Submitted:
03 March 2024
Posted:
05 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Symmetries
2.1. Symmetry
2.2. Noether’s Theory in Developmental Biology
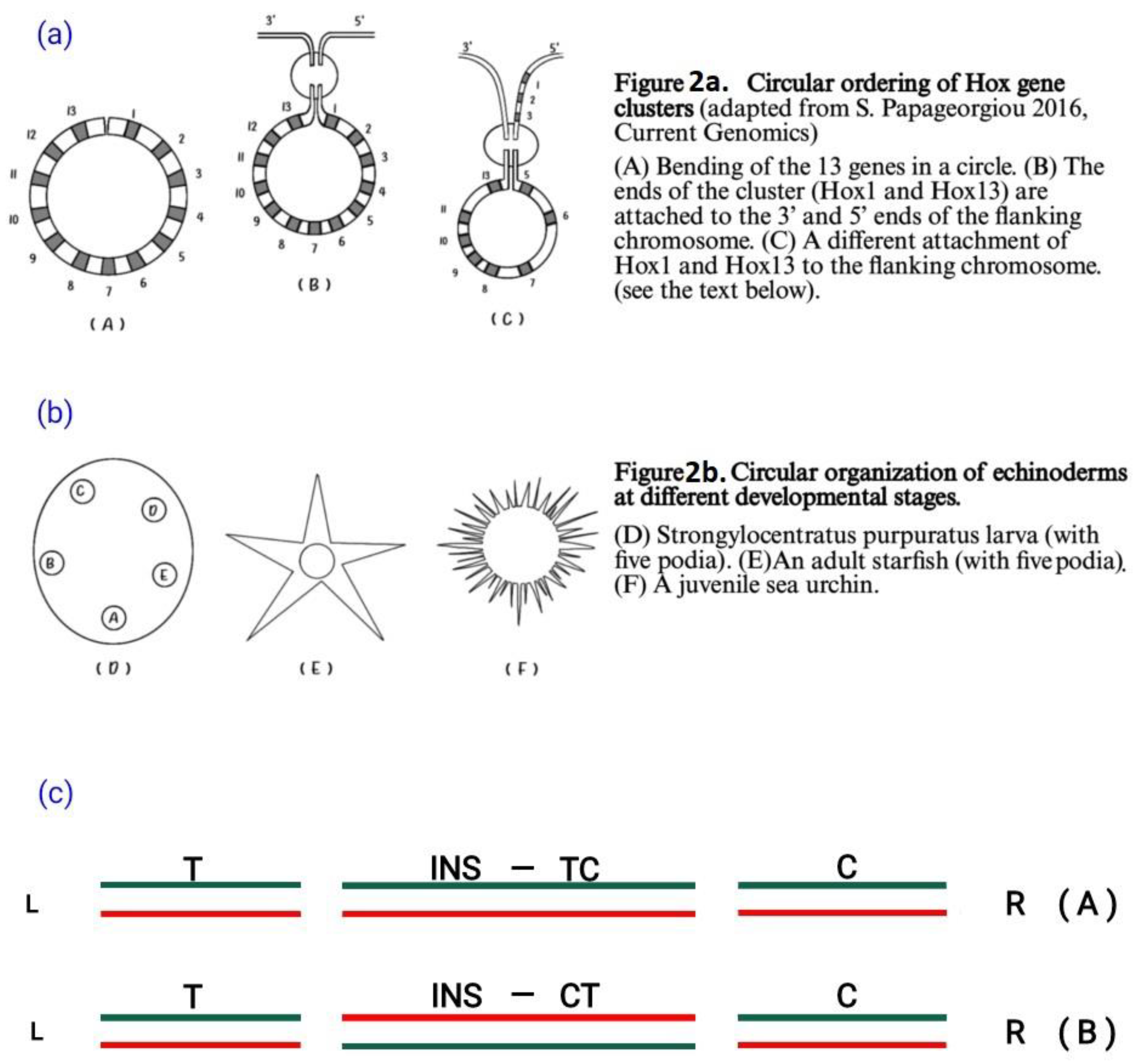
3. Insertion of a DNA Fragment in a Hosting DNA Sequence
3. Complete vs Split Hox Clusters
4. Discussion
4.1. Empirical Rule on Complete and Split Hox Clusters
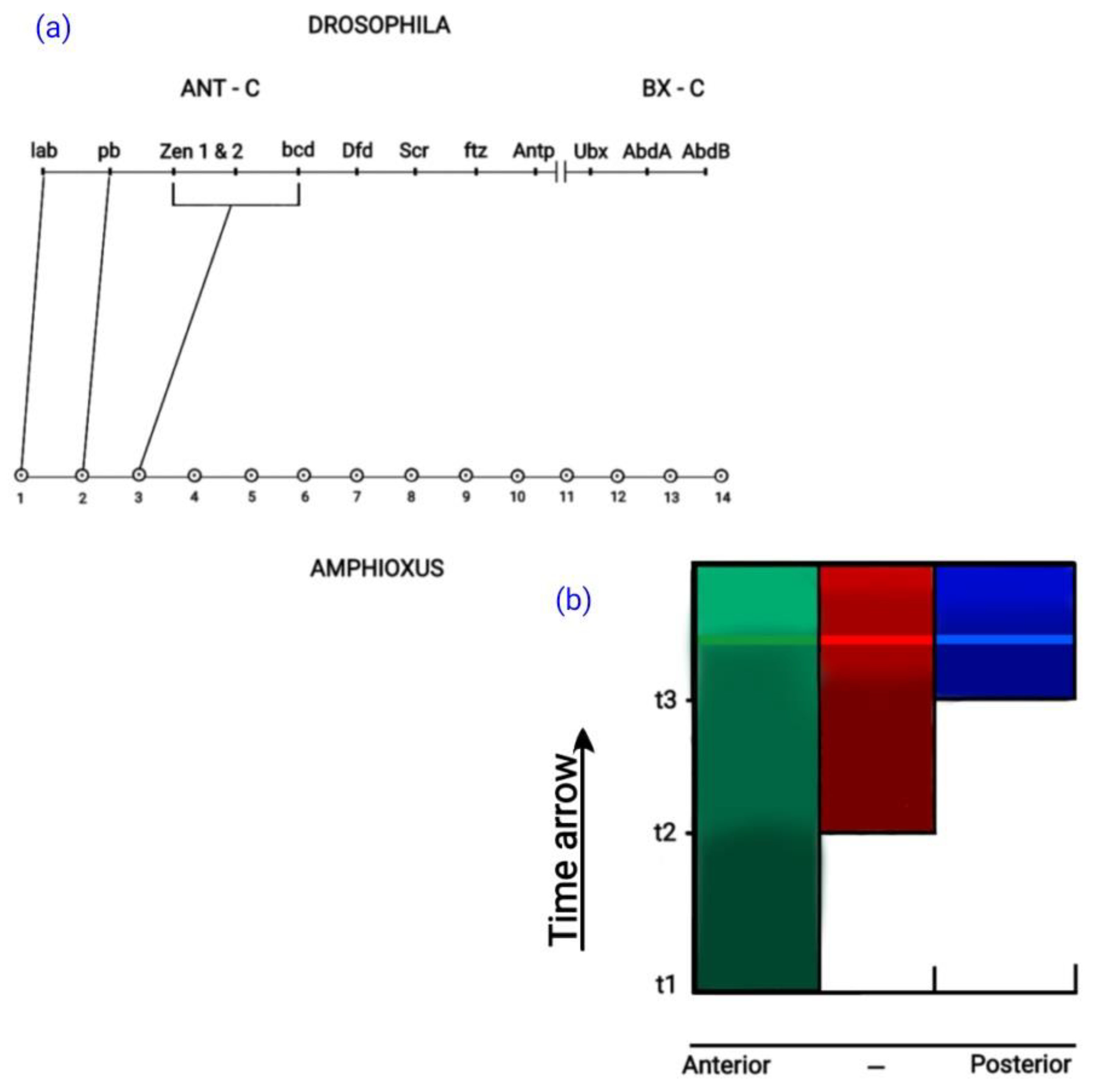
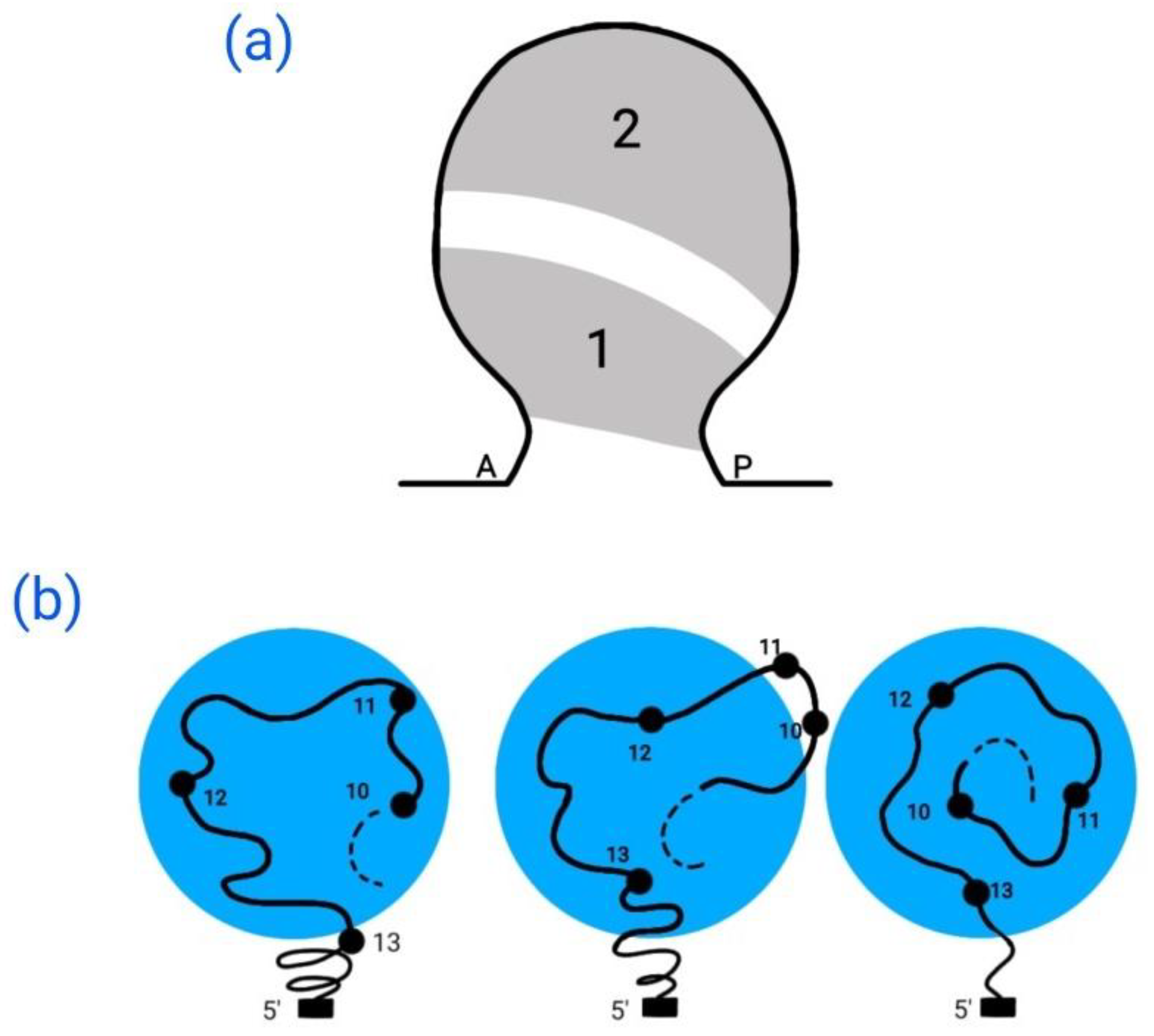
4.2. Development in the Secondary Developmental Axis
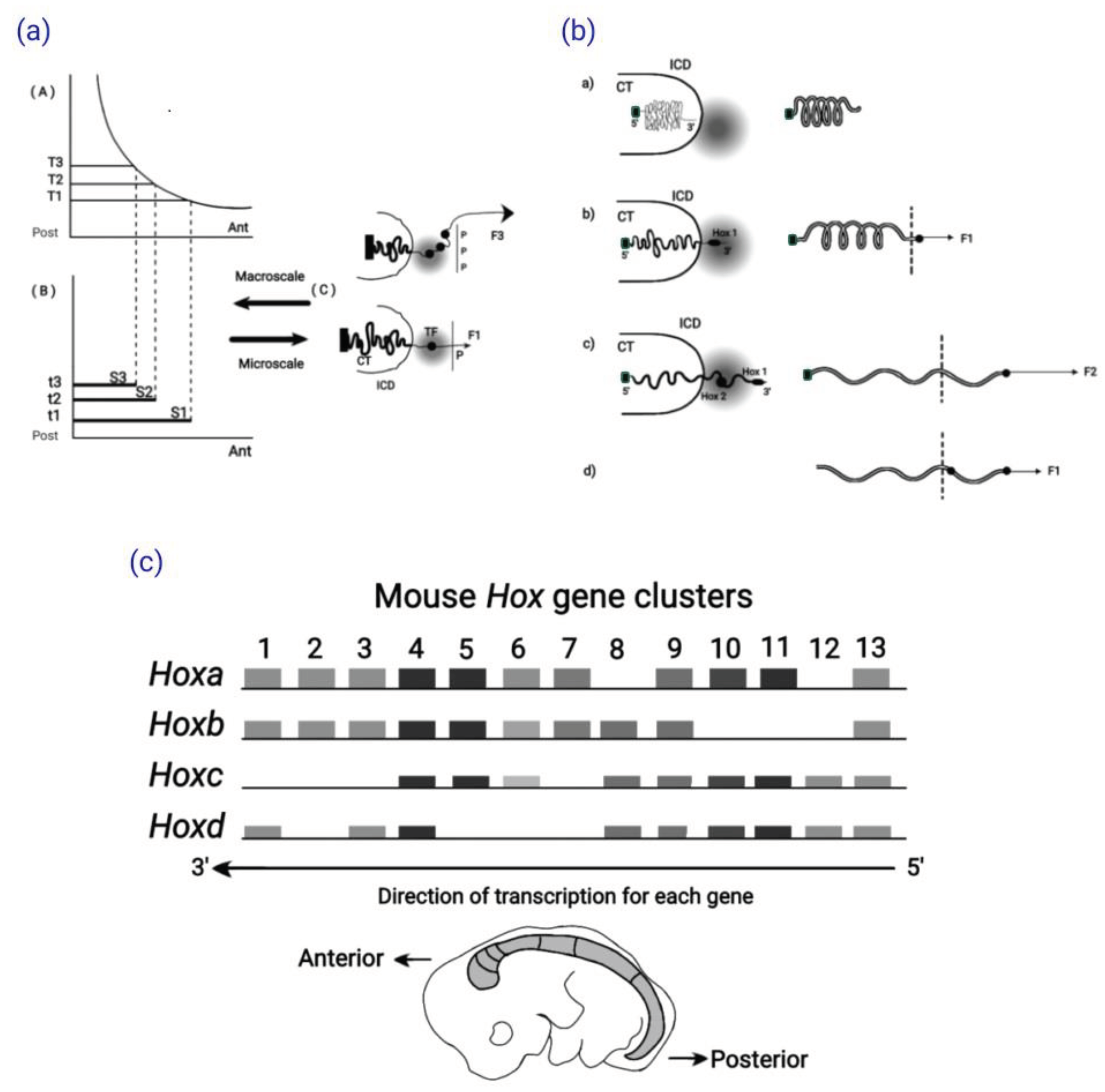
4.3. Development in the Mouse Primary Anterior-Posterior Axis
4.4. Quantitative Collinearity
4.5. A Spontaneous Mutation in the Drosophila Case
5. Complex Expression Patterns
6. Recent Findings Caused by BM Physical Forces
6.1.a Physical Forces May Cause a Tension in the Hox Clusters
6.1.b Proposal for a Test of 6.1a
6.2. Confirmation That FGF Causes the Necessary (but Not Sufficient) Condition for Limb Growth
7. Conclusions and Predictions
7.1
7.2
7.3
Epilogue
List of Abbreviations
| AER | Apical Ectodermal Ridge |
| BM | Biophysical Model |
| CF | Coulomb Force |
| CT | Chromatin territory |
| DSB | Double Strand Break |
| ER | Empirical Rule |
| FGF | Fibroblast Growth Factor |
| HGC | Hox Gene Collinearity |
| ICD | Interchromosome domain |
| Mya | Million years ago |
| PG | Paralogy Group |
| PP | Posterior Prevalence |
| QC | Quantitative Collinearity |
| Quasi Coulomb force | |
| SC | SpatialCollinearity |
| TC | TemporalCollinearity |
| TFD | Transcription Factory Domain |
| wt | wild type |
References
- Lewis E.B A gene complex controlling segmentation in Drosophila. Nature 1978 Hox genes 276, 565.
- Papageorgiou S. A physical force may expose express in a morphogenetic density gradient. Bull. Math.Biol. 2001, 63,185-200. [CrossRef] [PubMed]
- Dollé P et al., HOX-4 and the morphogenesis of mammalian genitalia. Genes and Development 1991,. [CrossRef]
- Papageorgiou S. Pulling forces acting on Hox gene clusters cause expression collinearity. Int.J. Dev.Biol.2006, 50, 301-308. [CrossRef] [PubMed]
- Tarchini B, Duboule D Control Hoxd genes collinearity during animal development. Dev Cell 2006, 10, 93-103. [CrossRef] [PubMed]
- Tschopp P et al. Uncoupling time and space in the collinear regulation of hox genes.PLOS Genetics 2009, 5(3). [CrossRef]
- Papageorgiou S. A biophysical mechanism may control the collinearity of hoxd genes duting the early phase of limb development. Dev Growth & Differ 2011, 3, 275-280. [CrossRef]
- Papageorgiou S Physical laws shape up Hox gene collinearity. J. Dev. Biol. 2021, 9, 17. [CrossRef]
- Duboule D The rise and fall of Hox gene clusters. Development 2007, 134, 2549-2560 I. [CrossRef]
- Shimizu K, Gurdon J.B. A quantitative analysis of signal transduction from activin receptor to nucleus. Proc. Nat. Academy of Sci.1999, 8;96(12): 6791-96. [CrossRef]
- Afzal Z, Krumlauf R. Transacriptional regulation and implications for controlling Hox gene expression J. Dev. Biol.2022, 10; 10(1):4. [CrossRef]
- Simeoni I , Gurdon J.B. Interpretation of BMP signaling in early Xenopus development Dev.Biol. 2007, 308 (1): 82-92. [CrossRef]
- Bourillot Y-P et al. A changing morphogen gradient is interpreted by continuous transduction flow. Development 2002, 129(9): 2167-80. [CrossRef]
- Papageorgiou S Comparison of models for the collinearity of Hox genes in the developmental axes of vertebrates. Curr. Genomics 2012, 13(3): 245-51. [CrossRef]
- NoordermeerD et al. Temporal dynamics and development memory of 3D chromatin architecture at hox gene loci., eLIFE2014, 3, e0255. [CrossRef]
- Fabre P et al, .Nanoscale spatial organization of the HoxD gene cluster organization in distinct transcriptional states Proc. Nat. Acad. Sci.2015, 112, 13964- 13969. [CrossRef]
- Fabre P et al,. Visualizing HoxD gene cluster at the nanoscale level. Cold Spring Harb.Symp. Quant. Biol. 2015, 80, 9-16. [CrossRef] [PubMed]
- Papantonis A, Cook P.R. Fixing the model for transcription: the DNA moves, not the polymerase. Transcription 2011, 2(1) :41-44. [CrossRef]
- Brackley C.A. et al, Complex small-world regulatory networks emerge from the 3D organization of the human genome. Nat. Commun. 2021, 12(1): 5756. [CrossRef]
- Wilczek F, A Beautiful Question, 2015, Allen Lane Editions .
- Marinho R.M., Noether’s theorem in Classical Mechanics revisited. Eur. J. Phys. 2006, 28, 37-43. [CrossRef]
- Papageorgiou S, Hox gene collinearity may be related to Noether Theory on Symmetry and its linked conserved quantity. J. Multidiscipl. Sci. J 2020, 3, 13. [CrossRef]
- Iliopoulos J, Aux origins de la masse: particules élémentaires et symmetries fondamentales, 2014, Editions EDPSciences, Paris, France.
- Mandelbrot B.B. ‘The Fractal Geometry of Nature’, 1982, Editions Freeman, New York.
- 25. Almirantis Y, Provata A, Li W, Noether’s Theory as a metaphor for Chargaff’s 2nd parity rule in Genomics. J. Mol. Evol.,2022,. [CrossRef]
- Hanscom T et al., Regulation of Error-Prone DNA Double-Strand Break Repair and its impact on genome evolution, 2020, Cells 9, 165. [CrossRef]
- Schiemann S.M., Heznol A. et al. Clustered brachiopod Hox genes are not expressed collinearly and are associated with lophotrochozoan. PNAS USA 2017, 114(10) E1913-E1922. [CrossRef]
- Ferrier D, Holland P. Cionaintestinalis ParaHox genes: Evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal collinearity. Mol. Phyl. Evol.2002, 24:412-417. [CrossRef]
- Ferrier D in ‘HOX GENE EXPRESSION’ Editor S. Papageorgiou, Landes Bioscience and Springer Science, 2007, USA.
- Duboule D The (unusual) heuristic value of Hox gene clusters: a mater of time? Dev. Biol.2022, 484:75-87. [CrossRef]
- Kondo T, Duboule D Breaking collinearity in the mouse HoxD complex. Cell 1999, 97, 407-417. [CrossRef]
- Vargesson N et al. Characterisation of HoxA gene expression in the chick limb bud in response to FGF. Dev. Dynamics 2001,. [CrossRef]
- 33. Papageorgiou S. The multiple roles of Temporal Collinearity in Hox gene clustering Preprints. [CrossRef]
- Papageorgiou S Disappearance of Temporal collinearity in Vertebrates and its eventual Reappearance Biology 2021, 10(10): 1018. [CrossRef]
- Durston A.J. Global Posterior Prev9alence is unique to Vertebrates: a dance to the music of Time? Dev. Dynamics 2012, 241: 179-1807. [CrossRef]
- Durston A, Some questions and answers about the role of Hox Temporal Collinearity. [CrossRef]
- Andrey G. et al. A switch between topological domains underlies HoxD genes colline arity in mouse limbs. Science 2013, 340/1234167. [CrossRef]
- Papageorgiou S. Biophysics precedes Biochemistry in Hox Gene Collinearity. http://webmedcentralplus.com/ article_ view/405.
- Sedas Perez S, et al. Fgf signalling triggers an intrinsic mesodermal timer that determines the duration of limb patterning. Nature Com. 2023, 14(1), 5841. [CrossRef]
- Amândio A.R. et al. Sequential in cis mutagenesis in vivo reveals various functions for CTCF sites at the mouse HoxD cluster. Genes & Development 2021, 35(21-22):1490-1509. [CrossRef]
- Papageorgiou S. In ‘Chaos, Information processing and Paradoxical Games’ Editors Nicolis G - Basios V, World Scientific, 2015.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



