Submitted:
04 February 2024
Posted:
12 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
Results
2.1. Phenotype Variations of Yield-Related Traits in Vegetable Soybean
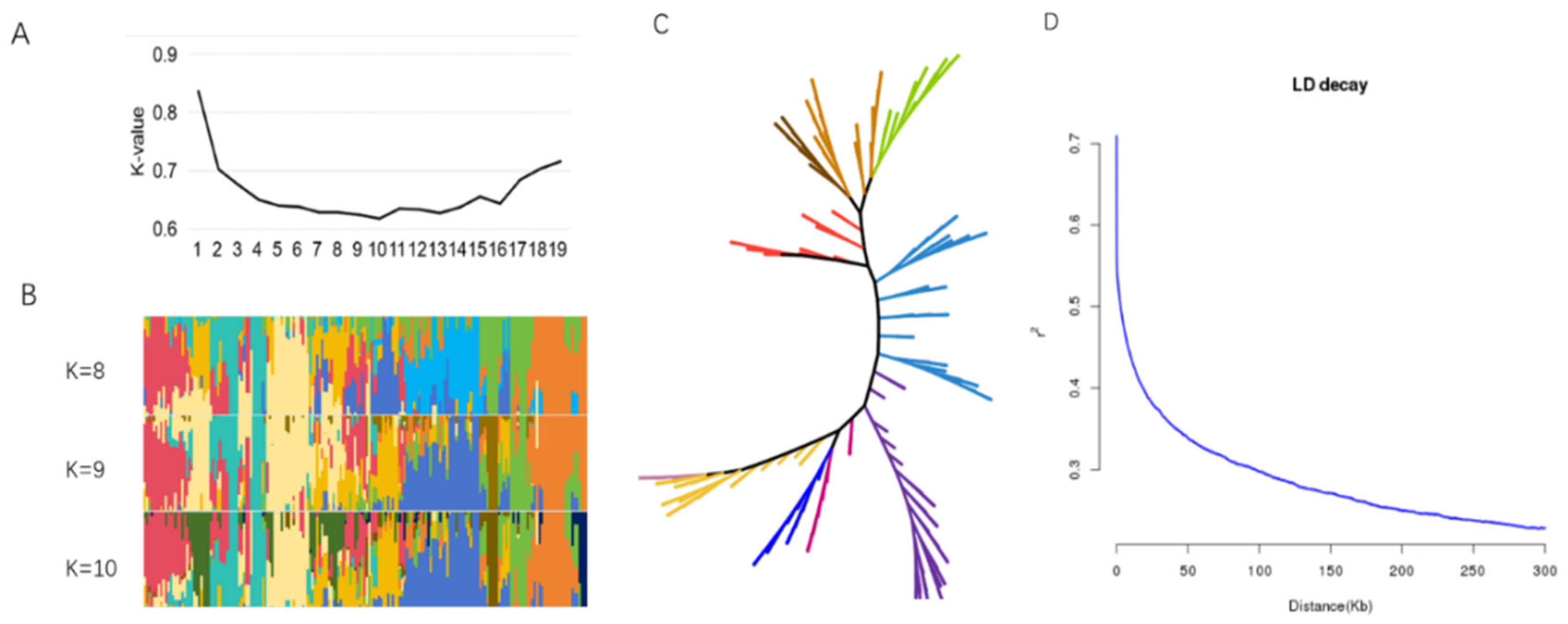
2.2. Population Structure Analysis of Vegetable Soybean
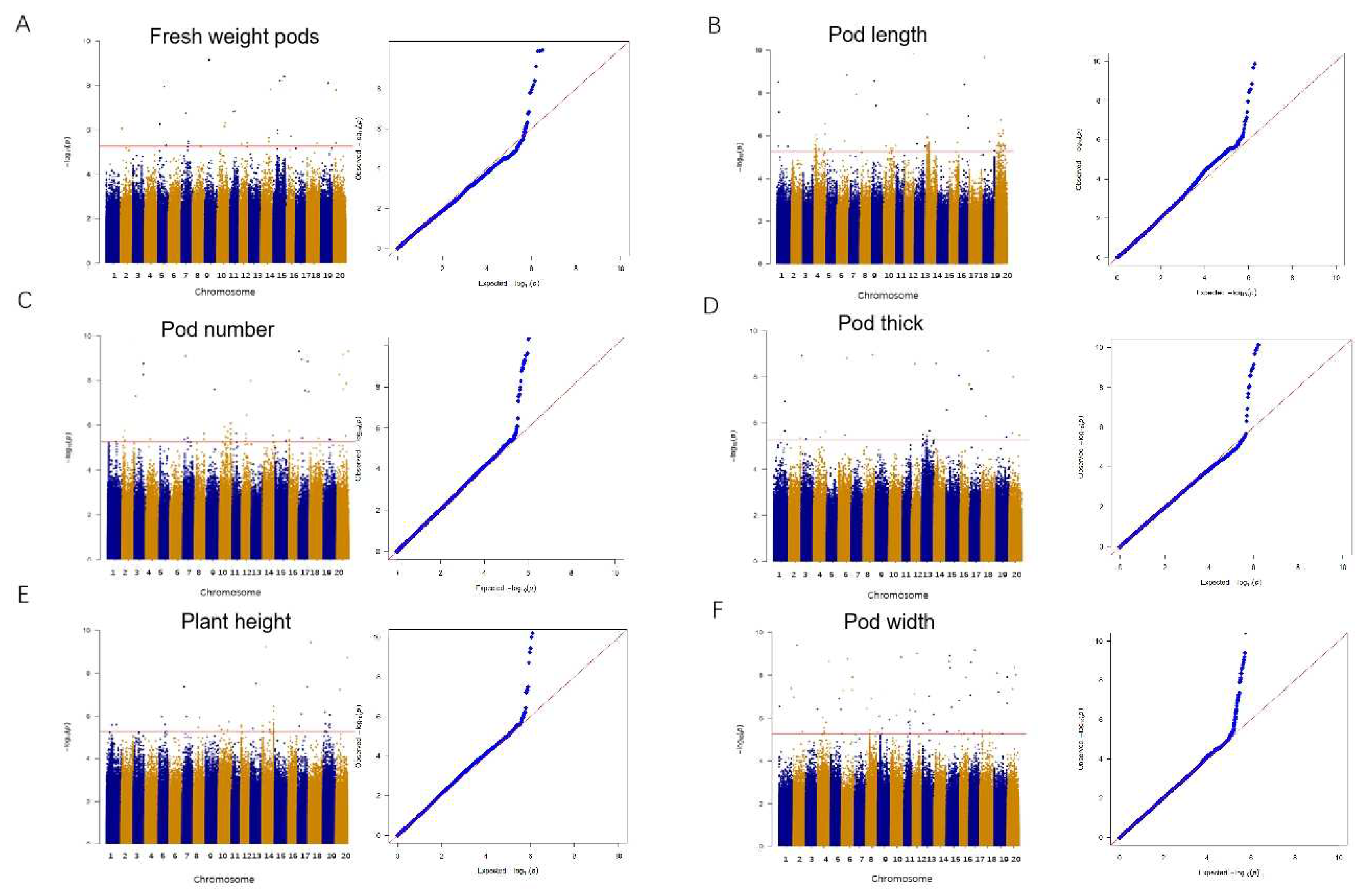
2.3. Yield-Related SNPs were Identified in Vegetable Soybean via GWAS
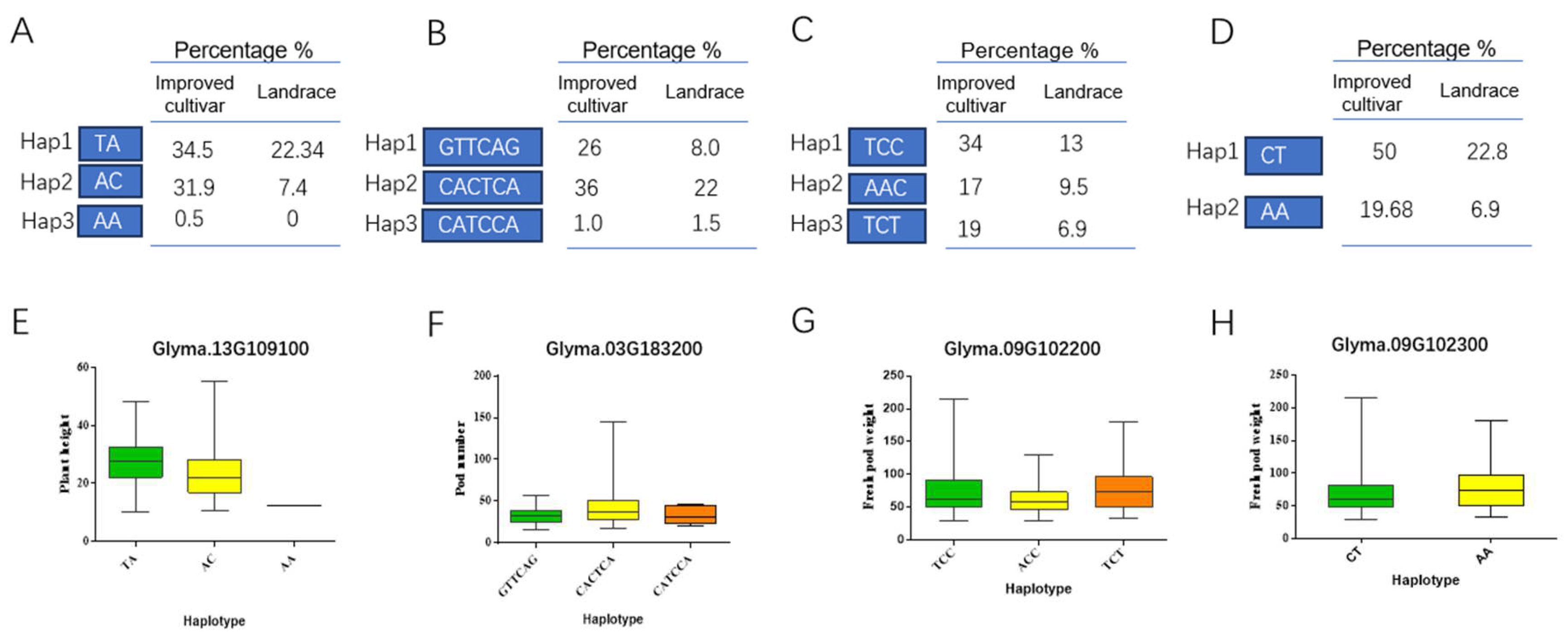
2.4. Candidate Genes Analysis Involved Ofyield Related Traits in Vegetable Soybean
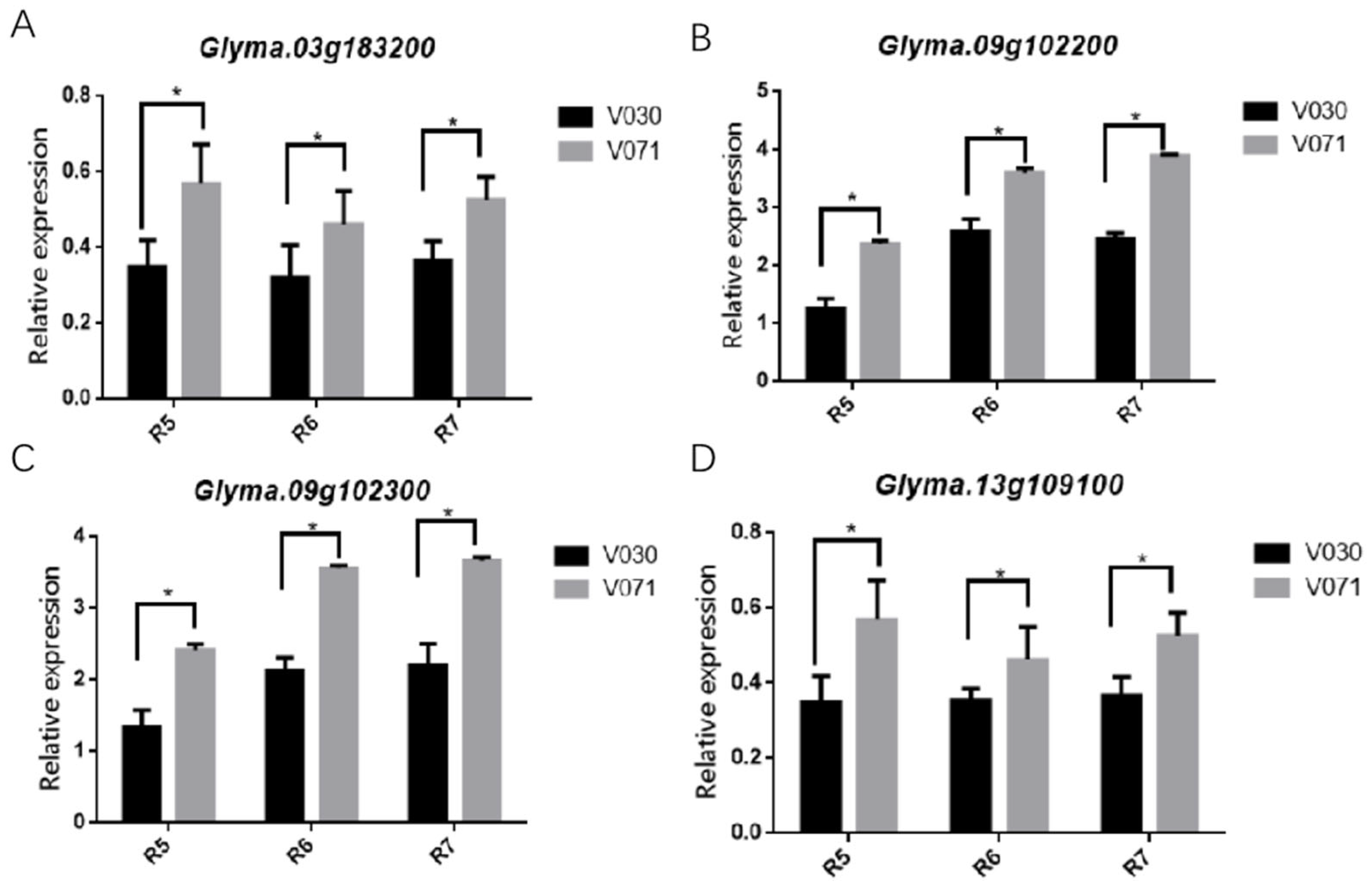
2.5. Different Expression Pattern of Candidate Genes were Observed in Pods and Stem
3. Discussion
4. Method
4.1. Plant Material and Field Experiments
4.2. Vegetable Soybean Yield-Related Trait Data Analysis
4.3. Population Analysis
4.4. Candidate Gene Analysis and Gene-Based Association Mapping
4.5. Real-Time Quantitative Reverse Transcription PCR Analysis for the Yield-Related Candidate Genes of Vegetable Soybean
5 Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu N., Niu Y., Zhang G., Feng Z., Bo Y., Lian J., Wang B., Gong Y. Genome sequencing and population resequencing provide insights into the genetic basis of domestication and diversity of vegetable soybean.Horticulture Research.2022;9. [CrossRef]
- Kao C., He S., Wang C., Lai Z., Lin D., Chen S. A modified roger’s distance algorithm for mixed quantitative-qualitative phenotypes to establish a core collection for Taiwanese vegetable soybeans.Frontiers in Plant Science.2021, 11:612106. [CrossRef]
- Zhang B., Lord N., Kuhar T., Duncan S., Huang H., Ross J., Rideout S., Arancibia R., Reiter M., Li S., et al. ‘VT Sweet’: A vegetable soybean cultivar for commercial edamame production in the mid-Atlantic USA.Journal of Plant Register.2022; 16:29–33. [CrossRef]
- Chen Z., Zhong W., Zhou Y., Ji P., Wan Y., Shi S., Yang Z., Gong Y., Mu F., Chen S. Integrative analysis of metabolome and transcriptome reveals the improvements of seed quality in vegetable soybean (Glycine max(L.) Merr.)Phytochemistry.2022, 200: 113216. [CrossRef]
- Xu W., Liu H., Li S., Zhang W., Wang Q., Zhang H., Liu X., Cui X., Chen X., Tang W., et al. GWAS and identification of candidate genes associated with seed soluble sugar content in vegetable soybean.Agronomy Journal.2022; 12:1470. [CrossRef]
- Nair RM, Boddepalli VN, Yan MR, Kumar V, Gill B, Pan RS, Wang C, Hartman GL, Silva E Souza R, Somta P. Global Status of Vegetable Soybean. Plants (Basel). 2023, 12(3):609. [CrossRef]
- Zhang H, Hao D, Sitoe HM, Yin Z, Hu Z, Zhang G, et al. Genetic dissection of the relationship between plant architecture and yield component traits in soybean (Glycine max) by association analysis across multiple environments.Plant Breed.2015; 134(5):564–572. [CrossRef]
- Zhao X, Dong H, Chang H, Zhao J, Teng W, Qiu L, Li W, Han Y. Genome wide association mapping and candidate gene analysis for hundred seed weight in soybean [Glycine max (L.) Merrill]. BMC Genomics. 2019 Aug 14;20(1):648. [CrossRef]
- Li X, Zhang X, Zhu L, Bu Y, Wang X, Zhang X, Zhou Y, Wang X, Guo N, Qiu L, Zhao J, Xing H. Genome-wide association study of four yield-related traits at the R6 stage in soybean. BMC Genetics. 2019, 29;20(1):39. [CrossRef]
- Cao Y, Jia S, Chen L, Zeng S, Zhao T, Karikari B. Identification of major genomic regions for soybean seed weight by genome-wide association study. Molecular Breed. 2022, 42(7):38. [CrossRef]
- Ayalew H, Schapaugh W, Vuong T, Nguyen HT. Genome-wide association analysis identified consistent QTL for seed yield in a soybean diversity panel tested across multiple environments. Plant Genome. 2022, 15(4): e20268. [CrossRef]
- Wang J, Hu B, Jing Y, Hu X, Guo Y, Chen J, Liu Y, Hao J, Li WX, Ning H. Detecting QTL and Candidate Genes for Plant Height in Soybean via Linkage Analysis and GWAS. Frontiers in Plant Science. 2022, 21; 12:803820. [CrossRef]
- Chang F, Guo C, Sun F, Zhang J, Wang Z, Kong J, He Q, Sharmin RA, Zhao T. Genome-Wide Association Studies for Dynamic Plant Height and Number of Nodes on the Main Stem in Summer Sowing Soybeans. Front Plant Sci. 2018,20;9:1184. [CrossRef]
- Liu Z, Li H, Fan X, Huang W, Yang J, Wen Z, Li Y, Guan R, Guo Y, Chang R, Wang D, Chen P, Wang S, Qiu LJ. Phenotypic characterization and genetic dissection of nine agronomic traits in Tokachi nagaha and its derived cultivars in soybean (Glycine max (L.) Merr.). Plant Science. 2017, 256:72-86. [CrossRef]
- Ayalew H, Schapaugh W, Vuong T, Nguyen HT. Genome-wide association analysis identified consistent QTL for seed yield in a soybean diversity panel tested across multiple environments. Plant Genome. 2022,15(4): e20268. [CrossRef]
- Wen Z, Boyse JF, Song Q, Cregan PB, Wang D. Genomic consequences of selection and genome-wide association mapping in soybean. BMC Genomics. 2015;16(1):671. [CrossRef]
- Zhang J, Song Q, Cregan PB, Jiang GL. Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max) Theor Appl Genet.2016;129(1):117–130. [CrossRef]
- Assefa T, Otyama PI, Brown AV, Kalberer SR, Kulkarni RS, Cannon SB. Genome-wide associations and epistatic interactions for internode number, plant height, seed weight and seed yield in soybean.BMC Genomics.2019; 20(1):527. [CrossRef]
- Zhao X, Li W, Zhao X, Wang J, Liu Z, Han Y, et al. Genome-wide association mapping and candidate gene analysis for seed shape in soybean (Glycine max). Crop Pasture Science.2019;70(8):684–693. [CrossRef]
- Zhang X, Ding W, Xue D, Li X, Zhou Y, Shen J, Feng J, Guo N, Qiu L, Xing H, Zhao J. Genome-wide association studies of plant architecture-related traits and 100-seed weight in soybean landraces. BMC Genom Data. 2021, 6;22(1):10. [CrossRef]
- Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci U S A. 2012;109(44):18198-18203. [CrossRef]
- Li YX, Li CH, Bradbury PJ, Liu XL, Lu F, Romay CM, et al. Identifcation of genetic variants associated with maize fowering time using an extremely large multi-genetic background population. Plant Journal. 2016; 86:391–402. [CrossRef]
- Lin C. Frozen edamame: global market conditions. USA: Second International Vegetable Soybean conference; 2001. pp. 93–97.
- Nguyen VQ. Edamame (vegetable green soybean)Austrália: Rural Industries Research & Development. The new rural industries: a handbook for farmers and investors; 2001. pp. 49–56.
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice.Nature Genetics.2016,48, 927. [CrossRef]
- Hao, H.; Li, Z.; Leng, C.; Lu, C.; Luo, H.; Liu, Y.; Wu, X.; Liu, Z.; Shang, L.; Jing, H.C. Sorghum breeding in the genomic era: Opportunities and challenges.TAG. Theor. Appl. Genetics. Theor. Und Angew. Genet.2021,134, 1899–1924. [CrossRef]
- Zeng T, Meng Z, Yue R, Lu S, Li W, Li W, Meng H, Sun Q. Genome wide association analysis for yield related traits in maize. BMC Plant Biology. 2022, 21;22(1):449. [CrossRef]
- Jiao, X.; Lyu, Y.; Wu, X.; Li, H.; Cheng, L.; Zhang, C.; Yuan, L.; Jiang, R.; Jiang, B.; Rengel, Z.; et al. Grain production versus resource and environmental costs: Towards increasing sustainability of nutrient use in China. Journal of Experiment Botany.2016,67, 4935–4949. [CrossRef]
- Eltaher S, Sallam A, Belamkar V, Emara HA, Nower AA, Salem KFM, Poland J, Baenziger PS. Genetic Diversity and Population Structure of F3:6Nebraska Winter Wheat Genotypes Using Genotyping-By-Sequencing. Frontiers Genetic. 2018; 9:76. [CrossRef]
- Li Y, Reif JC, Hong H, Li H, Liu Z, Ma Y, et al. Genome-wide association mapping of QTL underlying seed oil and protein contents of a diverse panel of soybean accessions.Plant Science.2018; 266:95–101. [CrossRef]
- Li S, Cao Y, Wang C, Yan C, Sun X, Zhang L, Wang W, Song S. Genome-wide association mapping for yield-related traits in soybean (Glycine max) under well-watered and drought-stressed conditions. Frontiers in Plant Science. 2023;14:1265574. [CrossRef]
- Paterne A. A., Norman P. E., Asiedu R., Asfaw A.Identification of quantitative trait nucleotides and candidate genes for tuber yield and mosaic virus tolerance in an elite population of white Guinea yam (Dioscorea rotundata) using genome-wide association scan.BMC Plant Biology.2021,21, 552. [CrossRef]
- Zhang H , Hao D ,Sitoe, Hélder Manuel,et al.Genetic dissection of the relationship between plant architecture and yield component traits in soybean (Glycine max) by association analysis across multiple environments[J]. Plant Breeding, 2015, 134(5):564-572. [CrossRef]
- Zhang J , Song Q , Cregan P B ,et al.Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genomics, 2015, 16(1):217. [CrossRef]
- Chen L, Yang H, Fang Y, Guo W, Chen H, Zhang X, Dai W, Chen S, Hao Q, Yuan S, Zhang C, Huang Y, Shan Z, Yang Z, Qiu D, Liu X, Tran LP, Zhou X, Cao D. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnology Journal. 2021,19(4):702-716. [CrossRef]
- Qi X, Tang W, Li W, He Z, Xu W, Fan Z, Zhou Y, Wang C, Xu Z, Chen J, Gao S, Ma Y, Chen M.ArabidopsisG-Protein β Subunit AGB1 Negatively Regulates DNA Binding of MYB62, a Suppressor in the Gibberellin Pathway. International Journal Molecular Science. 2021;22(15):8270. [CrossRef]
- Wang T, Jin Y, Deng L, Li F, Wang Z, Zhu Y, Wu Y, Qu H, Zhang S, Liu Y, Mei H, Luo L, Yan M, Gu M, Xu G. The transcription factor MYB110 regulates plant height, lodging resistance, and grain yield in rice. Plant Cell. 2023, 268. [CrossRef]
- Zhang J, Song Q, Cregan PB, Jiang GL. Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max). Theor Appl Genet. 2016, 129(1):117-30. [CrossRef]
- Li S, Cao Y, Wang C, Yan C, Sun X, Zhang L, Wang W, Song S. Genome-wide association mapping for yield-related traits in soybean (Glycine max) under well-watered and drought-stressed conditions. Frontiers in Plant Science. 2023;14:1265574. [CrossRef]
- Rani R, Raza G, Ashfaq H, Rizwan M, Razzaq MK, Waheed MQ, Shimelis H, Babar AD, Arif M. Genome-wide association study of soybean (Glycine max [L.] Merr.) germplasm for dissecting the quantitative trait nucleotides and candidate genes underlying yield-related traits. Frontiers in Plant Science. 2023;14:1229495. [CrossRef]
- Xu K, Zhao Y, Zhao Y, Feng C, Zhang Y, Wang F, Li X, Gao H, Liu W, Jing Y, Saxena RK, Feng X, Zhou Y, Li H. Soybean F-Box-Like Protein GmFBL144 Interacts With Small Heat Shock Protein and Negatively Regulates Plant Drought Stress Tolerance. Frontiers in Plant Science. 2022;13:823529. [CrossRef]
- Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH, Schroeder JI, Huq E. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiology. 2014,164(1):424-439. [CrossRef]
- Chen Y, Xu Y, Luo W, Li W, Chen N, Zhang D, Chong K. The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiology. 2013;163(4):1673-85. [CrossRef]
- Zhou S, Yang T, Mao Y, Liu Y, Guo S, Wang R, Fangyue G, He L, Zhao B, Bai Q, Li Y, Zhang X, Wang D, Wang C, Wu Q, Yang Y, Liu Y, Tadege M, Chen J. The F-box protein MIO1/SLB1 regulates organ size and leaf movement in Medicago truncatula. Journal of Experiment Botany. 2021;72(8):2995-3011. [CrossRef]
- Sun X, Xie Y, Xu K, Li J. Regulatory networks of the F-box protein FBX206 and OVATE family proteins modulate brassinosteroid pathway to regulate grain size and yield in rice. Journal of Experiment Botany. 2023, erad397. [CrossRef]
- Lee K, Han JH, Park YI, Colas des Francs-Small C, Small I, Kang H. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Physiologist. 2017, 215(1):202-216. [CrossRef]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant Journal. 2017, 91(1):132-144. [CrossRef]
- Zhao B, Dai A, Wei H, Yang S, Wang B, Jiang N, Feng X. Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Molecular Biology. 2016, 90(1-2):33-47. [CrossRef]
- Wang X, Li Y, Zhang H, Sun G, Zhang W, Qiu L. Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Molecular Biology Reports. 2015, 42(2):489-496. [CrossRef]
- Zhou C, Lin Q, Ren Y, Lan J, Miao R, Feng M, Wang X, Liu X, Zhang S, Pan T, Wang J, Luo S, Qian J, Luo W, Mou C, Nguyen T, Cheng Z, Zhang X, Lei C, Zhu S, Guo X, Wang J, Zhao Z, Liu S, Jiang L, Wan J. A CYP78As-small grain4-coat protein complex II pathway promotes grain size in rice. Plant Cell. 2023 Nov 30;35(12):4325-4346. [CrossRef]
- Guo L, Ma M, Wu L, Zhou M, Li M, Wu B, Li L, Liu X, Jing R, Chen W, Zhao H. Modified expression of TaCYP78A5 enhances grain weight with yield potential by accumulating auxin in wheat (Triticum aestivum L.). Plant Biotechnology Journal. 2022, 20(1):168-182. [CrossRef]
- Zhang S., Hao D., Zhang S., Zhang D., Wang H., Du H., et al.Genome-wide association mapping for protein, oil and water-soluble protein contents in soybean.Mol. Genet. Genomics, 2021, 296, 91–102. [CrossRef]
- Bolger, A.M., Lohse, M. & Usadel, B.Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014, 30, 2114–2120. [CrossRef]
- Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012, 9, 357–359. [CrossRef]
- Danecek, P., Auton, A., Abecasis, G., Albers, C.A., Banks, E., DePristo, M.A.et al. The variant call format and VCFtools. Bioinformatics, 2011, 27,2156–2158. [CrossRef]
- Earl, D.A. & vonHoldt, B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour, 2012, 4, 359–361. [CrossRef]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle.2005.
- Ivica Letunic, Peer Bork. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation, Nucleic Acids Research, 2021, 49(1):293–296. [CrossRef]
- Sabeti, P.C., et al. Detecting recent positive selection in the human genome from haplotype structure. Nature, 2002;419(6909):832-837. [CrossRef]
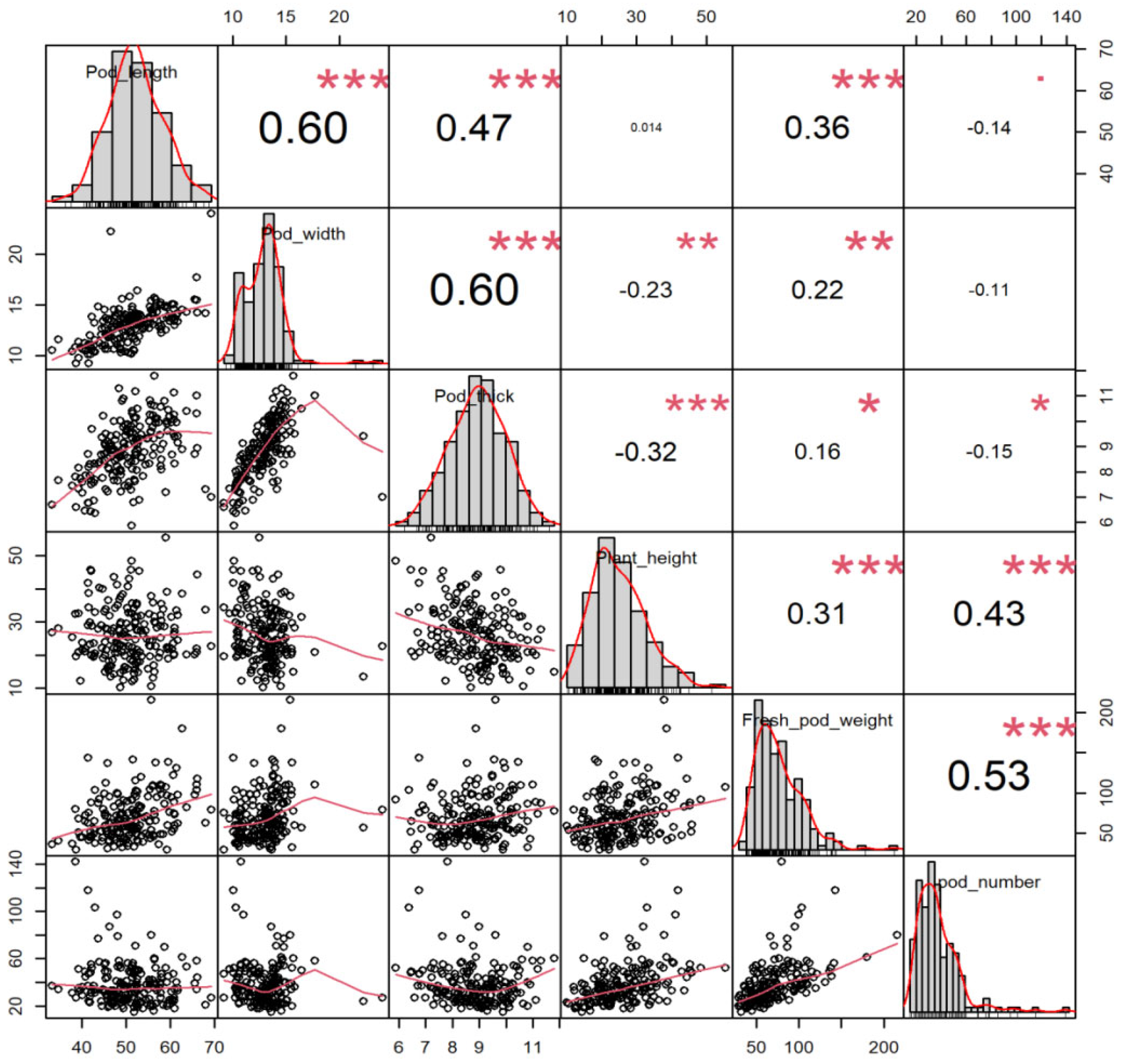
| Trait | Max | Min | Mean | SD | CV(%) |
| Pod length | 69.33 | 33.30 | 50.90 | 6.73 | 13.22% |
| Pod width | 24.04 | 9.19 | 12.97 | 1.90 | 14.65% |
| Pod thick | 11.80 | 5.87 | 8.84 | 1.13 | 12.75% |
| Plant height | 55.45 | 10.10 | 26.18 | 8.40 | 32.08% |
| Pods number | 15.00 | 4.50 | 8.57 | 1.66 | 19.41% |
| Fresh pod weight | 215.67 | 29.30 | 70.34 | 28.78 | 40.91% |
| Trait | SNP | -log10(P) | Candidate | Gene annotation |
|---|---|---|---|---|
| Plant height | Chr7:8350061 | 4.49E-08 | Glyma.07G089000 | Vernalization-insensitive protein3 |
| Plant height | Chr13:22150035 | 3.17E-08 | Glyma.13G107400 | Myosin-11-RELATED |
| Plant height | Chr13:22150035 | 3.17E-08 | Glyma.13G109100 | MYB-related transcription factors |
| Plant height | Chr14:44580744 | 1.29E-12 | Glyma.14G182400 | Hydroxyproline-rich glycoprotein family protein |
| Plant height | Chr14:14120295 | 5.18E-10 | Glyma.14G116600 | TPX2 protein family |
| Pod width | Chr03:24477441 | 1.8E-10 | Glyma.03G211200 | CW-type zinc-finger protein |
| Pod width | Chr04:45822312 | 2.28E-9 | Glyma.04G187000 | Histone deacetylase 2 |
| Pod length | Chr14:8677626 | 9.71E-08 | Glyma.14g093600 | Myc down regulated-like protein |
| Pod number | Chr03:39469452 | 1.94E-15 | Glyma.03G182100 | Small auxin-up RNA |
| Pod number | Chr03:39469452 | 1.94E-15 | Glyma.03G183200 | Auxin-responsive family protein |
| Pod number | Chr07:17801936 | 3.83E-11 | Glyma.07G148100 | Terminal domain phosphatase like 2 |
| Fresh pod weight | Chr09:18491673 | 3.00E-05 | Glyma.09G102300 | F-box protein |
| Fresh pod weight | Chr09:18491673 | 3.00E-05 | Glyma.09G101100 | WAT1-related protein |
| Fresh pod weight | Chr09:18491673 | 3.00E-05 | Glyma.09G101200 | Transcriptional regulator SNIP1 |
| Fresh pod weight | Chr09:18491673 | 3.00E-05 | Glyma.09G101300 | Solute carrier family 35, member C2 (SLC35C2) |
| Fresh pod weight | Chr09:18491673 | 3.00E-05 | Glyma.09G102200 | CYP72A154 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




