Submitted:
04 March 2024
Posted:
05 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cultures Employed
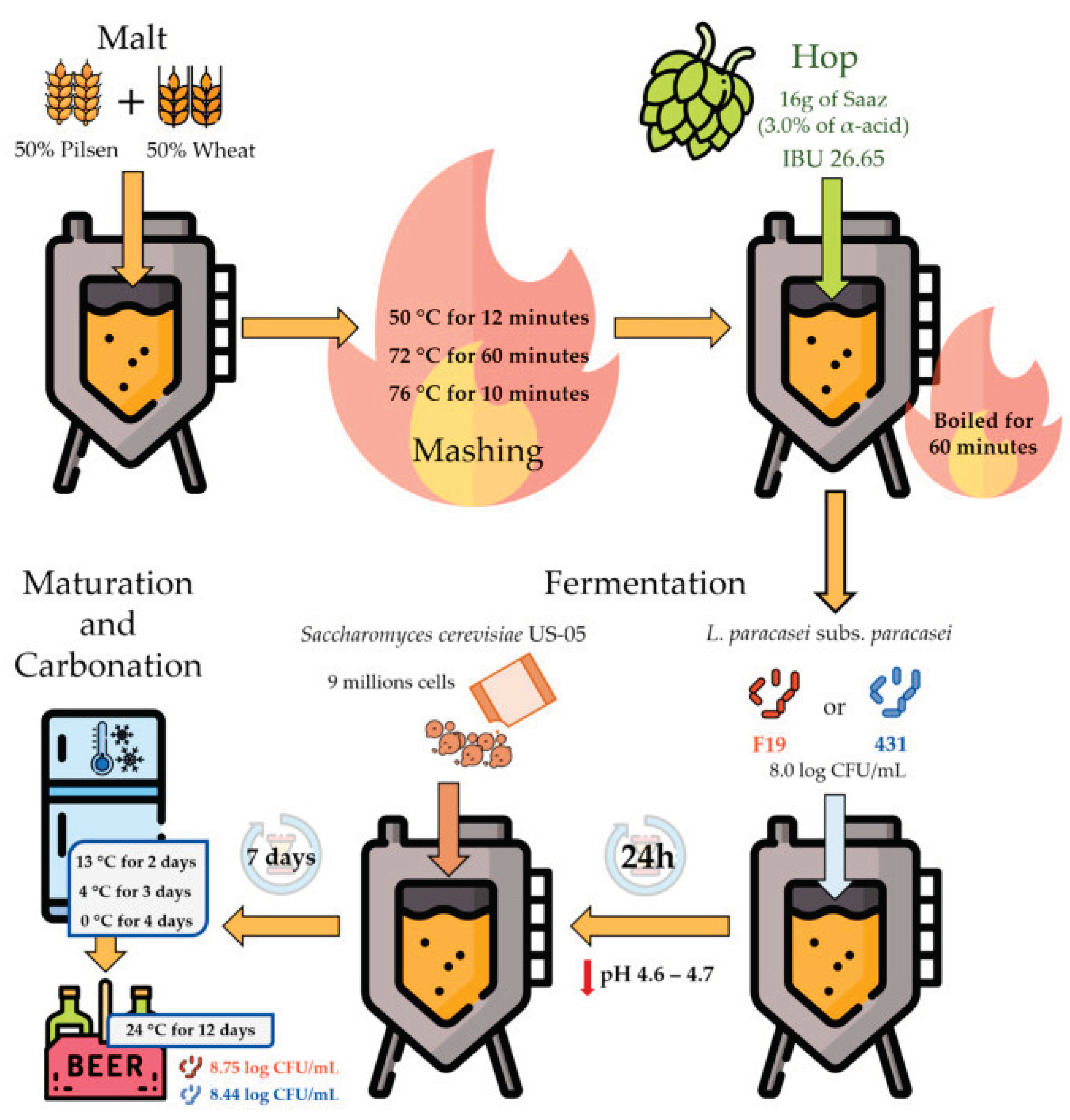
2.2. Formulation, and Brewery of the Sour Beer
2.3. Determination of pH and Microbiological Analyses
2.4. PMA Treatment and DNA and RNA Extraction
2.5. PMA-qPCR
2.6. RT-qPCR for Expression of Hop Resistance Genes
2.7. Statistical Analysis
3. Results and Discussion
3.1. pH and Microbial Populations
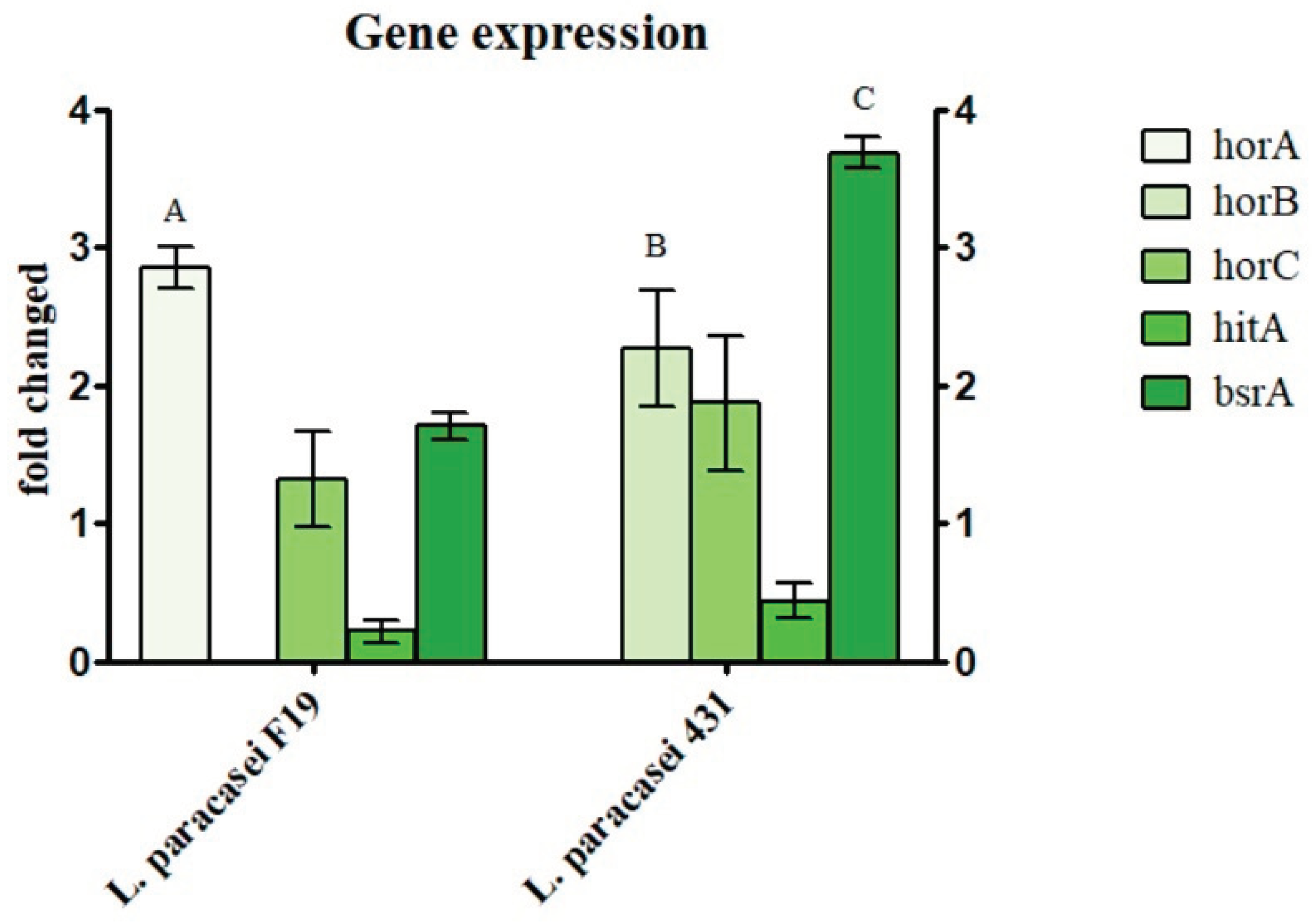
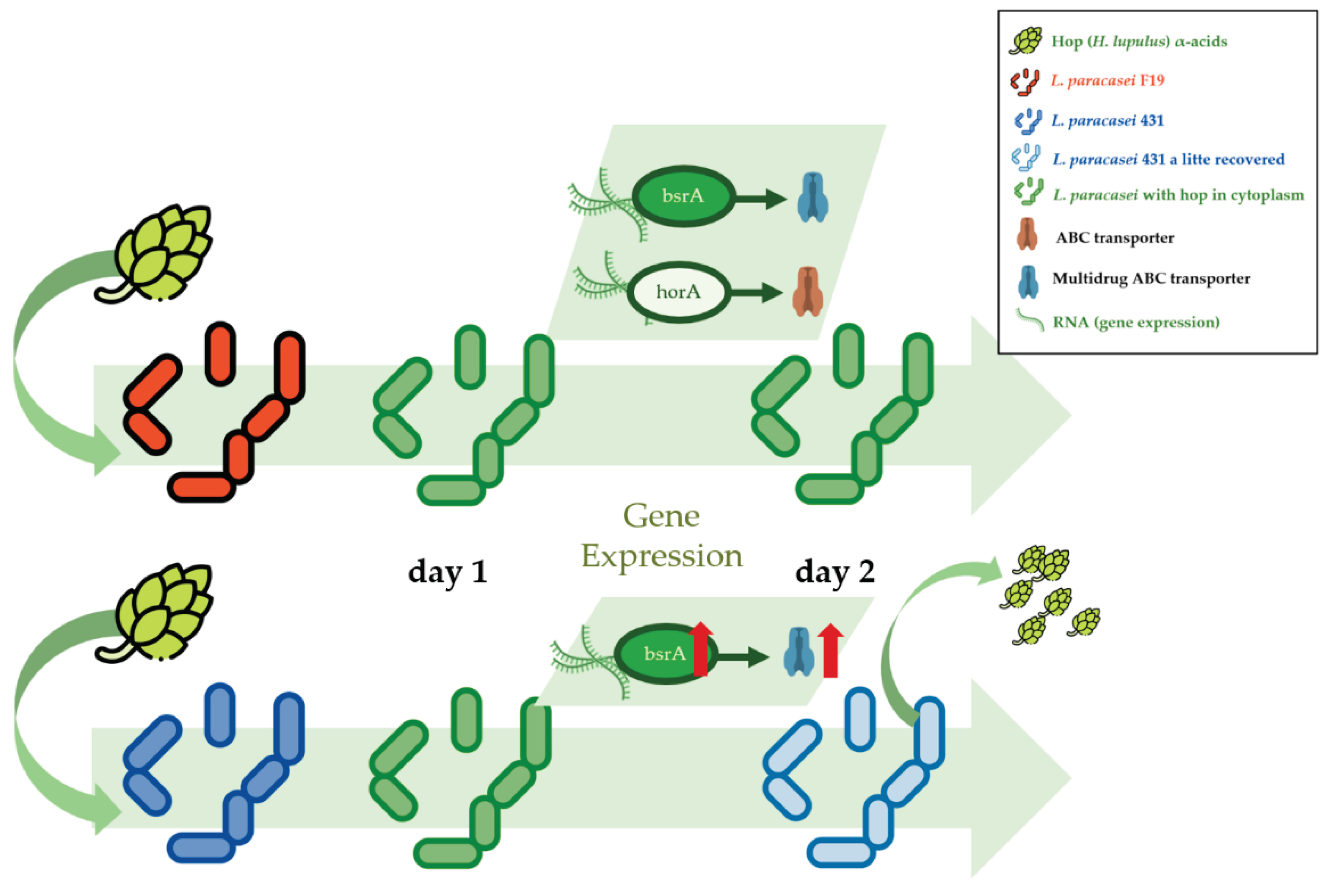
3.2. A-Acids Stress and Hop Resistance Genes Expression
3.3. A new Hop Sour Beer Style?
3.4. Final Considerations
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Callejo, M.J.; Tesfaye, W.; González, M.C.; Morata, A. Craft beers: Current situation and future trends. IntechOpen 2019. [Google Scholar] [CrossRef]
- Watson, B. State of the industry. 2020. Available online: https://www.brewersassociation.org/wp-content/uploads/2020/04/CBC-Online-2020-State-of-the-Craft-Brewing-Industry.pdf (accessed on 16 October 2023).
- Codifava, N. Craft beer revolution: Appropriability regime of the innovation and business model archetypes adopted by most growing craft breweries. 2018; Dissertation, Politecnico di Milano. [Google Scholar]
- Neves, V. Ramo de cervejarias artesanais cresceu 91% nos últimos três anos. Jornal da USP. 2019. Available online: https://jornal.usp.br/atualidades/ramo-de-cervejarias-artesanais-cresce-91-nos-ultimos-tres-anos (accessed on 16 October 2019).
- Dysvik, A.; La Rosa, S.L.; De Rouck, G.; Rukke, E.O.; Westereng, B.; Wicklund, T. Microbial Dynamics in Traditional and Modern Sour Beer Production. Appl Environ Microbiol. 2020, 86(14), e00566-20. [Google Scholar] [CrossRef]
- Ballan, R.; Battistini, C.; Xavier-Santos, D.; Saad, S.M.I. Interactions of probiotics and prebiotics with the gut microbiota. Prog Mol Biol Transl Sci. 2020, 171, 265–300. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H. J.; Salminen, S.; Calder, P.C.; Sanders, M.E. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Rev Gastroent Hepatol 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Knez Hrnčič, M.; Španinger, E.; Košir, I.; Knez, Ž.; Bren, U. Hop Compounds: Extraction Techniques, Chemical Analyses, Antioxidative, Antimicrobial, and Anticarcinogenic Effects. Nutrients 2019, 11(2), 257. [Google Scholar] [CrossRef]
- Aggarwal, D.; Upadhyay, S.K.; Singh, R.; Tuli, H.S. Recent patents on therapeutic activities of xanthohumol: a prenylated chalconoid from hops (Humulus lupulus L.). Pharm Pat Anal 2021. [Google Scholar] [CrossRef]
- Alcine Chan, M.Z.; Chua, J.Y.; Toh, M.; Liu, S.Q. Survival of probiotic strain Lactobacillus paracasei L26 during co-fermentation with S. cerevisiae for the development of a novel beer beverage. Food Microbiol. 2019, 82, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.C.; Schmidt, G.B.; Alves, L.G.O.; Oliveira, V.S.; Laureano-Melo, R.; Stutz, E.; Martins, J.F.P.; Paula, B.P.; Luchese, R.H.; Guerra, A.F.; Rodriguesa, R. Use of probiotic strains to produce beers by axenic or semi-separated co-culture system. Food Bioprod Process 2020, 124, 408–418. [Google Scholar] [CrossRef]
- Praia, A.B.; Herkenhoff, M.E.; Broedel, O.; Frohme, M.; Saad, S.M.I. Sour Beer with Lacticaseibacillus paracasei subsp. paracasei F19: Feasibility and Influence of Supplementation with Spondias mombin L. Juice and/or By-Product. Foods 2022, 11(24), 4068. [Google Scholar] [CrossRef] [PubMed]
- Herkenhoff, M.E.; Battistini, C.; Praia, A.B.; Rossini, B.C.; Dos Santos, L.D.; Brödel, O.; Frohme, M.; Saad, S.M.I. The combination of omics strategies to evaluate starter and probiotic strains in the Catharina sour Brazilian-style beer. Food Res Int. 2023, 167, 112704. [Google Scholar] [CrossRef] [PubMed]
- ASBC - American Society of Brewing Chemists - ASBC Methods of Analysis - Beer Bitterness. Published in March 1 2015st and updated in March 31st 2022. 2022. Available online: http://methods.asbcnet.org/about.aspx (accessed on 4 October 2022).
- Herkenhoff, M.E.; de Medeiros, I.U.D.; Garutti, L.H.G.; Salgaço, M.K.; Sivieri, K.; Saad, S.M.I. Cashew By-Product as a Functional Substrate for the Development of Probiotic Fermented Milk. Foods 2023, 12(18), 3383. [Google Scholar] [CrossRef]
- Shahrokhi, M.; Nagalli, S. Probiotics. In: StatPearls [Internet], Treasure Island (FL): StatPearls Publishing; 2020 Jan–. PMID: 31985927. [PubMed]
- Cazorla, S.I.; Maldonado-Galdeano, C.; Weill, R.; De Paul, J.; Perdigón, G.D.V. Oral Administration of Probiotics Increases Paneth Cells and Intestinal Antimicrobial Activity. Frontiers in Microbiology. 2018, 9, 736. [Google Scholar] [CrossRef]
- Kawa-Rygielska, J.; Adamenko, K.; Pietrzak, W.; Paszkot, J.; Głowacki, A.; Gasiński, A.; Leszczyński, P. The Potential of Traditional Norwegian KVEIK Yeast for Brewing Novel Beer on the Example of Foreign Extra Stout. Biomolecules 2021, 11(12), 1778. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Mazza, A.; Masson, L.; Camper, A.K.; Brousseau, R. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J Microbiol Methods 2009, 76((3)), 253–261. [Google Scholar] [CrossRef] [PubMed]
- Padilha, M.; Morales, M.L.V.; Vieira, A.D.S.; Costa, M.G.M.; Saad, S.M.I. A prebiotic mixture improved Lactobacillus acidophilus and Bifidobacterium animalis gastrointestinal in vitro resistance in petit-suisse. Food Funct 2016, 7, 2312–2319. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, A.; Håkansson, J. Development of a standardized method for the quantification of Lactobacillus paracasei F19 in stoll samples of various ages. EC Nutrition. 2016, 3, 633–642. [Google Scholar] [CrossRef]
- Byun, R.; Nadkarni, M.A.; Chhour, K.-L.; Martins, F.E.; Jacques, N.A.; Hunter, N. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. Journal of Clinical Microbiology. 2004, 42(7), 3128–3136. [Google Scholar] [CrossRef]
- Zott, K.; Claisse, O.; Lucas, P.; Coulon, J.; Lonvaud-Funel, A.; Masneuf Pomarede, I. Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol. 2010, 27, 559–567. [Google Scholar] [CrossRef]
- Bergsveinson, J.; Pittet, V.; Ziola, B. RT-qPCR analysis of putative beer-spoilage gene expression during growth of Lactobacillus brevis BSO 464 and Pediococcus claussenii ATCC BAA-344(T) in beer. Appl Microbiol Biotechnol. 2012, 96(2), 461–470. [Google Scholar] [CrossRef]
- Health Canada. Probiotic Claims. 2019. Available online: https://www.inspection.gc.ca/food-label-requirements/labelling/-f-for-industry/former-health-claims/eng/1514559099172/1514559100331?chap=9#s21c9 (accessed on 15 September 2020).
- Dysvik, A.; La Rosa, S.L.; Liland, K.H.; Myhrer, K.S.; Østlie, H.M.; De Rouck, G.; Rukke, E.O.; Westereng, B.; Wicklund, T. Co-fermentation Involving Saccharomyces cerevisiae and Lactobacillus Species Tolerant to Brewing-Related Stress Factors for Controlled and Rapid Production of Sour Beer. Front Microbiol. 2020, 11, 279. [Google Scholar] [CrossRef]
- Bergsveinson, J.; Ziola, B. Investigation of beer spoilage lactic acid bacteria using omic approaches, 2017. p 245–274. In Bokulich NA, Bamforth CW (ed), Brewing microbiology: current research, omics and microbial ecology. Caister Academic Press, Norfolk, UK. [CrossRef]
- Cuevas-González, P.F.; Aguilar-Toalá, J.E.; García, H.S.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Protective Effect of the Intracellular Content from Potential Probiotic Bacteria against Oxidative Damage Induced by Acrylamide in Human Erythrocytes. Probiotics and Antimicrobial Proteins. 2020, 12(4), 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yeh, C.; Jin, Z.; Ding, L.; Liu, B.Y.; Zhang, L.; Dannelly, H.K. Prospective study of probiotic supplementation results in immune stimulation and improvement of upper respiratory infection rate. Synthetic and Systems Biotechnology. 2018, 3(2), 113–120. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Margolles, A.; van Veen, H.W.; Konings, W.N. (2001). Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. Journal of Bacteriology 2001, 183, 5371–5375. [Google Scholar] [CrossRef] [PubMed]
- Iijima, K.; Suzuki, K.; Asano, S.; Ogata, T.; Kitagawa, Y. HorC, a hopresistance related protein, presumably functions in homodimer form. Bioscience, Biotechnology, Biochemistry 2009, 73, 1880–1882. [Google Scholar] [CrossRef]
- Haakensen, M.; Vickers, D.M.; Ziola, B. Susceptibility of Pediococcus isolates to antimicrobial compounds in relation to hop-resistance and beer spoilage. BMC Microbiol. 2009, 9, 190. [Google Scholar] [CrossRef]
- Feyereisen, M.; Mahony, J.; O'Sullivan, T.; Boer, V.; van Sinderen, D. (2020). A Plasmid-Encoded Putative Glycosyltransferase Is Involved in Hop Tolerance and Beer Spoilage in Lactobacillus brevis. Applied Environmental Microbiology. 2020, 86(3), e02268–19. [Google Scholar] [CrossRef]
- Feyereisen, M.; Mahony, J.; Kelleher, P.; Roberts, R.J.; O’Sullivan, T.; Geertman, J.-M.A.; van Sinderen, D. Comparative genome analysis of the Lactobacillus brevis species. BMC Genomics. 2019, 20, 416. [Google Scholar] [CrossRef]
- Schneiderbanger, J.; Jacob, F.; Hutzler, M. Genotypic and phenotypic diversity of Lactobacillus rossiae isolated from beer. J Appl Microbiol. 2019, 126(4), 1187–1198. [Google Scholar] [CrossRef]
- Yadav, P.; Ambudkar, S.V.; Rajendra Prasad, N. Emerging nanotechnology-based therapeutics to combat multidrug-resistant cancer. J Nanobiotechnology. 2022, 20(1), 423. [Google Scholar] [CrossRef]
- Tonsmeire, M. American sour beer: innovative techniques for mixed fermentations. Brewers Publications 2014. Maston. [Google Scholar]
- de Gaetano, G.; Costanzo, S.; Di Castelnuovo, A.; Badimon, L.; Bejko, D.; Alkerwi, A.; Chiva-Blanch, G.; Estruch, R.; La Vecchia, C.; Panico, S.; Pounis, G.; Sofi, F.; Stranges, S.; Trevisan, M.; Ursini, F.; Cerletti, C.; Donati, M.B.; Iacoviello, L. Effects of moderate beer consumption on health and disease: A consensus document. Nutr Metab Cardiovasc Dis. 2016; 26(6), 443–67. [Google Scholar] [CrossRef]
- Marhuenda, J.; Villaño, D.; Arcusa, R.; Zafrilla, P. (2021). Melatonin in Wine and Beer: Beneficial Effects. Molecules, 2021; 26, (2), E343. [Google Scholar] [CrossRef]
- Takase, T.; Toyoda, T.; Kobayashi, N.; Inoue, T.; Ishijima, T.; Abe, K.; Kinoshita, H.; Tsuchiya, Y.; Okada, S. Dietary iso-α-acids prevent acetaldehyde-induced liver injury through Nrf2-mediated gene expression. PLoS One. 2021, 16(2), e0246327. [Google Scholar] [CrossRef]
- Sánchez-Muniz, F.J.; Macho-González, A.; Garcimartín, A.; Santos-López, J.A.; Benedí, J.; Bastida, S.; González-Muñoz, M.J. The Nutritional Components of Beer and Its Relationship with Neurodegeneration and Alzheimer's Disease. Nutrients. 2019, 11(7), 1558. [Google Scholar] [CrossRef] [PubMed]
- Ponticelli, M.; Russo, D.; Faraone, I.; Sinisgalli, C.; Labanca, F.; Lela, L.; Milella, L.; The Promising Ability of Humulus lupulus, L. (2021) Iso-α-acids vs. Diabetes, Inflammation, and Metabolic Syndrome: A Systematic Review. Molecules 2021, 26(4), 954. [Google Scholar] [CrossRef] [PubMed]
| Period (day)\ Formulation | Control | with Lacticaseibacillus paracasei F19 | with Lacticaseibacillus paracasei 431 | ||||||||
| pH | pH | pour-plate | PMA-qPCR | Yeast count | pH | pour-plate | PMA-qPCR | Yeast count | |||
| 0 | 5.9 | 5.9 | 0 | 0 | 0 | 5.9 | 0 | 0 | 0 | ||
| 1 | 5.9 | 4.6 | 5.57 ±0.21 A | 8.15 ±0.08 A,a | 0 | 4.7 | 5.11 ±0.38 A | 8.60 ±0.03 A,a | 0 | ||
| 2 | 4.8 | 4.3 | 3.65 ±0.74 A | 8.91 ±0.09 A,a | 0 | 4.4 | 5.00 ±0.13 B | 8.55 ±0.11 A,a | 0 | ||
| 8 | 4.2 | 3.9 | 3.63 ±1.07 A | 8.93 ±0.07A,a | 7.21 ±0.20 A,b | 3.9 | 3.66 ±0.46 A | 8.24 ±0.34 B,b | 6.47 ±0.23 B,c | ||
| 17 | 4.2 | 3.9 | 5.51 ±0.45 A | 8.90 ±0.15 A,a | 6.22 ±0.24 A,c | 3.9 | 5.73 ±0.08 A | 8.47 ±0.06 A,a | 5.77 ±0.15A,b | ||
| 28 | 4.2 | 3.9 | 6.52 ±0.13 A | 8.80 ±0.13 A,b | 5.87 ±0.34 A,c | 3.9 | 6.46 ±0.13 A | 8.31 ±0.04 B,a | 5.81 ±0.05 A,b | ||
| 58 | 4.2 | 3.9 | 5.39 ±0.26 A | 8.75 ±0.14 A,a | 6.42 ±0.07 A,c | 3.9 | 4.91 ±0.30 A | 8.44 ±0.05 A,a | 5.19 ±0.06 A,a | ||
| Category | Beer style | IBU | SRM | OG | FG | ABV (%) |
| Sour | Berliner Weisse | 3.0 - 8.0 | 2.0 - 3.0 | 1.028 - 1.032 | 1.003 - 1.006 | 2.8 - 3.8 |
| Catharina sour | 2.0 - 8.0 | 2.0 - 6.0 | 1.039 - 1.048 | 1.002 - 1.008 | 4.0 - 5.5 | |
| Lambic | 0.0 - 10.0 | 3.0 - 6.0 | 1.040 - 1.054 | 1.001 - 1.010 | 5.0 - 6.5 | |
| Fruit Lambic | 0.0 - 10.0 | 3.0 - 7.0 | 1.040 - 1.060 | 1.000 - 1.010 | 5.0 - 7.0 | |
| Gueuze | 0.0 - 10.0 | 5.0 - 6.0 | 1.040 - 1.054 | 1.000 - 1.006 | 5.0 - 8.0 | |
| Gose | 5.0 - 12.0 | 3.0 - 4.0 | 1.036 - 1.056 | 1.006 - 1.010 | 4.2 - 4.8 | |
| Flanders Red Ale | 10.0 - 25.0 | 10.0 - 17.0 | 1.048 - 1.057 | 1.002 - 1.012 | 4.6 - 6.5 | |
| Oud Bruin | 20.0 - 25.0 | 17.0 - 22.0 | 1.040 - 1.074 | 1.008 - 1.074 | 4.0 - 8.0 | |
| Pilsen | German Pilsen | 25.0 - 45.0 | 2.0 - 5.0 | 1.044 - 1.050 | 1.008 - 1.013 | 4.4 - 5.2 |
| Bohemian Pilsen | 35.0 - 45.0 | 3.5 - 6.0 | 1.044 - 1.056 | 1.013 - 1.017 | 4.2 - 5.4 | |
| Indian Pale Ale (IPA) | Brut IPA | 20.0 - 30.0 | 2.0 - 4.0 | 1.046 - 1.057 | 0.990 - 1.004 | 6.0 - 7.5 |
| Hazy IPA | 25.0 - 60.0 | 3.0 - 7.0 | 1.060 - 1.085 | 1.010 - 1.015 | 6.0 - 9.0 | |
| American IPA | 40.0 - 70.0 | 6.0 - 14.0 | 1.056 - 1.070 | 1.008 - 1.014 | 5.5 - 7.5 | |
| Black IPA | 50.0 - 90.0 | 25.0 - 40.0 | 1.050 - 1.085 | 1.010 - 1.018 | 5.5 - 9.0 | |
| Belgian IPA | 50.0 - 100.0 | 5.0 - 8.0 | 1.058 - 1.080 | 1.008 - 1.016 | 6.2 - 9.5 | |
| Our study | Brazilian sour hop* | 29.65 | 3.9 | 1.045 | 1.008 | 4.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





