1. Introduction
Lynch syndrome (LS) is an inherited cancer-predisposing disorder caused by a germline pathogenic variant (PV) in one of the mismatch repair (MMR) genes or deletions in the 3′ region of the
EPCAM gene [
1]. LS is associated with a very high lifetime risk of developing primarily colorectal cancer (CRC) and extra-colonic cancers at a younger age, compared to the general population [
1,
2,
3]. The lifetime risk of developing cancer in LS variant heterozygotes (LSVH) ranges from 30% to 80% depending on the mutated gene, cancer type, and other factors such as lifestyle, environmental exposure, epigenetic changes, and genetic risk modifiers [
4,
5,
6]. This lifetime risk of developing cancer in LSVH differs in terms of age of cancer diagnosis (often used as a proxy for age of onset) and tumour site, even among individuals carrying the same PV [
7,
8,
9]. Thus, the identification of additional specific genetic risk modifiers contributing to phenotype variations in cancer risk and age at cancer diagnosis in LSVH may assist in the implementation of highly personalized surveillance and screening interventions to reduce morbidity and mortality related to cancer in this at-risk population.
In LS cancer microenvironments, carcinogenesis is driven by immunoediting through the counter-selection of cell clones presenting frameshift neoantigens (produced as a consequence of MMR-deficiency), which depends mainly on the Human Leukocyte Antigen (HLA) alleles [
10,
11]. HLA alleles are responsible for presenting cellular antigens and eliciting antigen-specific immune responses [
12]. While exogenous antigens are presented by HLA class II molecules, endogenous antigens are presented by HLA class I molecules, which interact with CD4-positive and CD8-positive T cells, respectively, which are among the most powerful mediators of anti-tumor immune responses [
11,
13,
14]. HLA alleles are highly diverse among different individuals and populations. Each HLA allele has a unique shape and chemical properties in its antigen-binding groove, which enables it to fit with a specific antigen to a varying degree [
11,
15,
16,
17]. Thus, an individual’s HLA typing is crucial in determining the binding of the antigens for presentation on the surface of the cell and eliciting antigen-specific immune response. However, it is not clear whether HLA allele variations may protect or influence cancer initiation in LSVH, hence the variability in cancer risk and age of cancer diagnosis/onset in these individuals [
11]. This is because the crucial presentation of cancer antigens to the immune system by the HLA alleles may have a positive or negative impact on tumor initiation and progression, which can modify an individual’s cancer onset risk [
10,
11,
13,
18,
19]. For example, effective presentation of cancer antigens by HLA to the immune system has been found to play a significant role in the effectiveness of cancer immunotherapy, which aims to reactivate the impaired HLA-mediated anti-tumor immune response, such as immune checkpoint blockade [
20,
21].
The unique characteristics of cancer pathogenesis in LSVH present an opportunity to study the possible influence of HLA variations on cancer risk and the age of cancer onset. By investigating the impact of HLA allele variations on cancer incidence, cancer onset, and mutation profile in LSVH, we can gain new insights into the role of HLA alleles as modifiers of cancer risk. Several studies have investigated the influence of HLA allele variations on cancer susceptibility [
22,
23,
24,
25,
26]. However, none of these studies investigated the influence of HLA allele variations on cancer risk or the age at cancer diagnosis in LSVH. We hypothesize that HLA allele variations may influence an individual’s age at cancer diagnosis in LSVH.
In this study, we investigated our hypothesis using a unique cohort of LSVH carrying the same PV in the hMLH1 gene (NM_000249.4(MLH1):c.1528C>T (p.Gln510Ter)) in South Africa.
2. Materials and Methods
2.1. Patients
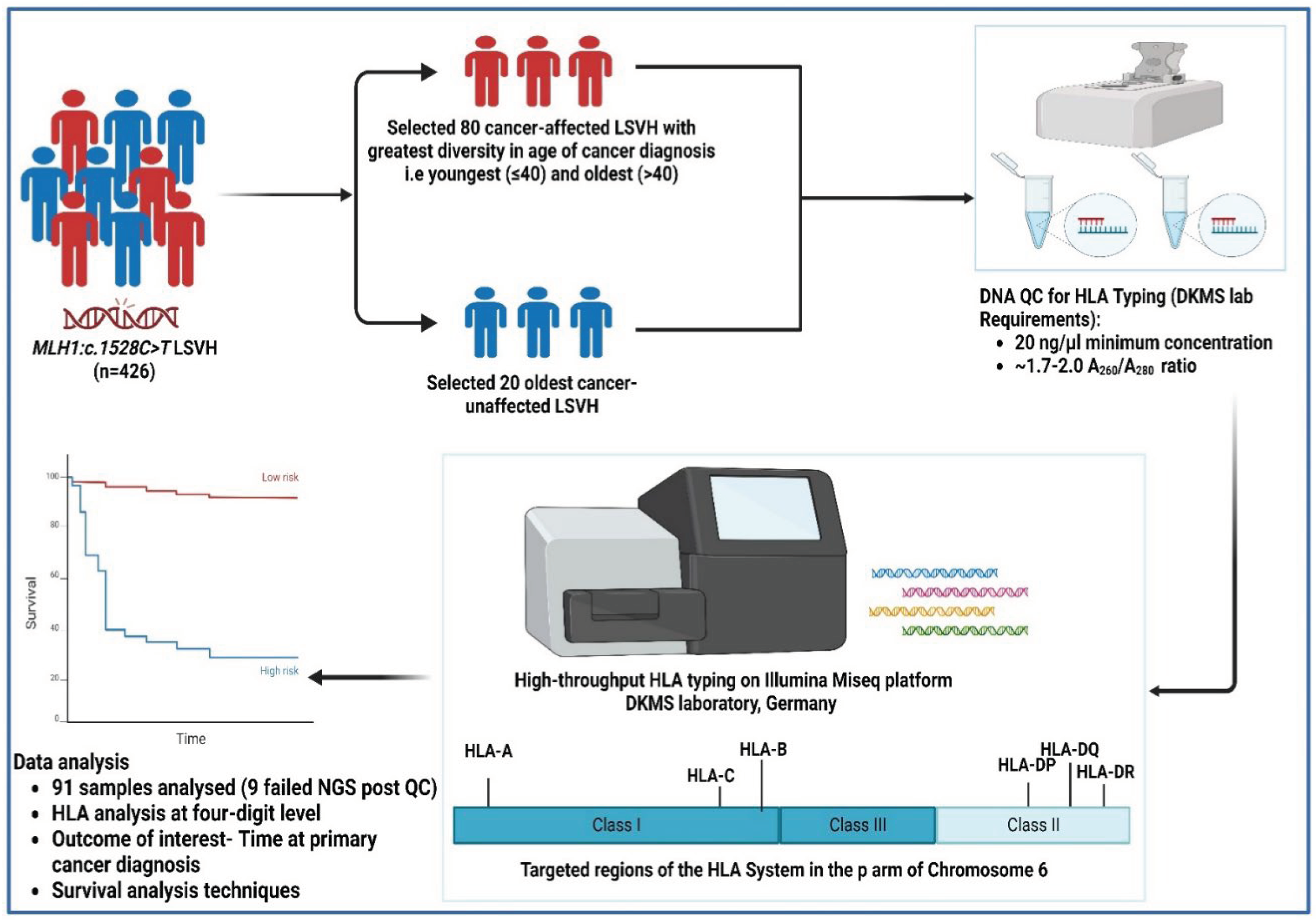
In our large homogenous cohort of 426 genetically confirmed LSVH carrying the same
MLH1:c.1528C>T South African founder PV, we selected 100 subjects for this study based on the following inclusion criteria: (i) patients exhibiting the greatest diversity in age at cancer diagnosis, i.e., patients who had 3SD below (youngest) and above (oldest) the mean age at cancer diagnosis from both extremities (n = 80, mean age 42.9, SD ±11.1 years), (ii) the oldest individuals who were not yet affected with cancer (n = 20), and (iii) the availability of blood genomic DNA sample with a minimum concentration of 20 ng/µl in our designated biorepository. All cancer patients were confirmed through pathology reports indicating the gender, age at the time of first diagnosis, presence or absence of malignancy, and the tumour site. Other demographics, such as ethnicity were retrieved from our in-house LS electronic database (
Figure 1).
2.2. DNA Samples
DNA samples of subjects meeting our inclusion criteria were retrieved from the -80°C biorepository at the Division of Human Genetics, University of Cape Town, South Africa. Genomic DNA was extracted from white blood cells (buffy coats). The DNA was quantified using the NanoDrop spectrophotometer and viewed using version 3.8.1 of the Nanodrop 1000 operating software (Thermo Fisher Scientific™, Johannesburg, South Africa). The integrity of the DNA samples was checked using 2.0% (w/v) gel electrophoresis, and the gel was visualized using the UVIpro Gold Transilluminator and through the UVPro software (UVItec, United Kingdom). DNA samples with the A
260/A
280 ratio of 1.7–2.0 and a minimum of 20ng/ul concentration were selected for the downstream HLA high-throughput typing as per requirement by the Deutsche Knochenmarkspenderdatei (DKMS) laboratory, Germany (
Figure 1).
2.3. HLA Typing
A high-throughput HLA genotyping targeting a total of twelve (12) HLA class I (-A, -B, -C, -E) and class II (DRB1, - DRB3, -DRB4, - DRB5, - DQA1, - DQB1, - DPA1, -DPB1) loci was performed at the DKMS Life Science Lab, Germany. The shotgun next-generation sequencing (NGS) technique was used on the Illumina MiSeq platform (Illumina, San Diego, California). All laboratory procedures and the sequencing strategy were performed similarly to a previous publication [
27]. The amino acid sequence of the HLA protein distinguishes the biological effects of different HLA alleles, therefore, we limited our downstream analysis to a four-digit level to investigate the influence of HLA alleles on the age at cancer diagnosis [
28] (
Figure 1). The potential novel allele in the
DRB3 locus was characterized using the NGS-engine NGS-HLA typing software package (Version 2.15, GenDX) and the IPD-IMGT/HLA database (Version 3.54).
2.4. Statistical Analysis
Statistical analysis was performed using the R statistical software (R Core Team, version 4.3.3). The outcome of interest was time at first cancer diagnosis (i.e., CRC or extra-colonic cancer). The risk of cancer diagnosis in one group relative to a reference group, at any age, was calculated using survival analysis techniques (Kaplan-Meier product limit method, with Logrank tests and Cox proportional hazards with 95% confidence intervals (CIs)), taking into account the fact that our research cohort included subjects who were cancer unaffected. Continuous data were presented as mean and standard deviation (SD) or median and interquartile range (IQR), whereas categorical data were presented as numbers (percentage). The immunotation R package (Version 1.10.0) was used to call the frequencies of HLA alleles from the IPD-IMGT/HLA Database (Version 3.54) [
29]. All tests were two-tailed and p values were corrected for multiple comparisons according to the Benjamini-Hochberg method. Associations were considered significant if both the p- and q-values (adjusted p-value) <0.05 (
Figure 1).
3. Results
Overall Demographic and Clinical Characteristics of the Patients
Demographics and clinical characteristics of two groups (cancer-affected and cancer-unaffected) of LSVH carrying the same PV (
MLH1:c.1528C>T) in the
hMLH1 gene are summarized in
Table 1. There were 78 cancer-affected and 13 cancer-unaffected LSVH. Of the 78 cancer-affected patients, 60 (78%) were diagnosed with CRC. As expected in LS, proximal colon tumours were the most common (58%) in this cohort (
Table 1).
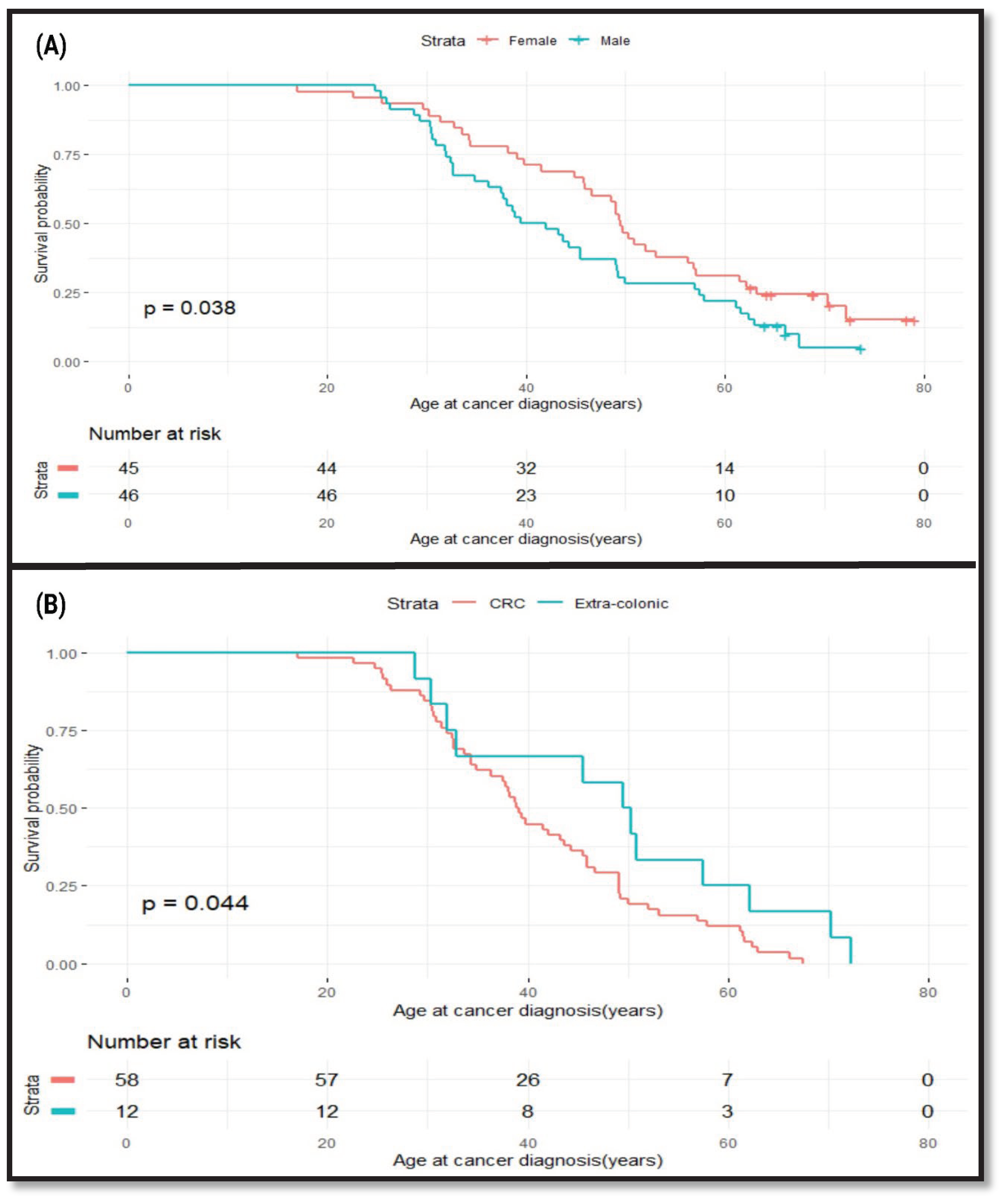
Effects of Gender and Cancer Site on Age at Cancer Diagnosis in LSVH
To find out whether the age at cancer diagnosis in this cohort was influenced by the gender of an individual and the type of cancer, we performed a survival analysis. We used age at cancer diagnosis as our outcome of interest to determine whether there is a difference in age at cancer diagnosis between males and females. We found that male LSVH were diagnosed with cancer at a younger age compared to female LSVH (Mean age: 40.7y [95% CI: 37.5-49.1] and 49.6y [95% CI: 46.0-57.1]), respectively, p=0.038) (
Figure 2A). Our observations are consistent with previous findings on the effect of gender on cancer risk in LSVH [
30,
31,
32]. We performed another analysis to compare the age at cancer diagnosis between CRC and extra-colonic cancers in this cohort. We found that LSVH with CRC were diagnosed ten-years younger than LSVH with extra-colonic cancers (Mean age: 39.0y [95% CI: 36.3 – 45.5] and 49.9y [95% CI: 32.8 -72], respectively, p=0.044) (
Figure 2B). As the majority of these patients were actively undergoing endoscopic surveillance, we anticipate an earlier detection of CRC events compared to other types of LS-associated cancers. This finding was also similar to other cohorts of LSVH carrying different PVs in that the risk of developing CRC was typically higher and at a younger age compared to most extra-colonic cancers [
33,
34].
Effects of HLA Alleles on the Age at Cancer Diagnosis in LSVH
In order to study the effects of HLA allele variations on the age at cancer diagnosis in this cohort, we performed a Cox regression analysis to investigate whether HLA allele variations can influence age at cancer diagnosis in LSVH. We further adjusted our analysis by gender, to avoid the potential confounding effects of gender bias. Of 1785 HLA alleles typed from 12 HLA class I and II loci (187 unique HLA alleles), 78 individuals had primary cancers, and 13 remained unaffected. We summarised the details of different HLA alleles showing significant associations with age at any cancer diagnosis (either CRC or extra-colonic cancer) in
Table 2.
The following HLA class II alleles in the
HLA-DPB1 gene were significantly associated with a younger age at cancer diagnosis:
HLA-DPBI*04:02,
HLA-DPBI*20:01,
HLA-DPBI*55:01 and
HLA-DPBI*296:01 (
Table 2). A potential novel allele in the
DRB3 locus detected in our youngest cancer patient diagnosed at the age of 17 years showed a significant association with young age at cancer diagnosis (Hazard ratio (HR) = 301.04, p<0.001, q<0.001). This potential novel allele bearing the
HLA-DRB3:g.7953C>T variant is most similar to the
HLA-DRB3*03:01 allele (
Supplementary Figure S1). There was no statistically significant association between all other HLA alleles observed and the age at cancer diagnosis in this cohort, as shown in a complete unadjusted and gender-adjusted comparison data for
HLA-A, -B, -C, -DRB1, - DRB3, -DRB4, - DRB5, - DQA1, - DQB1, - DPA1, -DPB1, and
-E alleles in
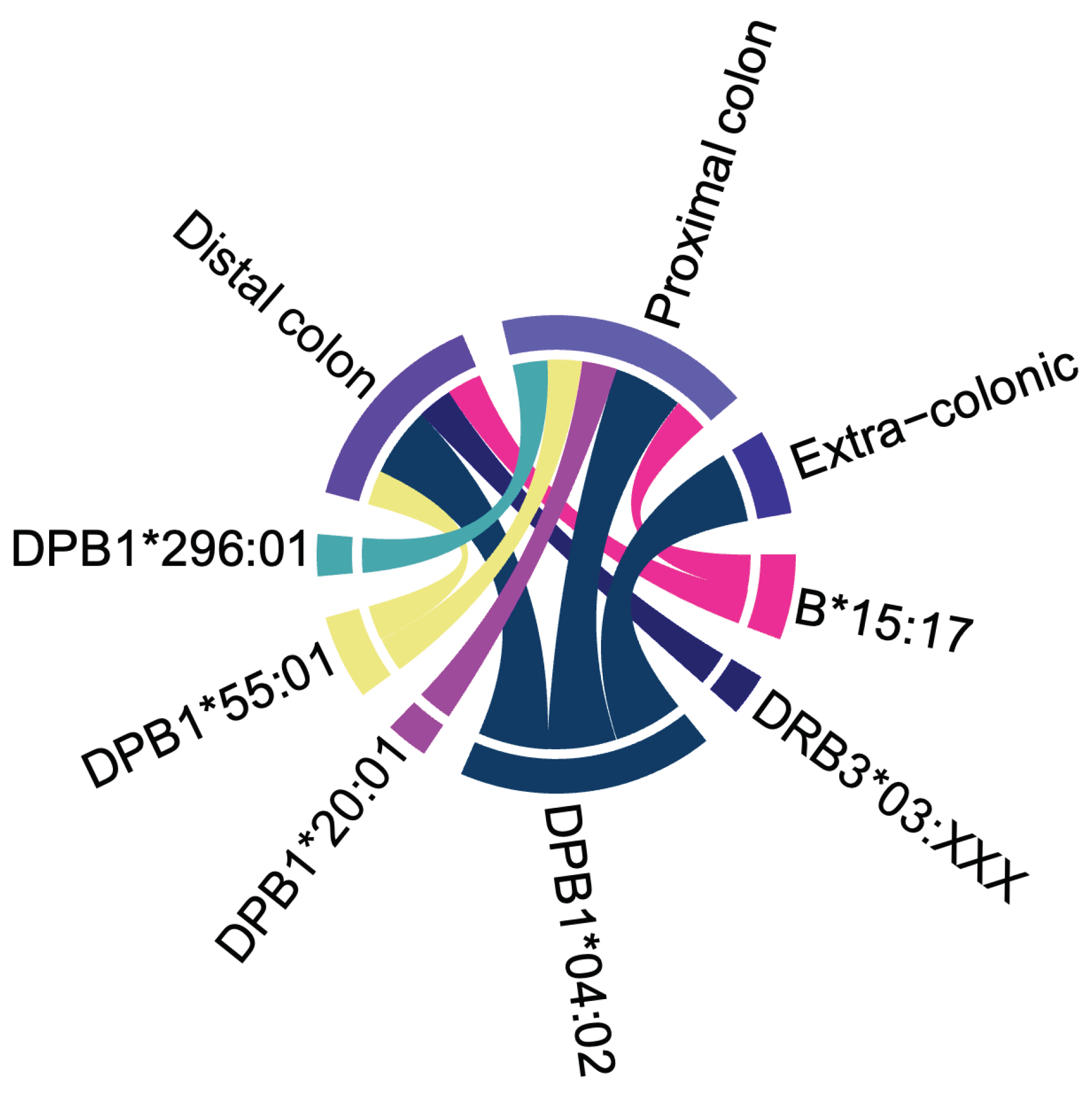
Supplementary Tables S1 and S2, respectively. The relationship between the six HLA alleles associated with young age at cancer diagnosis and the cancer sites in our LSVH is depicted in
Figure 3. The
HLA-DPB1*04:02 allele was linked to the age of CRC and extracolonic cancer diagnoses. Notably, it was the only allele associated with extracolonic cancer.
Effects of HLA Alleles on the Age at CRC Diagnosis in LSVH
Considering that CRC is the most diagnosed cancer in LSVH and the most observed cancer in this study cohort, we investigated whether HLA allele variations could impact age at CRC diagnosis. All LSVH affected with extra-colonic cancers (n =20) were excluded from this analysis. Three (3) HLA alleles were significantly associated with age at CRC diagnosis (
Table 3). Interestingly, two (2) LSVH carrying both
HLA-B*15:17 and
HLA-DPB1*55:10 developed CRC at a young age. There is no published evidence of linkage disequilibrium between these two HLA alleles [
35]. These two alleles were significantly associated with young age at CRC diagnosis, with the mean age at cancer diagnosis of 21 years (Range 17-25, p<0.001, q=0.003 for
HLA-B*15:17 and p<0.001, q<0.001 for
HLA-DPB1*55:10) (
Table 3). Unadjusted and gender-adjusted association data of one hundred eighty-four (184) HLA alleles that had no significant association with age at CRC diagnosis are presented in
Supplementary Tables S3 and S4, respectively.
Different HLA Allele Frequencies between LSVH and the Previously Studied South African General Populations
South Africa has a diverse population with a unique genetic background, (ranging from indigenous African subpopulations, and immigrant European and Asian populations) and varying disease prevalence. This diversity could present unique HLA allele variations that are not found in other populations within the country (and internationally) [
36,
37,
38]. Comparing HLA allele frequency (AF) between LSVH and different populations in South Africa, allows us to account for these population-specific factors. This information can aid in tailoring cancer screening and management approaches that are more effective and relevant to the South African context. Also, it can identify specific novel HLA alleles that may be associated with LS susceptibility or be likely protective, and unique amongst this cohort, compared to the general population. This information may contribute to early cancer risk assessments and highly personalized prevention strategies for LSVH in South Africa [
39].
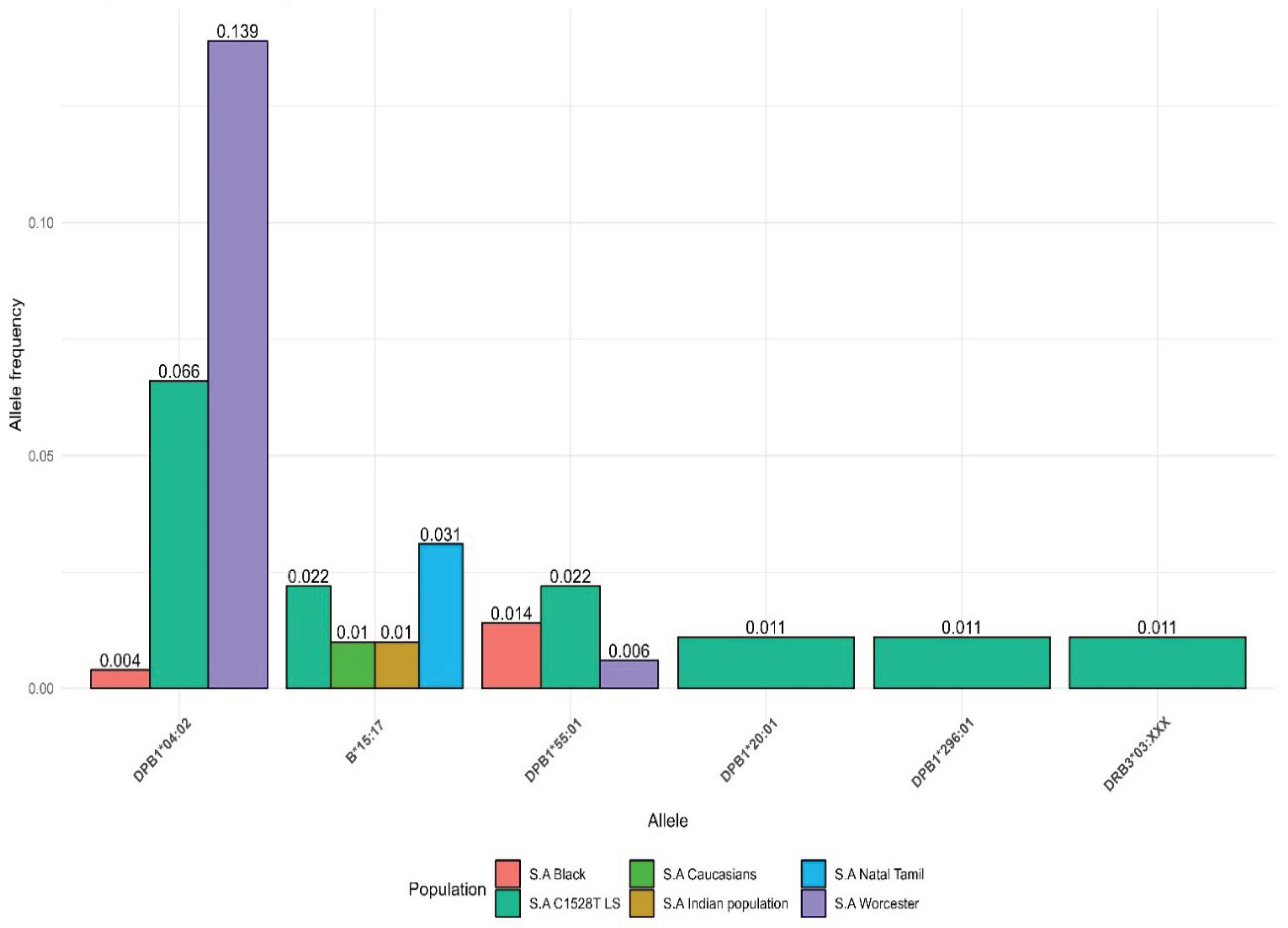
We compared the AF of the HLA alleles which were significantly associated with young age at cancer diagnosis in our LSVH cohort with the AF observed from other non-LS South African cohorts reported in the IPD-IMGT/HLA Database [
35]. The
HLA-DPB1*04:02 allele was common in the LSVH cohort and South African general population (i.e., with AF greater than 0.05). Interestingly,
HLA-DPB1*20:01, and
HLA-DPB1*296:01 alleles associated with a young age at cancer diagnosis in our LSVH cohort were observed for the first time in the South African study cohort (
Figure 4).
4. Discussion
This is the first study aimed at identifying HLA class I and II alleles that may influence the age at cancer diagnosis in LSVH carrying the same PV in the hMLH1 gene (MLH1:c.1528C>T). Our findings suggest that, once validated in a large cohort, the identification of high-risk HLA alleles could be factored into the risk prediction model calculations for offering tailored personalized cancer screening and surveillance strategies for LSVH.
Our study is part of an ongoing programme promoting the utility of personalized early cancer prevention in LSVH. In this instance, the strategy is to consider the effects of HLA allele variations as one of the potential genetic modifiers for cancer onset risk in a well-defined LSVH cohort. Importantly, personalized screening strategies will potentially reduce the overuse of invasive colonoscopies for CRC screening and (premature) cancer-preventive surgeries in LSVH [
7,
40].
The strongest association with a young age at cancer diagnosis in LSVH were conferred by the presence of the
HLA-DPB1*04:02 class II allele (
Table 2). The
HLA-DPB1*04:02 allele was common in both LSVH cohort and non-LS South African general population, with the reported allele frequencies of 0.066 (6.6%) and 0.139 (13.9%), respectively (
Figure 4). Therefore, once validated in a longitudinal study with a large cohort of LSVH, this potentially high-risk HLA allele may be considered as part of cancer risk assessment in LSVH, potentially promoting a more genetically-informed predictive testing and much more precisely targeted surveillance for cancer prevention strategies in South Africa.
Worldwide in non-LS populations, different HLA alleles have been reported to be associated with various cancers, such as cervical [
26], leukaemia [
41], hepatocellular [
42], lung squamous cell [
43], cutaneous T-Cell Lymphoma [
44], and gastric carcinomas [
45,
46]. For instance, the HLA-DP gene polymorphisms (
HLA-DPB1*03:01, and
HLA-DPB1*13:01) have been significantly associated with an increased risk of cervical cancer in Chinese populations [
47,
48,
49,
50]. Furthermore,
HLA- DPB1*04:02:01:21 has been recently reported as a novel HLA allele in a patient with acute leukaemia in the UK [
41]. In our LSVH cohort, the associations between HLA-DP alleles and cancer risk are consistent with previous associations in non-LS populations across various cancers, worldwide [
41,
47,
51]. The HLA-DP locus forms part of the highly polymorphic HLA class II molecules, and genetic variations of the HLA alleles may lead to variations in the antigen-presentation on the specialised antigen-presenting cells, such as dendritic cells, thus potentially influencing or likely protecting against the development of cancer in LSVH [
52,
53].
Although we found statistically significant associations between different HLA alleles (
HLA-B*15:17, -
DPB1*20:01, -
DPB1*55:01, -
DPB1*296:01 and the potential novel allele in the
DRB3 locus (most similar to
HLA-DRB3*03:01 allele)) and relatively young age at cancer diagnosis, the 95% confidence intervals were very wide due to our small sample size. However, additional investigation for these HLA alleles in patients with young age of cancer onset in a large cohort of LSVH can further complement and validate our findings. Generally,
HLA-B*15 has been widely reported to be likely protective against viral-associated cancers, such as cervical cancer ([OR], 0.64; p = 1.56× 10
−9) [
54] and Human papillomavirus-positive head and neck cancers ([OR], 0.31; p = 0.015) [
55]. The HLA class I alleles play a crucial role in early infection and the mechanisms underlying early viral clearance [
56], which may explain the protective effect against viral-associated cancers. However, in our cohort of LSVH, we found a significant association between the
HLA-B*15:17 allele and a young age at CRC diagnosis, which is a non-infection-attributable cancer. These different findings of
HLA-B*15 in infection and non-infection-attributable cancers, between the general population and our LSVH population, may be due to the low incidence of cancers attributable to infections in the LSVH. This was proposed to be due to a chronic hyper-immune response caused by the persistent production of neopeptides [
57,
58,
59], as a result of a pathogenic germline defect in the post-replication DNA mismatch repair system (as occurs in LS).
Additionally, it is worth taking into account that HLA class I also plays a role in the presentation of tumor-derived peptides in a complex with the /beta 2 microglobulin (B2M) for recognition by cytotoxic CD8+ T lymphocytes to eliminate transformed cells. In this regard, any variation in this gene could favor cancer initiation and progression due to poor presentation of the cancer cells to the immune system, explaining the observed increased cancer risk in our LSVH cohort [
10,
19].
Our findings suggest that HLA allelic (amongst other genomic) variations could be potential factors influencing the age at cancer diagnosis in LSVH individuals. In this regard, it is worth considering that these findings, once confirmed in a large cohort of LSVH, could be used to implement more precise presymptomatic cancer surveillance programs as follows: (i) integrating HLA allele testing into routine cancer screening for LSVH, as those with certain low or high risk HLA alleles may require decreased or increased highly specialized screening and surveillance respectively, (ii) use of individual HLA allele information to stratify LSVH with the same PV into different risk groups for age at cancer onset (early or late), which could inform the frequency and intensity of their colonoscopic surveillance, (iii) consider HLA allelic information when deciding which cancer prevention strategy to recommend to LSVH individuals with known PVs, as those with certain high risk HLA alleles may benefit more from specific lifestyle changes or prophylactic treatments such as hysterectomy for endometrial cancer prevention or the use of aspirin and resistant starch for CRC prevention [
40,
60], (iv) develop predictive mathematical models that could take into account of both PVs and different HLA alleles to estimate an individual’s life-time risk of developing cancer and the likely estimated age at cancer diagnosis in LSVH. However, HLA allelic variations could be just one of many genetic modifiers that can influence a person’s risk of developing cancer in LSVH. Other already known cancer risk modifiers can affect the overall cancer risk in LSVH. These risk modifiers include polymorphisms in xenobiotic metabolism genes, epigenetic changes, lifestyle factors and strong family history of cancer [
8,
61,
62,
63,
64,
65,
66,
67,
68].
The strengths of our study include: (a) the first research study to investigate the associations between HLA class I and II alleles and the age at cancer diagnosis using a genetically confirmed LS cohort, (b) the utilization of high-throughput HLA genotyping using NGS in this regard, (c) the homogenous nature of the study cohort as patients harbour the same LS-PV in the hMLH1 gene and are originating from a population of a common ethnicity, and (d) new evidence suggesting that HLA allele variations may influence the age at cancer diagnosis in LSVH. The main limitations of our study are: (i) the relatively small sample size, mainly due to financial constraints to perform high-throughput NGS-based HLA typing in the whole cohort of LSVH, and (ii) we only performed a cross-sectional association study in LSVH without taking into account other possible cancer risk genetic and epigenetic modifiers, cofounders and the proved causality (causal-effect relationship). We are making efforts to recruit a longitudinal cohort of LSVH to overcome this limitation in our next study.
5. Conclusions
This study provides valuable insights into the potential role of HLA allele variations and the age at cancer diagnosis in LSVH. HLA allelic variations may influence the age at cancer diagnosis in LSVH carrying the same PV in the hMLH1 gene (MLH1:c.1528C>T). As such, it is worth considering HLA allele information when recommending cancer prevention strategies for LSVH. High-throughput HLA allele typing can be included in LSVH routine cancer screening after it has been successfully validated in a large longitudinal cohort. This can be achieved by stratifying these individuals into different risk groups based on their at-risk HLA alleles. In this population of people at risk, all of these approaches have the potential to enhance personalized cancer screening.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: A snapshot of a potential novel allele in the HLA DRB3 locus.; Table S1: Unadjusted comparison of HLA alleles and age at any cancer diagnosis in LSVH; Table S2: Gender-adjusted comparison of HLA alleles and age at any cancer diagnosis in LSVH; Table S3: Unadjusted comparison of HLA alleles and age at CRC diagnosis in LSVH; Table S4: Gender-adjusted comparison of HLA alleles and age at CRC diagnosis in LSVH.
Author Contributions
Conceptualization, L.N., R.C., G.R. and R.R.; methodology, L.N., R.C., G.R. and R.R.; formal analysis, L.N., Z.V.-O. and R.R.; investigation, L.N., R.C., G.R. and R.R; resources, P.G., A.B., U.A. and R.R.; data curation, L.N., G.R., P.G., A.B., U.A. and R.R.; writing—original draft preparation, L.N.; writing—review and editing, L.N., R.C., Z.V.-O., G.R., P.G., A.B., U.A. and R.R.; visualization, L.N.; supervision, R.C., G.R. and R.R., project administration, R.R.; funding acquisition, R.R. All authors have read and agreed to the published version of the manuscript.
Funding
The work reported herein was made possible through funding by the South African Medical Research Council through its Division of Research: Capacity Development under the Internship Scholarship Programme and funds received from the South African Department of Science and Innovation. The content hereof is the sole responsibility of the authors and does not necessarily represent the official views of the SAMRC.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Human Research Ethics Committee (HREC) of the University of Cape Town (HREC 972/2021).
Informed Consent Statement
All patients gave informed consent for their data to be used for subsequent genetic studies linked to the previously approved study (HREC R022/2019).
Data Availability Statement
The data analyzed during the current study are available from the corresponding author upon request.
Acknowledgments
The authors sincerely thank the National Health Laboratory Services for providing access to patients pathological data and DNA samples, and Dr. Armin Deffur, Infectious diseases specialist and Bioinformatician at the Division of Human Genetics, University of Cape Town, for helping with data extraction and processing.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Lynch HT, Krush AJ. Cancer family “G” revisited: 1895-1970. Cancer 1971, 27, 1505–1511. [CrossRef]
- Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009, 76, 1–18. [CrossRef]
- Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet. 1999, 36, 801–818.
- Talseth-Palmer BA, Wijnen JT, Grice DM, Scott RJ. Genetic modifiers of cancer risk in Lynch syndrome: a review. Fam. Cancer 2013, 12, 207–216. [CrossRef] [PubMed]
- Bucksch K, Zachariae S, Aretz S, Buttner R, Holinski-Feder E, Holzapfel S, et al. Cancer risks in Lynch syndrome, Lynch-like syndrome, and familial colorectal cancer type X: a prospective cohort study. BMC Cancer 2020, 20, 460. [CrossRef]
- Dominguez-Valentin M, Sampson JR, Seppala TT, Ten Broeke SW, Plazzer JP, Nakken S, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020, 22, 15–25. [CrossRef]
- Ahadova A, Seppala TT, Engel C, Gallon R, Burn J, Holinski-Feder E, et al. The “unnatural” history of colorectal cancer in Lynch syndrome: Lessons from colonoscopy surveillance. Int. J. Cancer 2021, 148, 800–811. [CrossRef] [PubMed]
- Felix R, Bodmer W, Fearnhead NS, van der Merwe L, Goldberg P, Ramesar RS. GSTM1 and GSTT1 polymorphisms as modifiers of age at diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC) in a homogeneous cohort of individuals carrying a single predisposing mutation. Mutat. Res. 2006, 602, 175–181. [CrossRef]
- Stupart DA, Goldberg PA, Algar U, Ramesar R. Cancer risk in a cohort of subjects carrying a single mismatch repair gene mutation. Fam. Cancer 2009, 8, 519–523. [CrossRef]
- Ballhausen A, Przybilla MJ, Jendrusch M, Haupt S, Pfaffendorf E, Seidler F, et al. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat. Commun. 2020, 11, 4740. [CrossRef]
- Ahadova A, Witt J, Haupt S, Gallon R, Huneburg R, Nattermann J, et al. Is HLA type a possible cancer risk modifier in Lynch syndrome? Int. J. Cancer. 2023, 152, 2024–2031. [CrossRef]
- Holoshitz, J. The quest for better understanding of HLA-disease association: scenes from a road less travelled by. Discov. Med. 2013, 16, 93–101. [Google Scholar] [PubMed]
- Klein J, Sato A. The HLA system. Second of two parts. N. Engl. J. Med. 2000, 343, 782–786. [CrossRef]
- Li XC, Raghavan M. Structure and function of major histocompatibility complex class I antigens. Curr. Opin. Organ Transplant. 2010, 15, 499–504. [CrossRef]
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017, 199, 3360–3368. [CrossRef] [PubMed]
- Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 1991, 351, 290–296. [CrossRef]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 1987, 329, 512–518. [CrossRef] [PubMed]
- Wysocki T, Olesińska M, Paradowska-Gorycka A. Current Understanding of an Emerging Role of HLA-DRB1 Gene in Rheumatoid Arthritis–From Research to Clinical Practice. Cells 2020, 9. [CrossRef]
- Klein J, Sato A. The HLA system. First of two parts. N. Engl. J. Med. 2000, 343, 702–709. [CrossRef]
- Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018, 359, 582–587. [CrossRef]
- Naranbhai V, Viard M, Dean M, Groha S, Braun DA, Labaki C, et al. HLA-A*03 and response to immune checkpoint blockade in cancer: an epidemiological biomarker study. Lancet Oncol. 2022, 23, 172–184. [CrossRef]
- Li H, Liu D, Li X. HLA-DPB1 and Epstein-Barr virus gp42 protein jointly contribute to the development of Hodgkin lymphoma. Transl. Cancer Res. 2020, 9, 4424–4432. [CrossRef]
- Liu Z, Hildesheim A. Association Between Human Leukocyte Antigen Class I and II Diversity and Non-virus-associated Solid Tumors. Frontiers in Genetics 2021, 12. [CrossRef]
- Liu Z, Huang CJ, Huang YH, Pan MH, Lee MH, Yu KJ, et al. HLA Zygosity Increases Risk of Hepatitis B Virus-Associated Hepatocellular Carcinoma. J. Infect. Dis. 2021, 224, 1796–1805. [CrossRef]
- Hirata I, Murano M, Ishiguro T, Toshina K, Wang FY, Katsu K. HLA genotype and development of gastric cancer in patients with Helicobacter pylori infection. Hepatogastroenterology 2007, 54, 990–994.
- Chambuso R, Ramesar R, Kaambo E, Denny L, Passmore JA, Williamson AL, et al. Human Leukocyte Antigen (HLA) Class II -DRB1 and -DQB1 Alleles and the Association with Cervical Cancer in HIV/HPV Co-Infected Women in South Africa. J. Cancer 2019, 10, 2145–2152. [CrossRef]
- Albrecht V, Zweiniger C, Surendranath V, Lang K, Schofl G, Dahl A, et al. Dual redundant sequencing strategy: Full-length gene characterisation of 1056 novel and confirmatory HLA alleles. HLA 2017, 90, 79–87. [CrossRef]
- Listgarten J, Brumme Z, Kadie C, Xiaojiang G, Walker B, Carrington M, et al. Statistical resolution of ambiguous HLA typing data. PLoS Comput. Biol. 2008, 4, e1000016. [CrossRef]
- Katharina Imkeller [cre a. immunotation: Tools for working with diverse immune genes. R package version 1.8.0. 2023. Available online: https://bioconductor.org/packages/immunotation/.
- Cohen SA, Leininger A. The genetic basis of Lynch syndrome and its implications for clinical practice and risk management. Appl. Clin. Genet. 2014, 7, 147–158. [CrossRef]
- Schneider R, Schneider C, Jakobeit C, Furst A, Moslein G. Gender-Specific Aspects of Lynch Syndrome and Familial Adenomatous Polyposis. Viszeralmedizin 2014, 30, 82–88. [CrossRef]
- Vasen HFA, Moslein G, Alonso A, Bernstein I, Bertario L, Blanco I, et al. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer). Journal of Medical Genetics 2007, 44, 353–362. [CrossRef] [PubMed]
- Dominguez-Valentin M, Haupt S, Seppälä TT, Sampson JR, Sunde L, Bernstein I, et al. Mortality by age, gene and gender in carriers of pathogenic mismatch repair gene variants receiving surveillance for early cancer diagnosis and treatment: a report from the prospective Lynch syndrome database. EClinicalMedicine 2023, 58, 101909. [CrossRef]
- Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. Jama 2011, 305, 2304–2310. [CrossRef] [PubMed]
- Barker DJ, Maccari G, Georgiou X, Cooper MA, Flicek P, Robinson J, et al. The IPD-IMGT/HLA Database. Nucleic Acids Res. 2023, 51, D1053-D60. [CrossRef]
- Tshabalala M, Mellet J, Vather K, Nelson D, Mohamed F, Christoffels A, et al. High Resolution HLA ∼A, ∼B, ∼C, ∼DRB1, ∼DQA1, and ∼DQB1 Diversity in South African Populations. Front. Genet. 2023, 13, 711944. [CrossRef]
- Janse van Rensburg WJ, de Kock A, Bester C, Kloppers JF. HLA major allele group frequencies in a diverse population of the Free State Province, South Africa. Heliyon 2021, 7, e06850. [CrossRef] [PubMed]
- Lombard Z, Brune AE, Hoal EG, Babb C, Van Helden PD, Epplen JT, et al. HLA class II disease associations in southern Africa. Tissue Antigens 2006, 67, 97–110. [CrossRef]
- Yurgelun MB, Hampel H. Recent Advances in Lynch Syndrome: Diagnosis, Treatment, and Cancer Prevention. Am Soc Clin Oncol Educ Book 2018, 38, 101–109. [CrossRef]
- Seppala TT, Dominguez-Valentin M, Crosbie EJ, Engel C, Aretz S, Macrae F, et al. Uptake of hysterectomy and bilateral salpingo-oophorectomy in carriers of pathogenic mismatch repair variants: a Prospective Lynch Syndrome Database report. Eur. J. Cancer 2021, 148, 124–133. [CrossRef]
- Williams HB, Turner TR, Cambridge CA, Marsh SGE, Mayor NP. The novel HLA-DRB1*03:01:01:05 and -DPB1*04:02:01:21 alleles identified in patients with acute leukemia. HLA 2022, 99, 650–652. [CrossRef]
- Xin YN, Lin ZH, Jiang XJ, Zhan SH, Dong QJ, Wang Q, et al. Specific HLA-DQB1 alleles associated with risk for development of hepatocellular carcinoma: a meta-analysis. World J. Gastroenterol. 2011, 17, 2248–2254. [CrossRef]
- Kohno T, Kunitoh H, Mimaki S, Shiraishi K, Kuchiba A, Yamamoto S, et al. Contribution of the TP53, OGG1, CHRNA3, and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. J. Thorac. Oncol. 2011, 6, 813–817. [CrossRef] [PubMed]
- Jackow CM, McHam JB, Friss A, Alvear J, Reveille JR, Duvic M. HLA-DR5 and DQB1*03 class II alleles are associated with cutaneous T-cell lymphoma. J. Invest. Dermatol. 1996, 107, 373–376. [CrossRef] [PubMed]
- Wu MS, Hsieh RP, Huang SP, Chang YT, Lin MT, Chang MC, et al. Association of HLA-DQB1*0301 and HLA-DQB1*0602 with different subtypes of gastric cancer in Taiwan. Jpn. J. Cancer Res. 2002, 93, 404–410. [CrossRef] [PubMed]
- Magnusson PKE, Enroth H, Eriksson I, Held M, Nyren O, Engstrand L, et al. Gastric cancer and human leukocyte antigen: distinct DQ and DR alleles are associated with development of gastric cancer and infection by Helicobacter pylori. Cancer Res. 2001, 61, 2684–2689.
- Cheng L, Guo Y, Zhan S, Xia P. Association between HLA-DP Gene Polymorphisms and Cervical Cancer Risk: A Meta-Analysis. Biomed. Res. Int. 2018, 2018, 7301595. [CrossRef]
- Shi Y, Li L, Hu Z, Li S, Wang S, Liu J, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat. Genet. 2013, 45, 918–922. [CrossRef] [PubMed]
- Wu Y, Liu B, Lin W, Xu Y, Li L, Zhang Y, et al. Human leukocyte antigen class II alleles and risk of cervical cancer in China. Hum. Immunol. 2007, 68, 192–200. [CrossRef]
- Liang J, Xu A, Xie Y, Awonuga AO, Lin Z. Some but not all of HLA-II alleles are associated with cervical cancer in Chinese women. Cancer Genet Cytogenet. 2008, 187, 95–100. [CrossRef]
- Rivera-Pirela SE, Echeverría M, Salcedo P, Márquez G, Carrillo Z, Parra Y, et al. [HLA DRB1*, DQB1*, DPA1*, and DPB1* and their association with the pathogenesis of leukemia in the population of Venezuela]. Rev. Alerg. Mex. 2016, 63, 237–251. [CrossRef]
- Sivapalan L, Anagnostou V. Genetic variation in antigen presentation and cancer immunotherapy. Immunity 2022, 55, 3–6. [CrossRef] [PubMed]
- Seliger B, Kloor M, Ferrone S. HLA class II antigen-processing pathway in tumors: Molecular defects and clinical relevance. Oncoimmunology 2017, 6, e1171447. [CrossRef] [PubMed]
- Leo PJ, Madeleine MM, Wang S, Schwartz SM, Newell F, Pettersson-Kymmer U, et al. Defining the genetic susceptibility to cervical neoplasia-A genome-wide association study. PLoS Genet. 2017, 13, e1006866. [CrossRef]
- Ekanayake Weeramange C, Shu D, Tang KD, Batra J, Ladwa R, Kenny L, et al. Analysis of human leukocyte antigen associations in human papillomavirus-positive and -negative head and neck cancer: Comparison with cervical cancer. Cancer 2022, 128, 1937–1947. [CrossRef] [PubMed]
- Augusto DG, Murdolo LD, Chatzileontiadou DSM, Sabatino JJ, Jr., Yusufali T, Peyser ND, et al. A common allele of HLA is associated with asymptomatic SARS-CoV-2 infection. Nature 2023, 620, 128–136. [CrossRef] [PubMed]
- Pastor DM, Schlom J. Immunology of Lynch Syndrome. Curr. Oncol. Rep. 2021, 23, 96. [CrossRef] [PubMed]
- Lynch HT, Drescher KM, de la Chapelle A. Immunology and the Lynch syndrome. Gastroenterology 2008, 134, 1246–1249. [CrossRef]
- Chambuso R, Kaambo E, Rebello G, Ramesar R. Correspondence on “Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database” by Dominguez-Valentin et al. Genet Med. 2022, 24, 1148–1150. [CrossRef]
- Burn J, Bishop DT, Chapman PD, Elliott F, Bertario L, Dunlop MG, et al. A randomized placebo-controlled prevention trial of aspirin and/or resistant starch in young people with familial adenomatous polyposis. Cancer Prev Res (Phila) 2011, 4, 655–665. [CrossRef]
- Pande M, Amos CI, Osterwisch DR, Chen J, Lynch PM, Broaddus R, et al. Genetic variation in genes for the xenobiotic-metabolizing enzymes CYP1A1, EPHX1, GSTM1, GSTT1, and GSTP1 and susceptibility to colorectal cancer in Lynch syndrome. Cancer Epidemiol Biomarkers Prev. 2008, 17, 2393–2401. [CrossRef]
- Campbell PT, Edwards L, McLaughlin JR, Green J, Younghusband HB, Woods MO. Cytochrome P450 17A1 and catechol O-methyltransferase polymorphisms and age at Lynch syndrome colon cancer onset in Newfoundland. Clin. Cancer Res. 2007, 13, 3783–3788. [CrossRef]
- Hitchins MP, Lynch HT. Dawning of the epigenetic era in hereditary cancer. Clin. Genet. 2014, 85, 413–416. [CrossRef] [PubMed]
- Hitchins MP, Rapkins RW, Kwok CT, Srivastava S, Wong JJ, Khachigian LM, et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5’UTR. Cancer Cell 2011, 20, 200–213. [CrossRef] [PubMed]
- Watson P, Ashwathnarayan R, Lynch HT, Roy HK. Tobacco use and increased colorectal cancer risk in patients with hereditary nonpolyposis colorectal cancer (Lynch syndrome). Arch. Intern. Med. 2004, 164, 2429–2431. [CrossRef] [PubMed]
- Pande M, Lynch PM, Hopper JL, Jenkins MA, Gallinger S, Haile RW, et al. Smoking and colorectal cancer in Lynch syndrome: results from the Colon Cancer Family Registry and the University of Texas M.D. Anderson Cancer Center. Clin. Cancer Res. 2010, 16, 1331–1339. [CrossRef]
- Dashti SG, Buchanan DD, Jayasekara H, Ait Ouakrim D, Clendenning M, Rosty C, et al. Alcohol Consumption and the Risk of Colorectal Cancer for Mismatch Repair Gene Mutation Carriers. Cancer Epidemiol Biomarkers Prev. 2017, 26, 366–375. [CrossRef]
- Botma A, Vasen HF, van Duijnhoven FJ, Kleibeuker JH, Nagengast FM, Kampman E. Dietary patterns and colorectal adenomas in Lynch syndrome: the GEOLynch cohort study. Cancer 2013, 119, 512–521. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).