.E. Targeted intracellular delivery of trastuzumab using designer phage lambda nanoparticles alters cellular programs in human breast cancer cells. ACS nano 2021, 15, (7), 11789–11805. [Google Scholar] [CrossRef]
- Pardridge, W.M. The blood-brain barrier: bottleneck in brain drug development. NeuroRx 2005, 2, 3–14. [Google Scholar] [CrossRef]
- Anand, P.; O’Neil, A.; Lin, E.; Douglas, T.; Holford, M. Tailored delivery of analgesic ziconotide across a blood brain barrier model using viral nanocontainers. Scientific reports 2015, 5, (1), 12497. [Google Scholar] [CrossRef]
- Apawu, A.K.; Curley, S.M.; Dixon, A.R.; Hali, M.; Sinan, M.; Braun, R.D.; Castracane, J.; Cacace, A.T.; Bergkvist, M.; Holt, A.G. MRI compatible MS2 nanoparticles designed to cross the blood–brain-barrier: Providing a path towards tinnitus treatment. Nanomedicine: Nanotechnology, Biology and Medicine 2018, 14, (7), 1999–2008. [Google Scholar] [CrossRef]
- Tsedev, U.; Lin, C.-W.; Hess, G.T.; Sarkaria, J.N.; Lam, F.C.; Belcher, A.M. Phage particles of controlled length and genome for in vivo targeted glioblastoma imaging and therapeutic delivery. ACS nano 2022, 16, (8), 11676–11691. [Google Scholar] [CrossRef]
- Tuyen Ho, M.; Barrett, A.; Wang, Y.; Hu, Q. Bioinspired and Biomimetic Gene Delivery Systems. ACS Applied Bio Materials 2023. [CrossRef]
- Larocca, D.; Witte, A.; Johnson, W.; Pierce, G.F.; Baird, A. Targeting bacteriophage to mammalian cell surface receptors for gene delivery. Human gene therapy 1998, 9, (16), 2393–2399. [Google Scholar] [CrossRef]
- Lankes, H.; Zanghi, C.; Santos, K.; Capella, C.; Duke, C.; Dewhurst, S. In vivo gene delivery and expression by bacteriophage lambda vectors. Journal of applied microbiology 2007, 102, (5), 1337–1349. [Google Scholar] [CrossRef]
- Bedi, D.; Gillespie, J.W.; Petrenko Jr, V.A.; Ebner, A.; Leitner, M.; Hinterdorfer, P.; Petrenko, V.A. Targeted delivery of siRNA into breast cancer cells via phage fusion proteins. Molecular pharmaceutics 2013, 10, (2), 551–559. [Google Scholar] [CrossRef]
- Yata, T.; Lee, E.L.; Suwan, K.; Syed, N.; Asavarut, P.; Hajitou, A. Modulation of extracellular matrix in cancer is associated with enhanced tumor cell targeting by bacteriophage vectors. Molecular cancer 2015, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-Y.; Pan, Y.-C.; Hsiao, Y.-H.; Lim, S.-K.; Cheng, T.-W.; Huang, S.-W.; Wu, S.M.-Y.; Sun, C.-P.; Tao, M.-H.; Mou, K.Y. Improvement of Gene Delivery by Minimal Bacteriophage Particles. ACS nano 2023, 17, (15), 14532–14544. [Google Scholar] [CrossRef]
- Hajitou, A.; Trepel, M.; Lilley, C.E.; Soghomonyan, S.; Alauddin, M.M.; Marini, F.C.; Restel, B.H.; Ozawa, M.G.; Moya, C.A.; Rangel, R. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell 2006, 125, (2), 385–398. [Google Scholar] [CrossRef]
- Przystal, J.M.; Waramit, S.; Pranjol, M.Z.I.; Yan, W.; Chu, G.; Chongchai, A.; Samarth, G.; Olaciregui, N.G.; Tabatabai, G.; Carcaboso, A.M. Efficacy of systemic temozolomide-activated phage-targeted gene therapy in human glioblastoma. EMBO Molecular Medicine 2019, 11, (4), e8492. [Google Scholar] [CrossRef]
- Qazi, S.; Miettinen, H.M.; Wilkinson, R.A.; McCoy, K.; Douglas, T.; Wiedenheft, B. Programmed self-assembly of an active P22-Cas9 nanocarrier system. Molecular pharmaceutics 2016, 13, (3), 1191–1196. [Google Scholar] [CrossRef]
- Haque, F.; Hu, H.; Guo, P. Bacteriophage RNA Leading the Way in RNA Nanotechnology for Targeted Cancer Therapy. In RNA Nanotechnology and Therapeutics, CRC Press: 2022; pp 473-483. [CrossRef]
- Wang, G.; Jia, T.; Xu, X.; Chang, L.; Zhang, R.; Fu, Y.; Li, Y.; Yang, X.; Zhang, K.; Lin, G. Novel miR-122 delivery system based on MS2 virus like particle surface displaying cell-penetrating peptide TAT for hepatocellular carcinoma. Oncotarget 2016, 7, (37), 59402. [Google Scholar] [CrossRef]
- Sharifi, M.; Alizadeh, A.A.; Hamzeh-Mivehroud, M.; Dastmalchi, S. Construction of a Bacteriophage-Derived Vector with Potential applications in Targeted Drug Delivery and Cell Imaging. 2022. [CrossRef]
- Gallo, J.; Villasante, A. Recent Advances in Biomimetic Nanocarrier-Based Photothermal Therapy for Cancer Treatment. International Journal of Molecular Sciences 2023, 24, (20), 15484. [Google Scholar] [CrossRef]
- Yue, H.; Li, Y.; Yang, M.; Mao, C. T7 phage as an emerging nanobiomaterial with genetically tunable target specificity. Advanced Science 2022, 9, (4), 2103645. [Google Scholar] [CrossRef]
- Gandra, N.; Abbineni, G.; Qu, X.; Huai, Y.; Wang, L.; Mao, C. Bacteriophage bionanowire as a carrier for both cancer-targeting peptides and photosensitizers and its use in selective cancer cell killing by photodynamic therapy. Small 2013, 9, (2), 215–221. [Google Scholar] [CrossRef]
- Qu, X.; Qiu, P.; Zhu, Y.; Yang, M.; Mao, C. Guiding nanomaterials to tumors for breast cancer precision medicine: from tumor-targeting small-molecule discovery to targeted nanodrug delivery. NPG Asia materials 2017, 9, (12), e452–e452. [Google Scholar] [CrossRef]
- Cao, B.; Xu, H.; Yang, M.; Mao, C. Virus-based cancer therapeutics for targeted photodynamic therapy. Virus-Derived Nanoparticles for Advanced Technologies: Methods and Protocols 2018, 643-652. [CrossRef]
- Stephanopoulos, N.; Tong, G.J.; Hsiao, S.C.; Francis, M.B. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS nano 2010, 4, (10), 6014–6020. [Google Scholar] [CrossRef]
- Hou, X.-L.; Xie, X.-T.; Tan, L.-F.; Zhang, F.; Fan, J.-X.; Chen, W.; Hu, Y.-G.; Zhao, Y.-D.; Liu, B.; Xu, Q.-R. T4 phage display technology for enhanced photodynamic therapy of breast cancer. ACS Materials Letters 2023, 5, (8), 2270–2281. [Google Scholar] [CrossRef]
- Ulfo, L.; Cantelli, A.; Petrosino, A.; Costantini, P.E.; Nigro, M.; Starinieri, F.; Turrini, E.; Zadran, S.K.; Zuccheri, G.; Saporetti, R. Orthogonal nanoarchitectonics of M13 phage for receptor targeted anticancer photodynamic therapy. Nanoscale 2022, 14, (3), 632–641. [Google Scholar] [CrossRef]
- Bortot, B.; Apollonio, M.; Baj, G.; Andolfi, L.; Zupin, L.; Crovella, S.; di Giosia, M.; Cantelli, A.; Saporetti, R.; Ulfo, L. Advanced photodynamic therapy with an engineered M13 phage targeting EGFR: Mitochondrial localization and autophagy induction in ovarian cancer cell lines. Free Radical Biology and Medicine 2022, 179, 242–251. [Google Scholar] [CrossRef]
- Sioud, M.; Zhang, Q. Precision Killing of M2 Macrophages with Phage-Displayed Peptide-Photosensitizer Conjugates. Cancers 2023, 15, (7), 2009. [Google Scholar] [CrossRef]
- Hashiguchi, S.; Yamaguchi, Y.; Takeuchi, O.; Akira, S.; Sugimura, K. Immunological basis of M13 phage vaccine: Regulation under MyD88 and TLR9 signaling. Biochemical and biophysical research communications 2010, 402, (1), 19–22. [Google Scholar] [CrossRef]
- Paludan, S.R.; Bowie, A.G. Immune sensing of DNA. Immunity 2013, 38, (5), 870–880. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Frontiers in immunology 2014, 5, 461. [Google Scholar] [CrossRef]
- Popescu, M.; Van Belleghem, J.D.; Khosravi, A.; Bollyky, P.L. Bacteriophages and the immune system. Annual Review of Virology 2021, 8, 415–435. [Google Scholar] [CrossRef]
- Champagne-Jorgensen, K.; Luong, T.; Darby, T.; Roach, D.R. Immunogenicity of bacteriophages. Trends in Microbiology 2023. [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nature immunology 2013, 14, (10), 1014–1022. [Google Scholar] [CrossRef]
- Dor-On, E.; Solomon, B. Targeting glioblastoma via intranasal administration of Ff bacteriophages. Frontiers in Microbiology 2015, 6, 530. [Google Scholar] [CrossRef] [PubMed]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, (6434), eaat9691. [Google Scholar] [CrossRef]
- Ghambashidze, K.; Chikhladze, R.; Saladze, T.; Hoopes, P.; Shubitidze, F. E. coli Phagelysate: A Primer to Enhance Nanoparticles and Drug Deliveries in Tumor. Cancers 2023, 15, 2315, In 2023. [Google Scholar] [CrossRef]
- Garg, P. Filamentous bacteriophage: A prospective platform for targeting drugs in phage-mediated cancer therapy. Journal of Cancer Research & Therapeutics 2019, 15. [CrossRef]
- Gaubin, M.; Fanutti, C.; Mishal, Z.; Durrbach, A.; De Berardinis, P.; Sartorius, R.; Del Pozzo, G.; Guardiola, J.; Perham, R.N.; Piatier-Tonneau, D. Processing of filamentous bacteriophage virions in antigen-presenting cells targets both HLA class I and class II peptide loading compartments. DNA and cell biology 2003, 22, (1), 11–18. [Google Scholar] [CrossRef]
- Goracci, M.; Pignochino, Y.; Marchiò, S. Phage display-based nanotechnology applications in cancer immunotherapy. Molecules 2020, 25, (4), 843. [Google Scholar] [CrossRef]
- Wang, J.; Lamolinara, A.; Conti, L.; Giangrossi, M.; Cui, L.; Morelli, M.B.; Amantini, C.; Falconi, M.; Bartolacci, C.; Andreani, C. HER2-Displaying M13 Bacteriophages induce Therapeutic Immunity against Breast Cancer. Cancers 2022, 14, (16), 4054. [Google Scholar] [CrossRef]
- Barati, N.; Razazan, A.; Nicastro, J.; Slavcev, R.; Arab, A.; Mosaffa, F.; Nikpoor, A.R.; Badiee, A.; Jaafari, M.R.; Behravan, J. Immunogenicity and antitumor activity of the superlytic λF7 phage nanoparticles displaying a HER2/neu-derived peptide AE37 in a tumor model of BALB/c mice. Cancer letters 2018, 424, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Pouyanfard, S.; Bamdad, T.; Hashemi, H.; Bandehpour, M.; Kazemi, B. Induction of protective anti-CTL epitope responses against HER-2-positive breast cancer based on multivalent T7 phage nanoparticles. PLoS One 2012, 7, (11), e49539. [Google Scholar] [CrossRef] [PubMed]
- Bartolacci, C.; Andreani, C.; Curcio, C.; Occhipinti, S.; Massaccesi, L.; Giovarelli, M.; Galeazzi, R.; Iezzi, M.; Tilio, M.; Gambini, V. Phage-based anti-HER2 vaccination can circumvent immune tolerance against breast cancer. Cancer Immunology Research 2018, 6, (12), 1486–1498. [Google Scholar] [CrossRef]
- Razazan, A.; Nicastro, J.; Slavcev, R.; Barati, N.; Arab, A.; Mosaffa, F.; Jaafari, M.R.; Behravan, J. Lambda bacteriophage nanoparticles displaying GP2, a HER2/neu derived peptide, induce prophylactic and therapeutic activities against TUBO tumor model in mice. Scientific reports 2019, 9, (1), 2221. [Google Scholar] [CrossRef]
- Jung, E.; Chung, Y.H.; Steinmetz, N.F. TLR Agonists Delivered by Plant Virus and Bacteriophage Nanoparticles for Cancer Immunotherapy. Bioconjugate Chemistry 2023, 34, (9), 1596–1605. [Google Scholar] [CrossRef]
- Protective, V.-L.P.I. Nonmethylated CG Motifs Packaged into. J Immunol 2004, 172, 1777–1785. [Google Scholar] [CrossRef]
- Wang, H.Y.; Chang, Y.-C.; Hu, C.-W.; Kao, C.-Y.; Yu, Y.-A.; Lim, S.-K.; Mou, K.Y. Development of a novel cytokine vehicle using filamentous phage display for colorectal cancer treatment. ACS Synthetic Biology 2021, 10, (8), 2087–2095. [Google Scholar] [CrossRef]
- Shahrivarkevishahi, A.; Luzuriaga, M.A.; Herbert, F.C.; Tumac, A.C.; Brohlin, O.R.; Wijesundara, Y.H.; Adlooru, A.V.; Benjamin, C.; Lee, H.; Parsamian, P. PhotothermalPhage: a virus-based photothermal therapeutic agent. Journal of the American Chemical Society 2021, 143, (40), 16428–16438. [Google Scholar] [CrossRef]
- Dong, X.; Pan, P.; Zhang, Q.; Ye, J.-J.; Zhang, X.-Z. Engineered living bacteriophage-enabled self-adjuvanting hydrogel for remodeling tumor microenvironment and cancer therapy. Nano Letters 2023, 23, (4), 1219–1228. [Google Scholar] [CrossRef]
- Neal, R.; Tharmanathan, P.; France, B.; Din, N.; Cotton, S.; Fallon-Ferguson, J.; Hamilton, W.; Hendry, A.; Hendry, M.; Lewis, R. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. British journal of cancer 2015, 112, (1), S92–S107. [Google Scholar] [CrossRef]
- Min, J.; Jung, H.; Shin, H.-H.; Cho, G.; Cho, H.; Kang, S. Implementation of p22 viral capsids as intravascular magnetic resonance T 1 contrast conjugates via site-selective attachment of Gd (III)-chelating agents. Biomacromolecules 2013, 14, (7), 2332–2339. [Google Scholar] [CrossRef]
- Carrico, Z.M.; Farkas, M.E.; Zhou, Y.; Hsiao, S.C.; Marks, J.D.; Chokhawala, H.; Clark, D.S.; Francis, M.B. N-Terminal labeling of filamentous phage to create cancer marker imaging agents. ACS nano 2012, 6, (8), 6675–6680. [Google Scholar] [CrossRef]
- Robertson, K.L.; Soto, C.M.; Archer, M.J.; Odoemene, O.; Liu, J.L. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. Bioconjugate chemistry 2011, 22, (4), 595–604. [Google Scholar] [CrossRef]
- Aanei, I.L.; ElSohly, A.M.; Farkas, M.E.; Netirojjanakul, C.; Regan, M.; Taylor Murphy, S.; O’Neil, J.P.; Seo, Y.; Francis, M.B. Biodistribution of antibody-MS2 viral capsid conjugates in breast cancer models. Molecular pharmaceutics 2016, 13, (11), 3764–3772. [Google Scholar] [CrossRef]
- Asar, M.; Newton-Northup, J.; Deutscher, S.; Soendergaard, M. Ovarian cancer targeting phage for in vivo near-infrared optical imaging. Diagnostics 2019, 9, (4), 183. [Google Scholar] [CrossRef]
- El-Sayed, A.; Bernhard, W.; Barreto, K.; Gonzalez, C.; Hill, W.; Pastushok, L.; Fonge, H.; Geyer, C.R. Evaluation of antibody fragment properties for near-infrared fluorescence imaging of HER3-positive cancer xenografts. Theranostics 2018, 8, (17), 4856. [Google Scholar] [CrossRef]
- Lee, K.J.; Lee, J.H.; Chung, H.K.; Ju, E.J.; Song, S.Y.; Jeong, S.-Y.; Choi, E.K. Application of peptide displaying phage as a novel diagnostic probe for human lung adenocarcinoma. Amino Acids 2016, 48, 1079–1086. [Google Scholar] [CrossRef]
- Foglizzo, V.; Marchiò, S. Bacteriophages as therapeutic and diagnostic vehicles in cancer. Pharmaceuticals 2021, 14, (2), 161. [Google Scholar] [CrossRef]
- Ghosh, D.; Kohli, A.G.; Moser, F.; Endy, D.; Belcher, A.M. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS synthetic biology 2012, 1, (12), 576–582. [Google Scholar] [CrossRef]
- Yadav, D.; Sankaranarayanan, S.; Thanekar, A.; Rengan, A. Bioinspired gold coated phage nanosomes for anti-microbial and anti-cancer theranostics. Materials Today Nano 2023, 23, 100348. [Google Scholar] [CrossRef]
- Martel, B.; Moineau, S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic acids research 2014, 42, (14), 9504–9513. [Google Scholar] [CrossRef]
- Mitsunaka, S.; Yamazaki, K.; Pramono, A.K.; Ikeuchi, M.; Kitao, T.; Ohara, N.; Kubori, T.; Nagai, H.; Ando, H. Synthetic engineering and biological containment of bacteriophages. Proceedings of the National Academy of Sciences 2022, 119, (48), e2206739119. [Google Scholar] [CrossRef]
- Wang, C.; Xia, Q.; Zhang, Q.; Qu, Y.; Su, S.; Cheng, J.K.; Hughes, N.W.; Cong, L. CRISPR-Cas12a System With Synergistic Phage Recombination Proteins for Multiplex Precision Editing in Human Cells. Frontiers in Cell and Developmental Biology 2022, 9, 719705. [Google Scholar] [CrossRef]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic engineering of bacteriophages against infectious diseases. Frontiers in microbiology 2019, 10, 954. [Google Scholar] [CrossRef]
- Duong, M.M.; Carmody, C.M.; Ma, Q.; Peters, J.E.; Nugen, S.R. Optimization of T4 phage engineering via CRISPR/Cas9. Scientific Reports 2020, 10, (1), 18229. [Google Scholar] [CrossRef]
- Krishnamurthy, M.; Moore, R.T.; Rajamani, S.; Panchal, R.G. Bacterial genome engineering and synthetic biology: combating pathogens. BMC microbiology 2016, 16, (1), 1–11. [Google Scholar] [CrossRef]
- Shen, J.; Zhou, J.; Chen, G.-Q.; Xiu, Z.-L. Efficient genome engineering of a virulent Klebsiella bacteriophage using CRISPR-Cas9. Journal of Virology 2018, 92, (17), 10.1128/jvi. 00534–18. [Google Scholar] [CrossRef]
- Hoshiga, F.; Yoshizaki, K.; Takao, N.; Miyanaga, K.; Tanji, Y. Modification of T2 phage infectivity toward Escherichia coli O157: H7 via using CRISPR/Cas9. FEMS microbiology letters 2019, 366, (4), fnz041. [Google Scholar] [CrossRef]
- Ali, Y.; Inusa, I.; Sanghvi, G.; Mandaliya, V.; Bishoyi, A.K. The current status of phage therapy and its advancement towards establishing standard antimicrobials for combating multi drug-resistant bacterial pathogens. Microbial Pathogenesis 2023, 106199. [Google Scholar] [CrossRef]
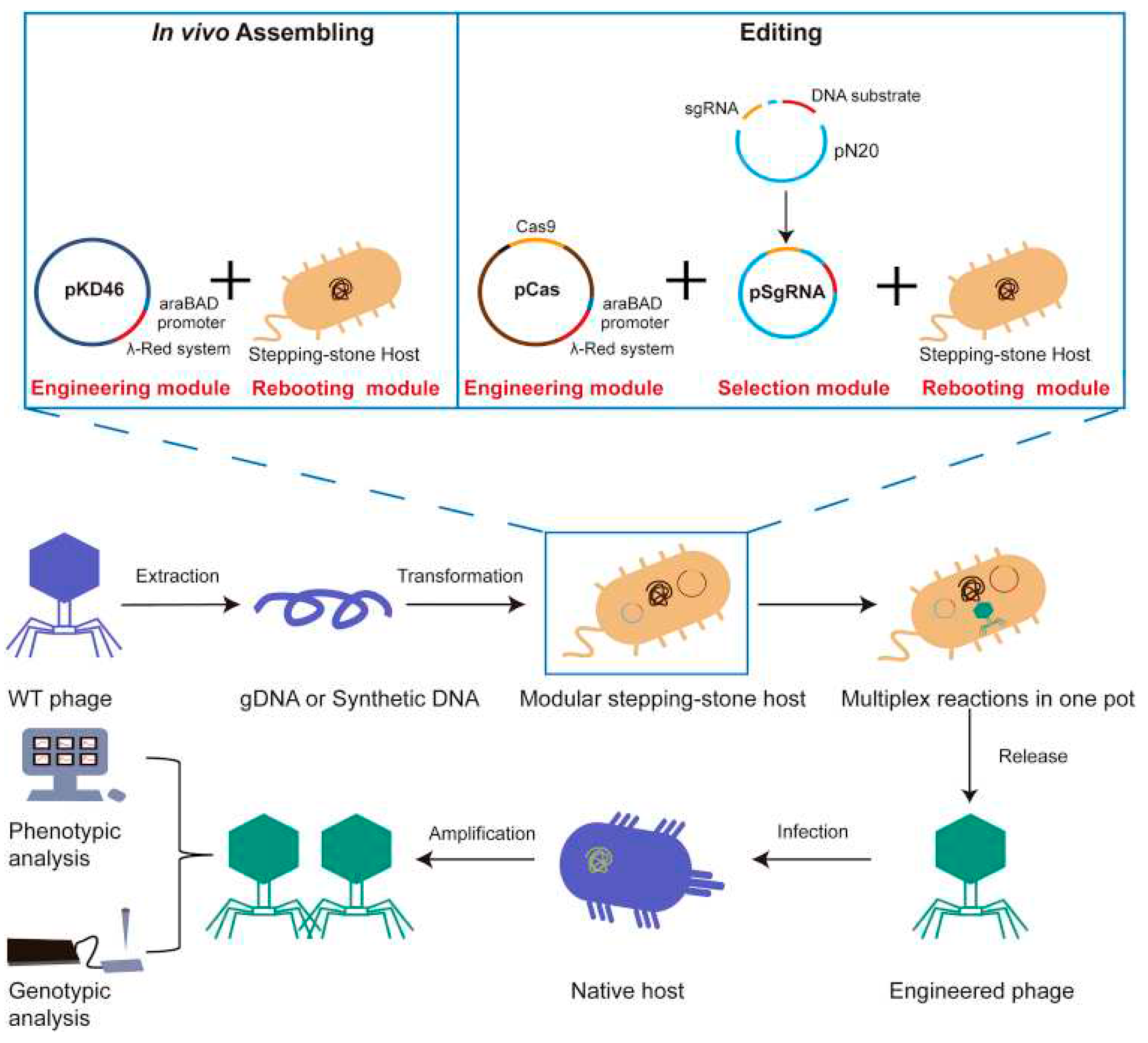
- Cheng, L.; Deng, Z.; Tao, H.; Song, W.; Xing, B.; Liu, W.; Kong, L.; Yuan, S.; Ma, Y.; Wu, Y. Harnessing stepping-stone hosts to engineer, select, and reboot synthetic bacteriophages in one pot. Cell Reports Methods 2022, 2, (5). [Google Scholar] [CrossRef]
- Kilcher, S.; Studer, P.; Muessner, C.; Klumpp, J.; Loessner, M.J. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proceedings of the National Academy of Sciences 2018, 115, (3), 567–572. [Google Scholar] [CrossRef]
- Smith, H.W.; Huggins, M. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. Microbiology 1982, 128, (2), 307–318. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, (2), 66–85. [Google Scholar] [CrossRef]
- El-Shibiny, A.; El-Sahhar, S. Bacteriophages: the possible solution to treat infections caused by pathogenic bacteria. Canadian journal of microbiology 2017, 63, (11), 865–879. [Google Scholar] [CrossRef]
- Dąbrowska, K. Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Medicinal research reviews 2019, 39, (5), 2000–2025. [Google Scholar] [CrossRef]
- Onsea, J.; Uyttebroek, S.; Chen, B.; Wagemans, J.; Lood, C.; Van Gerven, L.; Spriet, I.; Devolder, D.; Debaveye, Y.; Depypere, M. Bacteriophage therapy for difficult-to-treat infections: the implementation of a multidisciplinary phage task force (the PHAGEFORCE study protocol). Viruses 2021, 13, (8), 1543. [Google Scholar] [CrossRef]
- Khatami, A.; Foley, D.A.; Warner, M.S.; Barnes, E.H.; Peleg, A.Y.; Li, J.; Stick, S.; Burke, N.; Lin, R.C.; Warning, J. Standardised treatment and monitoring protocol to assess safety and tolerability of bacteriophage therapy for adult and paediatric patients (STAMP study): protocol for an open-label, single-arm trial. BMJ open 2022, 12, (12), e065401. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, (1), 17–31. [Google Scholar] [CrossRef]
- Verbeken, G.; Pirnay, J.-P.; De Vos, D.; Jennes, S.; Zizi, M.; Lavigne, R.; Casteels, M.; Huys, I. Optimizing the European regulatory framework for sustainable bacteriophage therapy in human medicine. Archivum immunologiae et therapiae experimentalis 2012, 60, 161–172. [Google Scholar] [CrossRef] [PubMed]

| NCT number | Conditions | Interventions | Phases | Locations |
|---|---|---|---|---|
| NCT05277350 | E. coli infections/bloodstream infection | Phages cocktail | Phase 1 | Denmark |
| NCT05272579 | Necrotizing enterocolitis/microbial substitution | Fecal phages transfer | Early phase 1 | Denmark |
| NCT05269134 | Prosthetic joint infection | Single phage/phages cocktail | Phase 2/phase 3 | United States |
| NCT05269121 | Prosthetic joint infection/bacterial infections | Single phage/phages cocktail | Phase 1/phase 2 | United States |
| NCT05240300 | Atopic dermatitis | Single phage/phages cocktail | Phase 1/phase 2 | Israel |
| NCT05184764 | Bacteremia/Staphylococcus aureus/Staphylococcus aureus bacteremia/bacteremia due to Staphylococcus aureus | Phages cocktail | Phase 1/phase 2 | United States |
| NCT05182749 | Shigellosis | Phages cocktail | Phase 1/phase 2 | United States |
| NCT05177107 | Osteomyelitis/diabetic foot osteomyelitis | Single phage/phages cocktail | Phase 2 | United States |
| Generic Name | Product Name | Company | Format | Target | First indication | Year approved |
|---|---|---|---|---|---|---|
| Moxetumomab pasudotox | Lumoxiti | AstraZeneca/Medimmune | dsFv-PE38 | CD22 | Hairy cell leukemia | 2018 |
| Caplacizumab | Cablivi | Ablynx | VHH | vWF | Acquired thrombotic thrombocytopenic purpura | 2018 |
| Emapalumab | Gamifant | Novimmune | IgG1 | INFγ | Hemophagocytic lymphohistiocytosis | 2018 |
| Lanadelumab | Takhzyro | Dyax, Shire | IgG1 | pKal | Hereditary angioedema | 2018 |
| Inebilizumab | Uplizna | AstraZeneca/Medimmune, Viela Bio | IgG1 | CD19 | Neuromyelitis optica spectrum disorder | 2020 |
| Tralokinumab | Adbry | AstraZeneca/Medimmune, Leo Pharma | IgG4 | IL-13 | Asthma | 2021 |
| Faricimab | Vabysmo | Roche | Bi-Fab | VEGFA, Ang2 | Neovascular age-related macular degeneration, Diabetic macular edema |
2022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





