1. Introduction
The accurate diagnosis of Primary Ciliary Dyskinesia (PCD), a genetic disorder characterized by dysfunctional motile cilia [
1], is crucial for the timely treatment and management of the disease [
2]. Motile cilia are essential for the proper functioning of various organ systems, and their impairment in PCD leads to a spectrum of clinical manifestations, notably chronic respiratory tract infections [
1]. Globally, the disease is underdiagnosed, with a reported prevalence of approximately 1 in 16,309 within Hispanic populations [
3], highlighting the need for improved diagnostic methodologies [
3,
4]. The field of PCD research is marked by significant controversies [
5], particularly regarding the diagnostic criteria and the heterogeneity of genetic mutations associated with the disease [
6,
7]. While nasal Nitric Oxide (nNO) levels have been a traditional diagnostic marker [
8], recent studies suggest that High-Speed Video Microscopy Analysis (HSVA) provides additional information about the ciliary function [
9,
10].
In Puerto Rico, the prevalence and specific microscopic characteristics of the cilia dynamics with the founder PCD mutation
RSPH4A [c.921+3_921+6del (intronic)] are not defined [
6], partly due to the limited access to diagnostic tools, including HSVA. This technology offers a detailed visualization of ciliary motion, enabling a deeper understanding of ciliary dynamics at a microstructural level [
11]. To date, its application in the diagnosis algorithm of PCD in Puerto Rico has not been explored.
This study addresses this gap by implementing a protocol for HSVA for the first time in Puerto Rico to investigate PCD in a Puerto Rican cohort with the RSPH4A [c.921+3_921+6del (intronic)] founder mutation. By describing HSVA findings for this specific genetic mutation, which accounts for over 70% of PCD cases on the island, this work aims to enhance the diagnostic accuracy for PCD in Puerto Rico significantly. Moreover, our research contributes to the broader understanding of PCD as a global effort to understand this disease in developing countries in Latin America.
2. Materials and Methods
2.1. Subjects
The study included well-characterized cohort of patients with PCD (
n=12) with confirmed bi-allelic pathogenic
RSPH4A [c.921+3_921+6del (intronic)] founder mutation and decreased nasal nitric oxide (nNO) levels, measured by a validated chemiluminescence technique (CLD 88sp Chemiluminescence Nitric Oxide Analyzer, Dürnten, Switzerland) as per protocol [
3]. Only one patient was classified as a compound heterozygous patient with two genetic variants in the
RSPH4A gene [c.921+3_921+6del (intronic)] and c.1103T>G (p.Val368Gly)]. Nasal ciliated epithelial samples from all subjects with PCD were collected at their baseline health status, without clinical history of viral or bacterial infection, for over two weeks.
2.2. Sample collection and preparation
Nasal ciliated epithelial samples were obtained via cytology brushing (Puritan Sterile Cytology Brushes). The nasal cytology brush was briefly inserted in one nostril and gently advanced to the inferior nasal turbinate with slight movements while performing internal rotation of the brush and slightly pressing on the lateral wall of the inferior turbinate [
12]. Once brushing was completed, immediate sample analysis was conducted. The nasal cytology brush sample was immersed in 3 mL of PneumaCult™-Ex Plus Medium to release any tissue that remained adherent to the bristles. The sample required to be placed in a sufficient media volume, allowing full immersion of the brush. Samples were washed in Dulbecco’s Phosphate Buffered Saline (D-PBS), without calcium and magnesium to remove mucus and debris and centrifuged at 400xg for seven minutes to pellet the airway epithelial cells. After resuspension in 500μL of PneumaCult™-Ex Plus Medium, the samples were incubated at 37⁰ degrees Celsius (°C) for 30 minutes before HSVA at 500 frames per second (fps), under a 40 and 60X lens magnification.
2.3. Highspeed Video Microscopy Analysis (HSVA)
The sample analysis was conducted using the Nikon Eclipse Ti2 inverted microscope with a long working distance 40X and a 60X objective lens. An AOS PROMON U750 mono-chrome high-speed camera was attached to the microscope to capture high-speed video recording data. The camera had a light sensitivity grade of ISO 3600 and a sensor size of 4.8 μm pixels. This setup recorded samples at a frame rate of 500 fps. The AOS Imaging Software Version 4 was used to process the footage, which was uploaded via a USB3 cable. A microscope air table was employed in conjunction with the HSVA to minimize movement during the recording. The optimal resolution setting for HSVA was recorded at 500 fps, as recommended by the AOS software in the camera suite, and set at 880 x 637 pixels. Following the European Respiratory Society (ERS) guidelines [
13], 2,200 frames were recorded for five seconds at the specified resolution and under 40X and 60X. The recorded HSVA footage was saved as unprocessed image data (RAW) files. During the analysis, the region of interest (ROI) was not selected if there was mucus or debris on the surface interface of the cell cluster. To ensure representative footage of the ciliary population, an ROI was only selected if 10-15 equally distant adjacent ciliated cells were present. A microscope temperature control system (Life Imaging Services, Efringerstrasse 79, CH-4057 Basel Switzerland) adapted to a Nikon Eclipse Ti2 Inverted microscope was used to maintain extended observation of motile cilia at 37°C to minimize variability in CBF. Intact cell lining was chosen over single cells to obtain a more representative sample [
14]. The entire HSVA process was completed in less than 30 minutes per sample at 37°C.
2.4. Manual method for CBF count
The CBF was manually calculated based on 10 complete beat cycles. This manual counting method adhered to protocols established in the literature, using the formula: Manual CBF (Hz) = (fps) / number of frames elapsed for 10 full ciliary beats) * 10. This formula facilitates the conversion to a per-beat-cycle basis [
15,
16]. To ensure precision in manual counting, each video was paused at the moment when cilia approached their maximal bend. The video was advanced frame by frame until the cilia reached full bent, marking this as the starting frame. A tally was made of the total number of complete ciliary beats—encompassing both the effective and recovery strokes—from this initial to the cycle’s final frame. The exact number of frames elapsed during 10 complete ciliary beats was carefully recorded for analysis.
2.5. Software analysis
The Cilialyzer (v1.2.1-b3098cb) and CiliarMove (v219) software were applied to the same set of recordings provided by the PCD patients and healthy controls. A reference picture of the ROI marked with a cursor was applied to ensure consistent analysis of the same ROI. For CBF computation using CiliarMove, the RAW format video files required one conversion step to be transformed into the tag image file format (TIFF) sequence file. After the file conversion, the image sequence was uploaded for analysis. Before analyzing the frequency, the ROI was manually selected using the cropping function. For Cilialyzer, the RAW format video files were converted into sequence images in bitmap (BMP) format and pre-processed as stated by the Cilialyzer manual [
15]. The pre-processing methods included initial manual image rotation, selecting ROI, image stabilization, cropping margins, and motion extraction. The CBF was then calculated by both CiliarMove and Cilialyzer, with the results displayed on a frequency distribution histogram and a power spectrum graph, respectively. These steps were followed to ensure accurate and consistent analysis of the CBF using Cilialyzer and CiliarMove for the respective software platforms. The Tracker Video Analysis and Modeling tool follows Brown (2023), was used for CBP analysis [
16].
2.6. Statistical Analysis
All statistical analyses were performed using the statistical software package GraphPad Prism version 10.1.1 for MacOS, developed by GraphPad Software, San Diego, CA, USA [
www.graphpad.com]. Descriptive statistics, including median and interquartile ranges (IQRs), were calculated to summarize the data, providing central tendency and dispersion measures.
3. Results
3.1. Subjects Characteristics and Demographics
In the study, 12 participants were recruited and underwent HSVA by the study protocol [
17]. The demographic details and clinical characteristics of PCD among the cohort are detailed in
Table 1.
The participant cohort comprised a slight majority of females, with 7 out of 12 subjects (58%) being female. The median age of the subjects was 21 years. 92% of the subjects exhibited homozygosity for the founder PCD mutation RSPH4A [c.921+3_921+6del (intronic)], while only one was compound heterozygous for the same gene RSPH4A [c.921+3_921+6del (intronic)] plus RSPH4A c.1103T>G (p.Val368Gly). All participants (100%) were found to have bronchiectasis on a chest computed tomography (CT) scan. None of the patients displayed laterality defects, accounting for 0% of the cohort. Chronic cough, defined as a persistent, year-round wet cough that begins before six months of age, was present in all subjects (100%). Neonatal respiratory distress and chronic otitis media were prevalent in half of the patients, with each condition affecting 50% of the cohort. Chronic sinusitis was a universal finding, present in 100% of the patients. Hearing loss was identified in 58% of the subjects. A notable 83% of the cohort showed colonization with Pseudomonas species, and 8% had a history of Burkholderia cepacia infection. All patients in the study exhibited nasal Nitric Oxide (nNO) levels below the diagnostic threshold of 77 nL/min. Only one patient was a recipient of a double lung transplant.
3.2. Ciliary Beat Frequency (CBF) Measurement and Ciliary Beat Pattern (CBP) Assessment
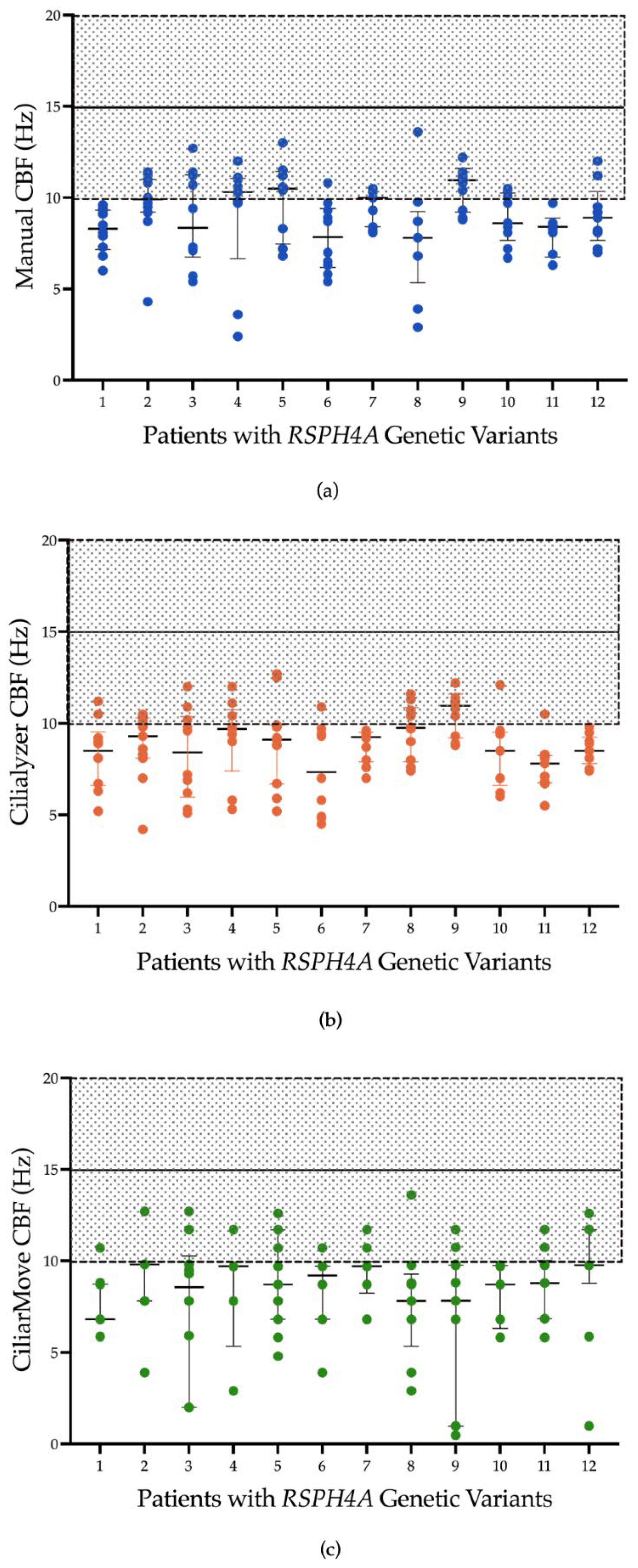
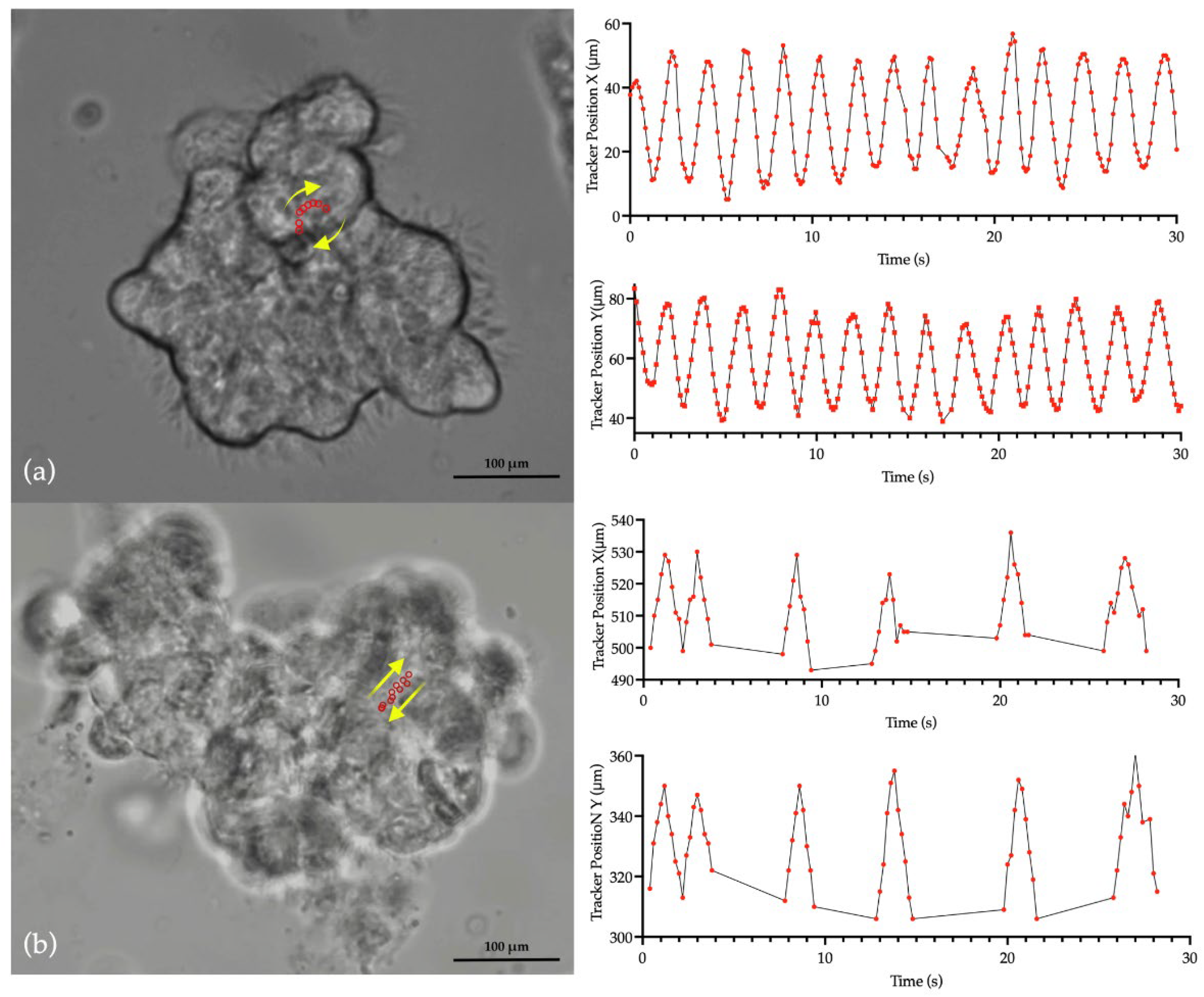
Figure 1 presents CBF measurements obtained from a sample set using three different methods in patients with PCD due to RSPH4A genetic variants: manual counting and two automated systems, Ciliayzer and CiliarMove. The manual method yielded CBF medians that ranged from 7.85 to 10.9 Hz. Ciliayzer’s median results range from 7 to 9.8 Hz. On the other hand, CiliarMove demonstrated a broader range of medians, from as low as 6.8 Hz to as high as 9.8 Hz. CBP, as analyzed using tracker software, exhibited a rotational motion in all samples (100%) when observed in top-view videos captured by HSVA,
Figure 2.
4. Discussion
This study represents a pioneering effort to advance the diagnosis of PCD in Puerto Rico by implementing, for the first time, the HSVA technique in Puerto Ricans with a PCD founder mutation. Daniels et al., 2013, represent the only study that has assessed CBF and CBP using HSVA at room temperature in a cohort of six patients of Puerto Rican Hispanic descent in the United States [
19]. While the average CBF reported in their study was slightly lower than we observed, both studies identified CBF values below the established normal threshold for healthy individuals [
18]. In contrast, our study analyzed each sample at 37°C to avoid temperature effects [
20]. Furthermore, their findings on CBP are consistent with ours, demonstrating a rotational movement in top-view analysis of the cilia. The
RSPH4A gene, which was the focus of our study, encodes a protein integral to the central apparatus and is implicated in the 9+2 microtubular organization [
21]. Defects in this central complex are known to be linked with atypical waveforms similar to those observed in the 9+0 nodal cilia, which exhibit a circular motion [
10].
In Europe, the introduction of HSVA has proven highly beneficial by providing a much-needed enhancement to the evaluation process, revealing critical insights into the ciliary dysfunction associated with several PCD-related genetic variants [
9]. By evaluating this technique in patients with the
RSPH4A [c.921+3_921+6del (intronic)] founder mutation, we were able to characterize the decreased CBF coupled with the rotational CBP, which aligns with the mutation’s expected physiological impact and PCD clinical outcomes. The consistency of the results obtained from manual and automated measurement methods reinforces the credibility of HSVA as an effective complementary diagnostic tool for PCD associated with the
RSPH4A founder mutation, independently of the method of analysis (Manual, Cililyzer, CiliarMove). The utility of HSVA lies in its capacity for ex-vivo assessment of ciliary motion, which provides an objective analysis of CBF and CBP. Moreover, HSVA minimizes the potential for human error typically associated with assessing ciliary function on conventional microscopy, thus providing valuable corroborative data to support clinical diagnoses of PCD. This is particularly beneficial in clinical environments where expedited patient evaluation is imperative, genetic testing may involve longer processing times, or other diagnostic tools are unavailable.
Furthermore, aligning results obtained from well-established and standardized protocols to perform HSVA attests to its accuracy, providing a complementary tool to the algorithm of PCD diagnosis. Ensuring consistency in applying HSVA across various PCD centers is essential when adopting new technologies within clinical practices. Such standardization is key to maintaining and enhancing the quality of patient care and minimizing the risk of misdiagnosing PCD as we advance diagnostic techniques. By adopting HSVA, Puerto Rico elevates its local healthcare standards and joins the international PCD community in adopting cutting-edge techniques for PCD diagnosis. This initiative is committed to improving patient outcomes and contributes to the global endeavor to understand and treat PCD more effectively. Moreover, the presence of ciliary dysfunction with reduced nNO levels below the established diagnostic threshold further substantiates the clinical utility of HSVA in the identification and characterization of PCD, as 100% of our patients had low nNO levels, low CBF and rotational CBP on HSVA.
Given the current diagnostic resources in Puerto Rico and following guidelines from the American Thoracic Society (ATS), ERS, and the PCD Foundation, we have developed a population-targeted diagnostic algorithm for PCD in Puerto Rico. The algorithm specifically addresses the high prevalence of the RSPH4A founder mutation within the Puerto Rican population and our available diagnostic modalities. This focused algorithm for Puerto Rico complements, rather than replaces, existing guidelines. The diagnostic process initiates with a comprehensive clinical assessment aimed at detecting symptoms and physical indicators of PCD, when Cystic Fibrosis has been excluded. All patients should undergo evaluation at center accredited by the PCD Foundation able to perform nNO levels. Evaluation is complemented by nNO levels as a non-invasive screening, during the initial visit and a follow-up measurement two weeks later, provided the patient’s respiratory symptoms are at baseline. Subsequent steps involve collecting a sample for HSVA to assess CBF and CBP, followed by an in-depth genetic panel test that specifically targets the RSPH4A founder mutation to confirm PCD diagnosis.
The combination of nNO levels with low CBF and rotational CBP on HSVA, and genetic testing showing pathogenic variants on RSPH4A gene, confirm the most common phenotype associated with PCD in Puerto Rico.
If patient is unable to complete nNO, HSVA results are inconclusive, or genetic testing is negative, a more comprehensive evaluation should be pursued, including repeat HSVA, nasal biopsy for transmission electron microscopy (TEM), and/or broader genetic testing (whole exome sequencing) if resources allow. The algorithm promotes a multidisciplinary approach to management and encourages regular follow-up for ongoing patient care. It is vital to update the algorithm periodically, reflecting new scientific insights, de implementation of new diagnostic tools, and ensuring it remains applicable to the healthcare context of Puerto Rico. The algorithm (
Figure 3) not only serves the immediate need for PCD diagnosis but also contributes to the broader goal of improving rare disease management on the island.
Our findings echo the existing body of research that underscores the heterogeneity of PCD presentations and the corresponding need for comprehensive diagnostic approaches [
22]. Integrating HSVA as part of the evaluation for patients with PCD could significantly enhance our comprehension of the disease, especially for populations like Puerto Ricans that present with unique founder mutations exhibiting rotational CBP. This study highlights the pairing of genetics to clinical and functional observations to increase our understanding of PCD across different populations and comprehensively address the challenges in securing a definitive diagnosis. As the field moves towards implementing advanced tools like HSVA to offer deeper insight into specific PCD phenotypes and ciliary behaviors, it raises the necessity of following rigorous protocols and ensuring standardization across diagnostic platforms. Using open-source software such as Cilialyzer and CiliarMove for PCD provides a standardized approach to measuring CBF [
15,
23]. Although manual counting demonstrated comparable efficacy in detecting reduced CBF in our cohort, proving reliable, objective, and accessible, software tools offer a standardized measurement system, which will facilitate comparison and translatability of the findings across different cohorts.
While our study provides a significant leap forward in PCD diagnostics in Puerto Rico, there are limitations of the study to be considered. The sample size, though sufficient for preliminary analysis in rare diseases, necessitates expansion in future studies to validate and generalize our findings. However, few studies have evaluated HSVA in cohorts with a specific and unique founder mutation [
19]. Our study did not extend to assessing the CBF of ciliated epithelial cells within air-liquid interface (ALI) cultures [
24]. This decision was strategically made, as our primary objective was to establish a rapid and cost-efficient diagnostic protocol that could be readily implemented in a clinical setting. While ALI cultures represent a more physiologically relevant model [
25], they require significant time and resources to establish. Our approach, focusing on direct sampling methods, allowed for immediate analysis and offered a balance between clinical efficiency and CBF accuracy both manually and using open-source software. Future studies incorporating ALI culture assessments are needed to complement and expand upon our findings, further enriching the understanding of ciliary function in PCD within the context of native epithelial architecture.
The successful application of HSVA for PCD diagnosis in Puerto Rico represents a significant stride in addressing the diagnostic challenges associated with this rare disease. Our work not only enhances the diagnostic capabilities on the island but also contributes to the global effort to understand and manage PCD in diverse populations, specifically those with founder mutations.
5. Conclusions
Our research has fundamentally advanced diagnosing PCD by integrating several diagnostic modalities in Puerto Rico. The HSVA was implemented for the first time in a Puerto Rican cohort in this study. This diagnostic approach has successfully identified the characteristic decreased CBF and distinctive rotational CBP associated with the RSPH4A founder mutation. Our approach emphasizes the importance of these diagnostic markers in evaluating PCD. The implications of our study are important, considering HSVA as a valuable tool that could reshape the identification, management, and research of this rare disease in Puerto Rico and throughout Latin America.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Video S1: Dynamic visualization of the cilia’s rotational motion in nasal ciliated epithelium from a patient homozygous for the
RSPH4A [c.921+3_921+6del (intronic)], with tracker software capturing the pattern characterized by extensive oscillations at the cilia tips.
Video S2: Dynamic visualization of the typical bidirectional ciliary motion seen in nasal ciliated epithelium from a healthy control subject.
Author Contributions
Conceptualization, W.D.J.-R., Z.J.D., and R.A.M.; methodology, W.D.J.-R. and Z.J.D.; software, Z.J.D.; validation, W.D.J.-R., Z.J.D. and G.R.-O.; formal analysis, Z.J.D. and F.M.Q.; investigation, W.D.J.-R, and G.R.-O.; resources, F.M.Q. and M.J.R.-B.; data curation, J.M.-H.; writing—original draft preparation, W.D.J.-R.; writing—review and editing, M.J.R.-B., F.M.Q., G.R.-O., and J.M.-H. ; visualization, G.R.-O. and J.M.-H.; supervision, R.A.M.; funding acquisition, W.D.J.-R., R.A.M. and M.J.R.-B. All authors have read and agreed to the published version of the manuscript.
Funding
The Puerto Rico Science, Technology, and Research Trust under the agreement number PRSTRT: 2022-00012A. This content is only the authors’ responsibility and does not necessarily represent the official views of The Puerto Rico Science, Technology, and Research Trust. UPRMSC Hispanics-In-Research Capability (HiREC) Endowment, Partnership between the School of Health Professions and School of Medicine, University of Puerto Rico, Funded (S21MD001830) by the NIH National Institute of Minority Health and Health Disparities. The National Institute of Health: Award Number: HCTRECD R25MD007607 from the National Institute on Minority Health and Health Disparities.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Ponce Health Sciences University (Protocol number: 2301128951, Date: May 4, 2023).
Informed Consent Statement
All subjects gave their informed consent for inclusion before they participated in the study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to privacy restrictions but are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leigh, M.W.; Horani, A.; Kinghorn, B.; O'Connor, M.G.; Zariwala, M.A.; Knowles, M.R. Primary Ciliary Dyskinesia (PCD): A genetic disorder of motile cilia. Transl Sci Rare Dis 2019, 4, 51–75. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.J.; Zariwala, M.A.; Ferkol, T.; Davis, S.D.; Sagel, S.D.; Dell, S.D.; Rosenfeld, M.; Olivier, K.N.; Milla, C.; Daniel, S.J.; et al. Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr Pulmonol 2016, 51, 115–132. [Google Scholar] [CrossRef] [PubMed]
- De Jesus-Rojas, W.; Alvarado-Huerta, F.; Melendez-Montanez, J.M.; Muniz-Hernandez, J.; Santos-Lopez, A.; Mosquera, R.A. Nasal Nitric Oxide Levels: Improving the Diagnosis of Primary Ciliary Dyskinesia in Puerto Rico. Adv Respir Med 2022, 90, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Hannah, W.B.; Seifert, B.A.; Truty, R.; Zariwala, M.A.; Ameel, K.; Zhao, Y.; Nykamp, K.; Gaston, B. The global prevalence and ethnic heterogeneity of primary ciliary dyskinesia gene variants: a genetic database analysis. Lancet Respir Med 2022, 10, 459–468. [Google Scholar] [CrossRef]
- Shapiro, A.J.; Davis, S.D.; Polineni, D.; Manion, M.; Rosenfeld, M.; Dell, S.D.; Chilvers, M.A.; Ferkol, T.W.; Zariwala, M.A.; Sagel, S.D.; et al. Diagnosis of Primary Ciliary Dyskinesia. An Official American Thoracic Society Clinical Practice Guideline. Am J Respir Crit Care Med 2018, 197, e24–e39. [Google Scholar] [CrossRef]
- De Jesus-Rojas, W.; Reyes-De Jesus, D.; Mosquera, R.A. Primary Ciliary Dyskinesia Diagnostic Challenges: Understanding the Clinical Phenotype of the Puerto Rican RSPH4A Founder Mutation. Diagnostics (Basel) 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, W.; Geng, G.; Dai, J.; Fu, Z.; Tian, D. Clinical and genetic features of primary ciliary dyskinesia in a cohort of consecutive clinically suspect children in western China. BMC Pediatr 2022, 22, 402. [Google Scholar] [CrossRef]
- Shapiro, A.J.; Dell, S.D.; Gaston, B.; O'Connor, M.; Marozkina, N.; Manion, M.; Hazucha, M.J.; Leigh, M.W. Nasal Nitric Oxide Measurement in Primary Ciliary Dyskinesia. A Technical Paper on Standardized Testing Protocols. Ann Am Thorac Soc 2020, 17, e1–e12. [Google Scholar] [CrossRef]
- Shoemark, A.; Rubbo, B.; Haarman, E.; Hirst, R.A.; Hogg, C.; Jackson, C.L.; Nielsen, K.G.; Papon, J.F.; Robinson, P.; Walker, W.T.; et al. The Controversies and Difficulties of Diagnosing Primary Ciliary Dyskinesia. Am J Respir Crit Care Med 2020, 201, 120–122. [Google Scholar] [CrossRef]
- De Jesus-Rojas, W.; Melendez-Montanez, J.; Muniz-Hernandez, J.; Marra-Nazario, A.; Alvarado-Huerta, F.; Santos-Lopez, A.; Ramos-Benitez, M.J.; Mosquera, R.A. The RSPH4A Gene in Primary Ciliary Dyskinesia. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Peabody, J.E.; Shei, R.J.; Bermingham, B.M.; Phillips, S.E.; Turner, B.; Rowe, S.M.; Solomon, G.M. Seeing cilia: imaging modalities for ciliary motion and clinical connections. Am J Physiol Lung Cell Mol Physiol 2018, 314, L909–L921. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, L.K.; Turgutoglu, N.; Allan, K.M.; Belessis, Y.; Widger, J.; Jaffe, A.; Waters, S.A. Comparing Cytology Brushes for Optimal Human Nasal Epithelial Cell Collection: Implications for Airway Disease Diagnosis and Research. J Pers Med 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Shoemark, A.; Dell, S.; Shapiro, A.; Lucas, J.S. ERS and ATS diagnostic guidelines for primary ciliary dyskinesia: similarities and differences in approach to diagnosis. European Respiratory Journal 2019, 54, 1901066. [Google Scholar] [CrossRef]
- Muller, L.; Savas, S.T.; Tschanz, S.A.; Stokes, A.; Escher, A.; Nussbaumer, M.; Bullo, M.; Kuehni, C.E.; Blanchon, S.; Jung, A.; et al. A Comprehensive Approach for the Diagnosis of Primary Ciliary Dyskinesia-Experiences from the First 100 Patients of the PCD-UNIBE Diagnostic Center. Diagnostics (Basel) 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Schneiter, M.; Tschanz, S.A.; Escher, A.; Muller, L.; Frenz, M. The Cilialyzer - A freely available open-source software for the analysis of mucociliary activity in respiratory cells. Comput Methods Programs Biomed 2023, 241, 107744. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.H., Robert; Christian, Wolfgang Tracker Video Analysis and Modeling Tool, 2023.
- Jackson, C.L.; Bottier, M. Methods for the assessment of human airway ciliary function. Eur Respir J 2022, 60. [Google Scholar] [CrossRef]
- Satir, P.; Christensen, S.T. Overview of structure and function of mammalian cilia. Annu Rev Physiol 2007, 69, 377–400. [Google Scholar] [CrossRef]
- Daniels, M.L.; Leigh, M.W.; Davis, S.D.; Armstrong, M.C.; Carson, J.L.; Hazucha, M.; Dell, S.D.; Eriksson, M.; Collins, F.S.; Knowles, M.R.; et al. Founder mutation in RSPH4A identified in patients of Hispanic descent with primary ciliary dyskinesia. Hum Mutat 2013, 34, 1352–1356. [Google Scholar] [CrossRef]
- Clary-Meinesz, C.F.; Cosson, J.; Huitorel, P.; Blaive, B. Temperature effect on the ciliary beat frequency of human nasal and tracheal ciliated cells. Biol Cell 1992, 76, 335–338. [Google Scholar] [CrossRef]
- Zhao, Y.; Pinskey, J.; Lin, J.; Yin, W.; Sears, P.R.; Daniels, L.A.; Zariwala, M.A.; Knowles, M.R.; Ostrowski, L.E.; Nicastro, D. Structural insights into the cause of human RSPH4A primary ciliary dyskinesia. Mol Biol Cell 2021, 32, 1202–1209. [Google Scholar] [CrossRef]
- Leigh, M.W.; Ferkol, T.W.; Davis, S.D.; Lee, H.S.; Rosenfeld, M.; Dell, S.D.; Sagel, S.D.; Milla, C.; Olivier, K.N.; Sullivan, K.M.; et al. Clinical Features and Associated Likelihood of Primary Ciliary Dyskinesia in Children and Adolescents. Ann Am Thorac Soc 2016, 13, 1305–1313. [Google Scholar] [CrossRef]
- Sampaio, P.; da Silva, M.F.; Vale, I.; Roxo-Rosa, M.; Pinto, A.; Constant, C.; Pereira, L.; Quintao, C.M.; Lopes, S.S. CiliarMove: new software for evaluating ciliary beat frequency helps find novel mutations by a Portuguese multidisciplinary team on primary ciliary dyskinesia. ERJ Open Res 2021, 7. [Google Scholar] [CrossRef]
- Hirst, R.A.; Jackson, C.L.; Coles, J.L.; Williams, G.; Rutman, A.; Goggin, P.M.; Adam, E.C.; Page, A.; Evans, H.J.; Lackie, P.M.; et al. Culture of primary ciliary dyskinesia epithelial cells at air-liquid interface can alter ciliary phenotype but remains a robust and informative diagnostic aid. PLoS One 2014, 9, e89675. [Google Scholar] [CrossRef]
- Jackson, A.D.; Rayner, C.F.; Dewar, A.; Cole, P.J.; Wilson, R. A human respiratory-tissue organ culture incorporating an air interface. Am J Respir Crit Care Med 1996, 153, 1130–1135. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).