Submitted:
22 February 2024
Posted:
22 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study plan and sample collections
2.2. Isolation and identification of bacteria
2.3. Antimicrobial susceptibility test
2.4. Determining minimum inhibitory concentrations (MIC)
2.5. Molecular detection of metallo-beta-lactamase (blaNDM-1 and blaVIM) genes
2.6. Data analysis
2.7. Ethical clearance
3. Results
3.1. Study population
3.2. Bacterial prevalence
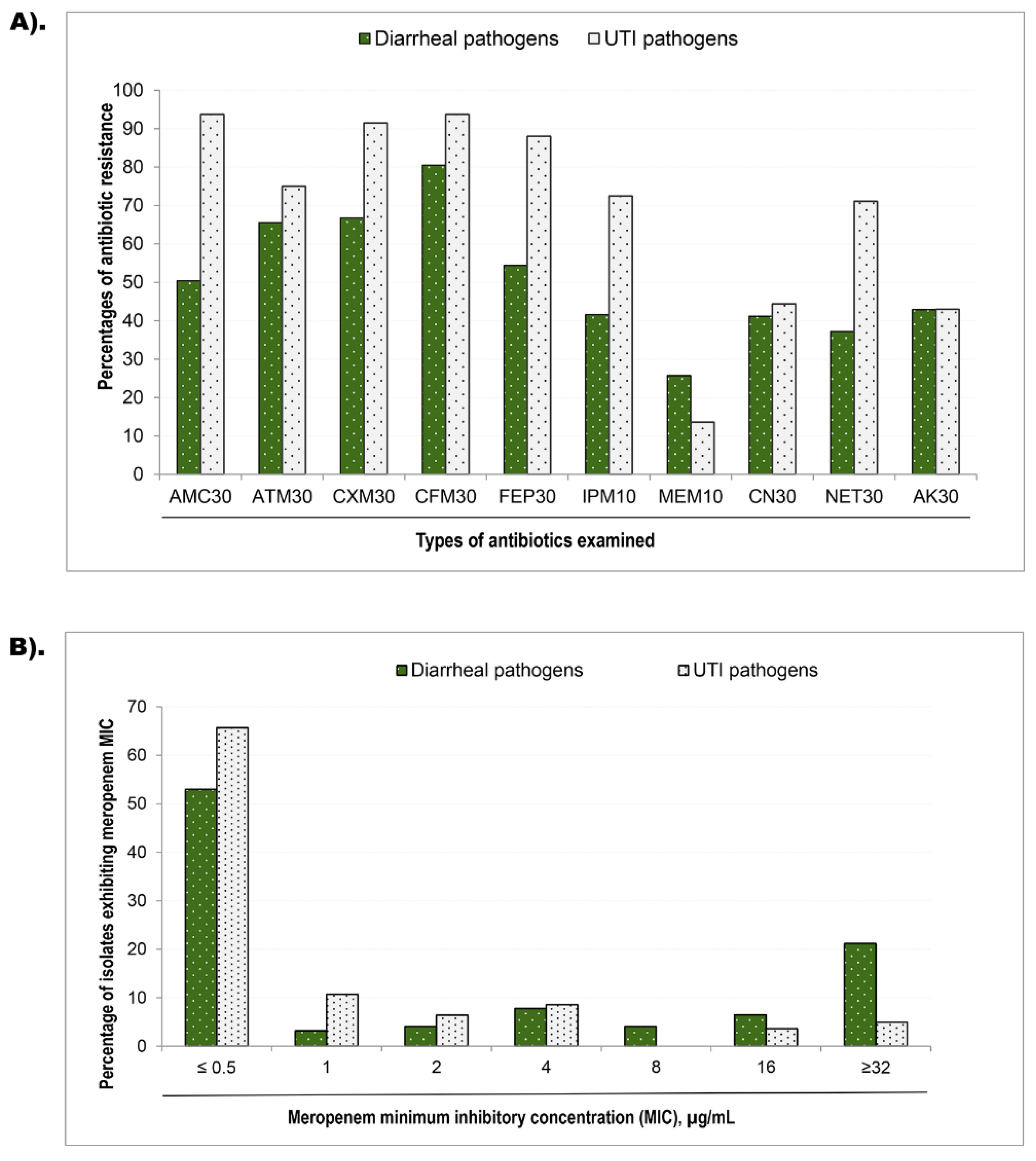
3.3. Phenotypic resistance and minimum inhibitory concentration
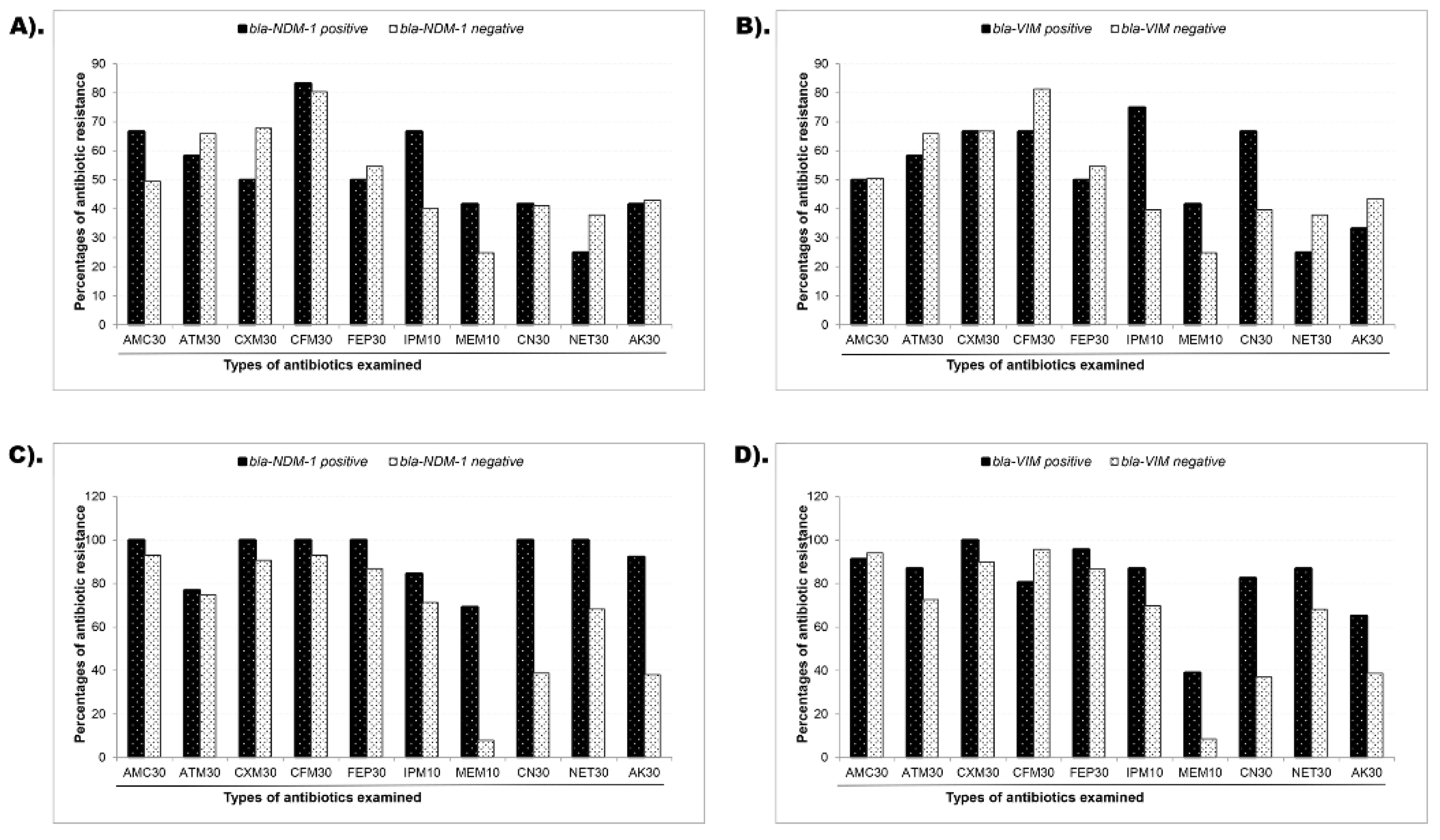
3.4. Prevalence of MBL Genes
3.5. Association of Phenotypic and Genotypic (blaNDM-1 and blaVIM gene) Resistance
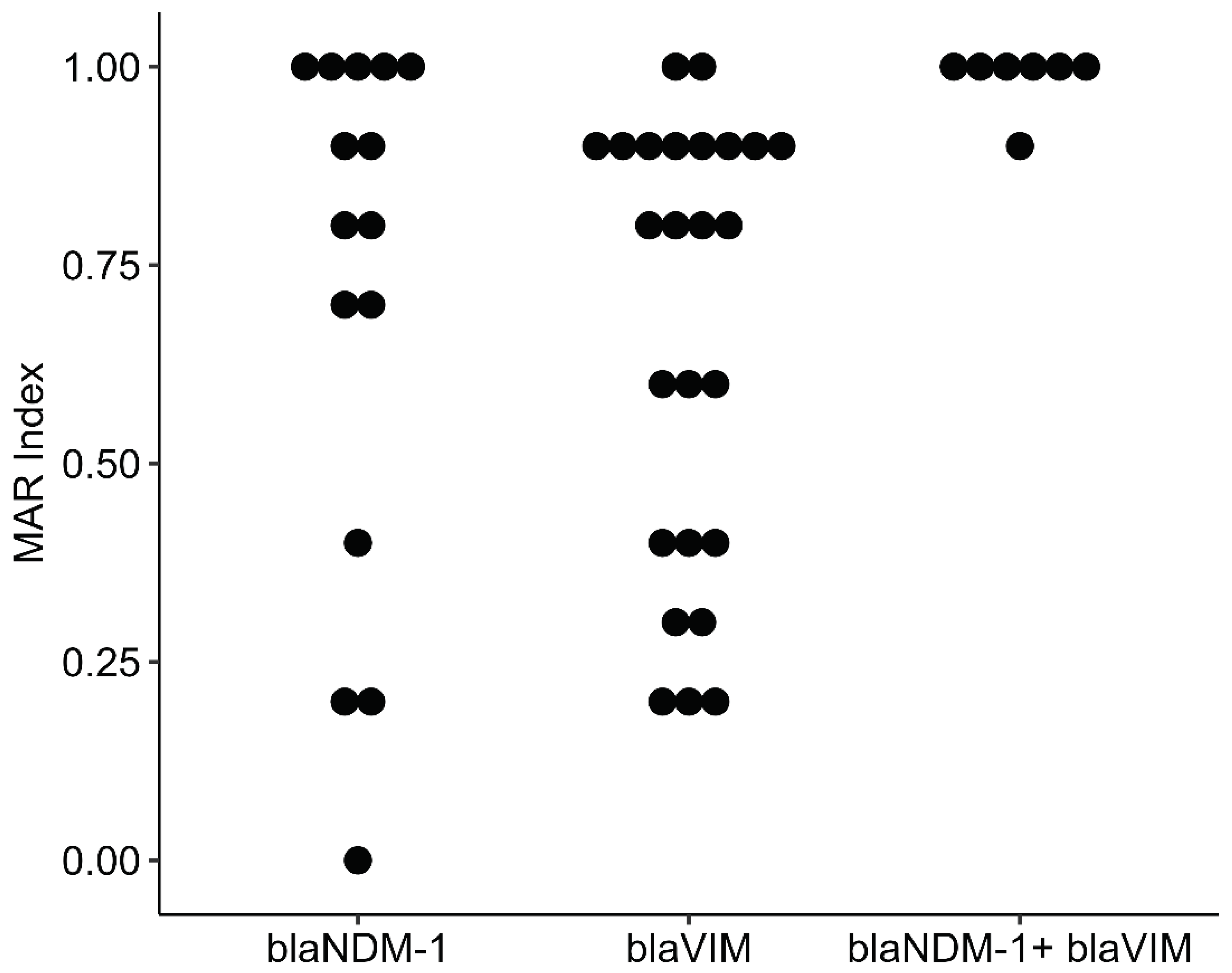
3.6. Co-Resistance Phenotype and Multiple Antibiotic Resistance (MAR) Index
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: past, present, and future. Antimicrobial agents and chemotherapy 2011, 55, 4943-4960.
- Sharland, M.; Zanichelli, V.; Ombajo, L.A.; Bazira, J.; Cappello, B.; Chitatanga, R.; Chuki, P.; Gandra, S.; Getahun, H.; Harbarth, S. The WHO essential medicines list AWaRe book: From a list to a quality improvement system. Clinical Microbiology and Infection 2022, 28, 1533-1535. [CrossRef]
- Sharland, M.; Gandra, S.; Huttner, B.; Moja, L.; Pulcini, C.; Zeng, M.; Mendelson, M.; Cappello, B.; Cooke, G.; Magrini, N. Encouraging AWaRe-ness and discouraging inappropriate antibiotic use—the new 2019 Essential Medicines List becomes a global antibiotic stewardship tool. The Lancet Infectious Diseases 2019, 19, 1278-1280. [CrossRef]
- Ahmed, I.; Rabbi, M.B.; Sultana, S. Antibiotic resistance in Bangladesh: A systematic review. International Journal of Infectious Diseases 2019, 80, 54-61. [CrossRef]
- Islam, S.; Urmi, U.L.; Rana, M.; Sultana, F.; Jahan, N.; Hossain, B.; Iqbal, S.; Hossain, M.M.; Mosaddek, A.S.M.; Nahar, S. High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Scientific Reports 2020, 10, 17292. [CrossRef]
- Hoque, R.; Ahmed, S.M.; Naher, N.; Islam, M.A.; Rousham, E.K.; Islam, B.Z.; Hassan, S. Tackling antimicrobial resistance in Bangladesh: A scoping review of policy and practice in human, animal and environment sectors. PloS one 2020, 15, e0227947. [CrossRef]
- Monira, S.; Shabnam, S.A.; Ali, S.I.; Sadique, A.; Johura, F.-T.; Rahman, K.Z.; Alam, N.H.; Watanabe, H.; Alam, M. Multi-drug resistant pathogenic bacteria in the gut of young children in Bangladesh. Gut pathogens 2017, 9, 1-8. [CrossRef]
- Haque, T.A.; Urmi, U.L.; Islam, A.B.M.M.K.; Ara, B.; Nahar, S.; Mosaddek, A.S.M.; Lugova, H.; Kumar, S.; Jahan, D.; Rahman, N.A.A. Detection of qnr genes and gyrA mutation to quinolone phenotypic resistance of UTI pathogens in Bangladesh and the implications. Journal of Applied Pharmaceutical Science 2022, 12, 185-198. [CrossRef]
- Begum, N.; Shamsuzzaman, S. Emergence of carbapenemase-producing urinary isolates at a tertiary care hospital in Dhaka, Bangladesh. Tzu Chi Medical Journal 2016, 28, 94-98. [CrossRef]
- Islam, M.A.; Akhtar, Z.; Hassan, M.Z.; Chowdhury, S.; Rashid, M.M.; Aleem, M.A.; Ghosh, P.K.; Mah-E-Muneer, S.; Parveen, S.; Ahmmed, M.K. Pattern of antibiotic dispensing at pharmacies according to the WHO Access, Watch, Reserve (AWaRe) classification in Bangladesh. Antibiotics 2022, 11, 247. [CrossRef]
- Orubu, E.; Samad, M.; Rahman, M.; Zaman, M.; Wirtz, V. Mapping the antimicrobial supply chain in Bangladesh: a scoping-review-based ecological assessment approach. Global Health: Science and Practice 2021, 9, 532-547. [CrossRef]
- Nizame, F.A.; Shoaib, D.M.; Rousham, E.K.; Akter, S.; Islam, M.A.; Khan, A.A.; Rahman, M.; Unicomb, L. Barriers and facilitators to adherence to national drug policies on antibiotic prescribing and dispensing in Bangladesh. Journal of Pharmaceutical Policy and Practice 2021, 14, 1-13. [CrossRef]
- Kabir, H.; Hasan, M.K.; Akter, N.; Tassdik, H.; Islam, M.F.; Jannat, H.; Tutul, A.H.; Akter, O.; Ara, R.; Islam, M.D. Antibiotics administration without prescription in Bangladesh. IJID regions 2023, 7, 11-17. [CrossRef]
- Sulis, G.; Sayood, S.; Katukoori, S.; Bollam, N.; George, I.; Yaeger, L.H.; Chavez, M.A.; Tetteh, E.; Yarrabelli, S.; Pulcini, C. Exposure to World Health Organization's AWaRe antibiotics and isolation of multidrug resistant bacteria: a systematic review and meta-analysis. Clinical Microbiology and Infection 2022, 28, 1193-1202.
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; Segre, J.A.; Westblade, L.F. Carbapenemase-producing organisms: a global scourge. Clinical infectious diseases 2018, 66, 1290-1297. [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011, 17, 1791-1798, doi:10.3201/eid1710.110655. [CrossRef]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460-469.
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis 2017, 215, S28-S36, doi:10.1093/infdis/jiw282. [CrossRef]
- Wilson, H.; Török, M.E. Extended-spectrum β-lactamase-producing and carbapenemase-producing Enterobacteriaceae. Microbial genomics 2018, 4. [CrossRef]
- Van Duin, D.; Arias, C.A.; Komarow, L.; Chen, L.; Hanson, B.M.; Weston, G.; Cober, E.; Garner, O.B.; Jacob, J.T.; Satlin, M.J. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. The Lancet Infectious Diseases 2020, 20, 731-741. [CrossRef]
- Goodman, K.; Simner, P.; Tamma, P.; Milstone, A. Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert review of anti-infective therapy 2016, 14, 95-108. [CrossRef]
- Livingstone, D.; Gill, M.; Wise, R. Mechanisms of resistance to the carbapenems. Journal of Antimicrobial Chemotherapy 1995, 35, 1-5.
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clinical Infectious Diseases 2017, 64, 257-264. [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem resistance: a review. Medical Sciences 2017, 6, 1. [CrossRef]
- Macesic, N.; Hawkey, J.; Vezina, B.; Wisniewski, J.A.; Cottingham, H.; Blakeway, L.V.; Harshegyi, T.; Pragastis, K.; Badoordeen, G.Z.; Dennison, A. Genomic dissection of endemic carbapenem resistance reveals metallo-beta-lactamase dissemination through clonal, plasmid and integron transfer. Nature Communications 2023, 14, 4764. [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008, 3, 163-175. [CrossRef]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-β-lactamases: the quiet before the storm? Clinical microbiology reviews 2005, 18, 306-325.
- Reddy, K.S. Global Burden of Disease Study 2015 provides GPS for global health 2030. The Lancet 2016, 388, 1448-1449. [CrossRef]
- Mokomane, M.; Kasvosve, I.; Melo, E.d.; Pernica, J.M.; Goldfarb, D.M. The global problem of childhood diarrhoeal diseases: emerging strategies in prevention and management. Therapeutic advances in infectious disease 2018, 5, 29-43. [CrossRef]
- WHO. Diarrhoeal disease: WHO. 2017.
- Troeger, C.E.; Khalil, I.A.; Blacker, B.F.; Biehl, M.H.; Albertson, S.B.; Zimsen, S.R.; Rao, P.C.; Abate, D.; Ahmadi, A.; brahim Ahmed, M.L.C. Quantifying risks and interventions that have affected the burden of diarrhoea among children younger than 5 years: an analysis of the Global Burden of Disease Study 2017. The Lancet Infectious Diseases 2020, 20, 37-59. [CrossRef]
- Wang, W.; Zhao, L.; Hu, Y.; Dottorini, T.; Fanning, S.; Xu, J.; Li, F. Epidemiological study on prevalence, serovar diversity, multidrug resistance, and CTX-M-type extended-spectrum β-lactamases of Salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrobial agents and chemotherapy 2020, 64. [CrossRef]
- Zheng, B.; Xu, H.; Lv, T.; Guo, L.; Xiao, Y.; Huang, C.; Zhang, S.; Chen, Y.; Han, H.; Shen, P. Stool samples of acute diarrhea inpatients as a reservoir of ST11 hypervirulent KPC-2-producing Klebsiella pneumoniae. MSystems 2020, 5, 10.1128/msystems. 00498-00420. [CrossRef]
- Ba, X.; Guo, Y.; Moran, R.A.; Doughty, E.L.; Liu, B.; Yao, L.; Li, J.; He, N.; Shen, S.; Li, Y. Global emergence of a hypervirulent carbapenem-resistant Escherichia coli ST410 clone. Nature Communications 2024, 15, 494. [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 2022, 399, 629-655. [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. The Lancet infectious diseases 2019, 19, 56-66. [CrossRef]
- Dadgostar, P. Antimicrobial resistance: implications and costs. Infection and drug resistance 2019, 3903-3910. [CrossRef]
- Hofer, U. The cost of antimicrobial resistance. Nature Reviews Microbiology 2019, 17, 3-3. [CrossRef]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Therapeutic advances in urology 2019, 11, 1756287219832172. [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature reviews microbiology 2015, 13, 269-284. [CrossRef]
- Foxman, B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. The American journal of medicine 2002, 113, 5-13. [CrossRef]
- Eshetie, S.; Unakal, C.; Gelaw, A.; Ayelign, B.; Endris, M.; Moges, F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrobial resistance and infection control 2015, 4, 1-8. [CrossRef]
- Dasgupta, C.; Rafi, M.A.; Salam, M.A. High prevalence of multidrug resistant uropathogens: A recent audit of antimicrobial susceptibility testing from a tertiary care hospital in Bangladesh. Pakistan journal of medical sciences 2020, 36, 1297.
- Urmi, U.L.; Nahar, S.; Rana, M.; Sultana, F.; Jahan, N.; Hossain, B.; Alam, M.S.; Mosaddek, A.S.M.; McKimm, J.; Rahman, N.A.A. Genotypic to Phenotypic Resistance Discrepancies Identified Involving β-Lactamase Genes, bla KPC, bla IMP, bla NDM-1, and bla VIM in Uropathogenic Klebsiella pneumoniae. Infection and drug resistance 2020, 2863-2875.
- Tompkins, K.; van Duin, D. Treatment for carbapenem-resistant Enterobacterales infections: recent advances and future directions. European Journal of Clinical Microbiology & Infectious Diseases 2021, 40, 2053-2068.
- Clancy, C.J.; Potoski, B.A.; Buehrle, D.; Nguyen, M.H. Estimating the treatment of carbapenem-resistant Enterobacteriaceae infections in the United States using antibiotic prescription data. In Proceedings of the Open forum infectious diseases, 2019; p. ofz344. [CrossRef]
- Hansen, G.T. Continuous evolution: perspective on the epidemiology of carbapenemase resistance among Enterobacterales and other Gram-negative bacteria. Infectious diseases and therapy 2021, 10, 75-92. [CrossRef]
- Löfmark, S.; Sjöström, K.; Mäkitalo, B.; Edquist, P.; Wisell, K.T.; Giske, C.G. Carbapenemase-producing Enterobacteriaceae in Sweden 2007–2013: experiences from seven years of systematic surveillance and mandatory reporting. Drug Resistance Updates 2015, 20, 29-38. [CrossRef]
- Miriagou, V.; Cornaglia, G.; Edelstein, M.; Galani, I.; Giske, C.; Gniadkowski, M.; Malamou-Lada, E.; Martinez-Martinez, L.; Navarro, F.; Nordmann, P. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clinical microbiology and infection 2010, 16, 112-122. [CrossRef]
- WHO. Antimicrobial stewardship programs in health-care facilities in low-and middle-income countries: a WHO practical toolkit. 2019.
- Pierce, J.; Apisarnthanarak, A.; Schellack, N.; Cornistein, W.; Al Maani, A.; Adnan, S.; Stevens, M.P. Global Antimicrobial Stewardship with a Focus on Low-and Middle-Income Countries: A position statement for the international society for infectious diseases. International Journal of Infectious Diseases 2020, 96, 621-629. [CrossRef]
- Nathwani, D.; Varghese, D.; Stephens, J.; Ansari, W.; Martin, S.; Charbonneau, C. Value of hospital antimicrobial stewardship programs [ASPs]: a systematic review. Antimicrobial Resistance & Infection Control 2019, 8, 1-13. [CrossRef]
- Cox, J.A.; Vlieghe, E.; Mendelson, M.; Wertheim, H.; Ndegwa, L.; Villegas, M.V.; Gould, I.; Hara, G.L. Antibiotic stewardship in low-and middle-income countries: the same but different? Clinical microbiology and infection 2017, 23, 812-818.
- Harun, M.G.D.; Anwar, M.M.U.; Sumon, S.A.; Hassan, M.Z.; Mohona, T.M.; Rahman, A.; Abdullah, S.A.H.M.; Islam, M.S.; Kaydos-Daniels, S.C.; Styczynski, A.R. Rationale and guidance for strengthening infection prevention and control measures and antimicrobial stewardship programs in Bangladesh: A study protocol. BMC health services research 2022, 22, 1-11. [CrossRef]
- Akpan, M.R.; Isemin, N.U.; Udoh, A.E.; Ashiru-Oredope, D. Implementation of antimicrobial stewardship programmes in African countries: a systematic literature review. Journal of global antimicrobial resistance 2020, 22, 317-324. [CrossRef]
- Otieno, P.A.; Campbell, S.; Maley, S.; Obinju Arunga, T.; Otieno Okumu, M. A Systematic Review of Pharmacist-Led Antimicrobial Stewardship Programs in Sub-Saharan Africa. International Journal of Clinical Practice 2022, 2022. [CrossRef]
- Sumon, S.; Anwar, M.; Akther, F.; Priyanka, A.; Tamanna, T.; Rahman, A.; Islam, M.; Harun, M.D. Perceptions of antibiotic stewardship programmes and determinants of antibiotic prescribing patterns among physicians in tertiary hospitals in Bangladesh: implications for future policy and practice. Journal of Hospital Infection 2024, 144, 56-65. [CrossRef]
- Haque, M.; Godman, B. Potential strategies to improve antimicrobial utilisation in hospitals in Bangladesh building on experiences across developing countries. Bangladesh Journal of Medical Science 2021, 20, 469-477. [CrossRef]
- M. Kurdi Al-Dulaimi, M.; Abd. Mutalib, S.; Abd. Ghani, M.; Mohd. Zaini, N.A.; Ariffin, A.A. Multiple antibiotic resistance (MAR), plasmid profiles, and DNA polymorphisms among Vibrio vulnificus isolates. Antibiotics 2019, 8, 68.
- Bulik, C.C.; Fauntleroy, K.A.; Jenkins, S.G.; Abuali, M.; LaBombardi, V.J.; Nicolau, D.P.; Kuti, J.L. Comparison of meropenem MICs and susceptibilities for carbapenemase-producing Klebsiella pneumoniae isolates by various testing methods. Journal of clinical microbiology 2010, 48, 2402-2406. [CrossRef]
- Humphries, R.M.; Hindler, J.A.; Epson, E.; Horwich-Scholefield, S.; Miller, L.G.; Mendez, J.; Martinez, J.B.; Sinkowitz, J.; Sinkowtiz, D.; Hershey, C. Carbapenem-resistant Enterobacteriaceae detection practices in California: what are we missing? Clinical Infectious Diseases 2018, 66, 1061-1067.
- Barbosa, C.; Nogueira, S.; Gadanho, M.; Chaves, S. DNA extraction: finding the most suitable method. In Molecular microbial diagnostic methods; Elsevier: 2016; pp. 135-154.
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagnostic microbiology and infectious disease 2011, 70, 119-123. [CrossRef]
- Gupta, V.; Datta, P.; Chander, J. Prevalence of metallo-β lactamase (MBL) producing Pseudomonas spp. and Acinetobacter spp. in a tertiary care hospital in India. Journal of Infection 2006, 52, 311-314.
- Mirbagheri, S.Z.; Meshkat, Z.; Naderinasab, M.; Rostami, S.; Nabavinia, M.S.; Rahmati, M. Study on imipenem resistance and prevalence of blaVIM1 and blaVIM2 metallo-beta lactamases among clinical isolates of Pseudomonas aeruginosa from Mashhad, Northeast of Iran. Iranian Journal of Microbiology 2015, 7, 72.
- Ministry of Health and Family Welfare (MoHFW), G.o.B. National Action Plan: Antimicrobial Resistance Containment in Bangladesh 2017-22.; 2017.
- Mishra, S.; Acharya, J.; Kattel, H.; Koirala, J.; Rijal, B.; Pokhrel, B. Metallo-beta-lactamase producing gram-negative bacterial isolates. 2012.
- Emeraud, C.; Escaut, L.; Boucly, A.; Fortineau, N.; Bonnin, R.A.; Naas, T.; Dortet, L. Aztreonam plus Clavulanate, Tazobactam, or Avibactam for Treatment of Infections Caused by Metallo-beta-Lactamase-Producing Gram-Negative Bacteria. Antimicrob Agents Chemother 2019, 63, doi:10.1128/AAC.00010-19. [CrossRef]
- Canton, R.; Ruiz-Garbajosa, P. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol 2011, 11, 477-485, doi:10.1016/j.coph.2011.07.007. [CrossRef]
- Rice, L.B. Antimicrobial stewardship and antimicrobial resistance. Medical Clinics 2018, 102, 805-818. [CrossRef]
- de Carvalho, F.R.T.; Telles, J.P.; Tuon, F.F.B.; Rabello Filho, R.; Caruso, P.; Correa, T.D. Antimicrobial stewardship programs: A review of strategies to avoid polymyxins and carbapenems misuse in low middle-income countries. Antibiotics 2022, 11, 378. [CrossRef]
- Sono, T.M.; Yeika, E.; Cook, A.; Kalungia, A.; Opanga, S.A.; Acolatse, J.E.E.; Sefah, I.A.; Jelić, A.G.; Campbell, S.; Lorenzetti, G. Current rates of purchasing of antibiotics without a prescription across sub-Saharan Africa; rationale and potential programmes to reduce inappropriate dispensing and resistance. Expert review of anti-infective therapy 2023, 21, 1025-1055. [CrossRef]
- Al Jarousha, A.M.K.; El Jarou, M.A.; El Qouqa, I.A. Bacterial enteropathogens and risk factors associated with childhood diarrhea. The Indian Journal of Pediatrics 2011, 78, 165-170. [CrossRef]
- Zhou, Y.; Zhu, X.; Hou, H.; Lu, Y.; Yu, J.; Mao, L.; Mao, L.; Sun, Z. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: a hospital based study. BMC infectious diseases 2018, 18, 1-10. [CrossRef]
- Meyer, E.; Schwab, F.; Schroeren-Boersch, B.; Gastmeier, P. Dramatic increase of third-generation cephalosporin-resistant E. coli in German intensive care units: secular trends in antibiotic drug use and bacterial resistance, 2001 to 2008. Critical care 2010, 14, 1-9. [CrossRef]
- Zanichelli, V.; Sharland, M.; Cappello, B.; Moja, L.; Getahun, H.; Pessoa-Silva, C.; Sati, H.; van Weezenbeek, C.; Balkhy, H.; Simão, M. The WHO AWaRe (Access, Watch, Reserve) antibiotic book and prevention of antimicrobial resistance. 2023. [CrossRef]
- Ansari, M.; Aryal, S.C.; Rai, G.; Rai, K.R.; Pyakurel, S.; Bhandari, B.; Sah, A.K.; Rai, S.K. Prevalence of multidrug-resistance and bla VIM and bla IMP genes among gram-negative clinical isolates in tertiary care hospital, Kathmandu, Nepal. Iranian journal of microbiology 2021, 13, 303.
- Chander, A.; Shrestha, C.D. Prevalence of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae urinary isolates in a tertiary care hospital in Kathmandu, Nepal. BMC Research notes 2013, 6, 1-6. [CrossRef]
- Ronald, A. The etiology of urinary tract infection: traditional and emerging pathogens. The American journal of medicine 2002, 113, 14-19. [CrossRef]
- Duffy, E.; Ritchie, S.; Metcalfe, S.; Van Bakel, B.; Thomas, M. Antibacterials dispensed in the community comprise 85%-95% of total human antibacterial consumption. Journal of clinical pharmacy and therapeutics 2018, 43, 59-64. [CrossRef]
| Age group (Years) | Diarrheal pathogens (n=228) |
Urinary tract infection pathogens (n=142) | |||
|---|---|---|---|---|---|
| Frequency | Percentage | Frequency | Percentage | ||
| 1-10 | 191 a | 83.8 | 5 | 3.5 | |
| 11-20 | 5 | 2.2 | 22 | 15.5 | |
| 21-30 | 8 | 3.5 | 50 b | 35.2 | |
| 31-40 | 10 | 4.4 | 19 b | 13.4 | |
| 41-50 | 0 | 0 | 27 b | 19.0 | |
| 51-60 | 5 | 2.2 | 8 | 5.6 | |
| 61-70 | 0 | 0 | 9 | 6.3 | |
| 71-80 | 9 | 3.9 | 2 | 1.4 | |
| Total | 228 | 100 | 142 | 100 | |
| MIC values (µg/mL) | Diarrheal pathogens | Urinary tract infection pathogen | |||||||
|---|---|---|---|---|---|---|---|---|---|
| bla-NDM-1 carriage a | bla-VIM carriage b | bla-NDM-1 carriage c | bla-VIM carriage d | ||||||
| Yes (n=12) ∆ | No (n=216) | Yes (n=12) | No (n=216) | Yes (n=13) | No (n=129) | Yes (n=23) | No (n=119) | ||
| ≤0.5 | 6 (50) | 109 (55) | 7 (58) | 108 (55) | 2 (15) | 90 (71) | 12 (52) | 80 (68) | |
| 1.0 | 1 (8) | 7 (3.5) | 1 (8) | 7 (3.6) | 1 (7.7) | 14 (11) | 1 (4.3) | 14 (12) | |
| 2.0 | 0 (0) | 9 (4.5) | 0 (0) | 9 (4.6) | 0 (0) | 9 (7.1) | 0 (0) | 9 (7.7) | |
| 4.0 | 0 (0) | 17 (8.6) | 1 8) | 16 (8.1) | 6 (46) | 6 (4.7) | 7 (30) | 5 (4.3) | |
| 8.0 | 1 (8) | 14 (7.1) | 0 (0) | 14 (7.1) | 1 (7.7) | 4 (3.1) | 0 (0) | 5 (4.3) | |
| 16 | 1 (8) | 1 (0.5_ | 0 (0) | 2 (1) | 0 (0) | 2 (1.6) | 0 (0) | 2 (1.7) | |
| ≥32 | 3 (25) | 41 (21) | 3 (25) | 41 (21) | 3 (23) | 2 (1.6) | 3 (13) | 2 (1.7) | |
| MBL genes a | Isolate ID | Phenotypic susceptibility assessment b | MAR index c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC 30 | CXM 30 | CFM 30 | FEP 30 | IMP 10 | MRP 10 | CN 30 | NET 30 | AK 30 | |||
|
blaNDM-1 |
PBD20 | R | R | R | R | R | R | S | S | R | 0.8 |
| PBD24 | R | S | S | S | S | S | S | S | R | 0.2 | |
| PBD78 | R | S | R | S | S | S | S | S | S | 0.2 | |
| PBD78C1 | R | R | R | R | R | R | S | S | S | 0.7 | |
| PBD78C2 | R | R | R | R | R | R | S | S | S | 0.7 | |
| PBD86 | S | S | S | S | S | S | S | S | S | 0 | |
| PBD86C1 | R | R | R | R | R | R | R | R | R | 1 | |
| PBD86C2 | R | R | R | R | R | R | R | R | R | 1 | |
| PBD103 | S | R | R | S | S | S | R | S | R | 0.4 | |
| UJ8 | R | R | R | R | R | S | R | R | S | 0.8 | |
| UJ18C2 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ29 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ49 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ56C1 | R | R | R | R | S | R | R | R | R | 0.9 | |
| UJ73 | R | R | R | R | R | S | R | R | R | 0.9 | |
|
blaVIM |
PBD5 | R | S | R | R | R | R | S | S | S | 0.6 |
| PBD8 | R | R | R | S | R | S | S | S | S | 0.4 | |
| PBD40 | S | R | R | S | R | S | S | S | S | 0.3 | |
| PBD53C2 | R | R | S | S | R | S | S | S | R | 0.4 | |
| PBD55 | S | S | S | S | S | R | R | S | S | 0.2 | |
| PBD77 | S | S | S | S | S | S | R | R | S | 0.2 | |
| PBD80C2 | S | R | R | R | S | S | R | S | S | 0.4 | |
| PBD81 | R | R | R | S | S | S | S | S | S | 0.3 | |
| PBD87 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ15 | R | R | R | R | R | R | R | R | S | 0.9 | |
| UJ19 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ39 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ57 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ59 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ62 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ75 | R | R | R | R | R | S | R | R | S | 0.8 | |
| UJ76C1 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ77 | R | R | R | R | R | S | R | R | S | 0.8 | |
| UJ78C2 | R | R | R | R | R | S | R | R | S | 0.8 | |
| UJ98 | R | R | R | R | S | R | S | S | S | 0.6 | |
| UJ108C1 | S | R | S | S | R | S | S | S | S | 0.2 | |
| UJ112 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ116 | R | R | R | R | S | S | R | R | R | 0.8 | |
| UJ120 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ123 | R | R | R | R | R | S | S | S | S | 0.6 | |
| blaNDM-1+ blaVIM | UJ48 | R | R | R | R | R | R | R | R | R | 1 |
| UJ79C1 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ79C2 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ87C1 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ87C2 | R | R | R | R | R | S | R | R | R | 0.9 | |
| UJ88 | R | R | R | R | R | R | R | R | R | 1 | |
| UJ90 | R | R | R | R | R | R | R | R | R | 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





