Submitted:
07 March 2024
Posted:
08 March 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
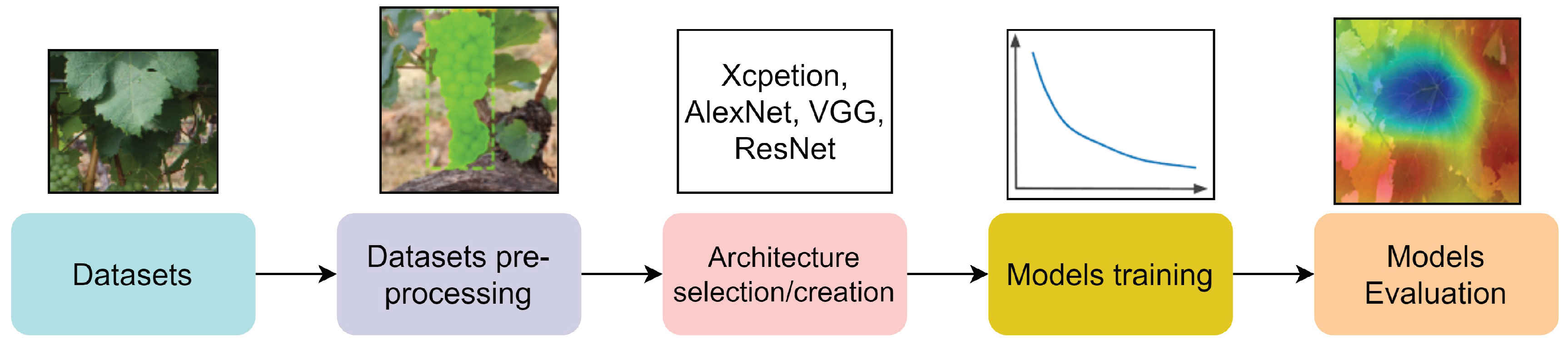
2. Methods
2.1. Research Questions
- (RQ1) How Deep-Learning techniques have been used for automatic identification of grapevine varieties?
- (RQ2) Which are the best DL-based models for Automatic Grapevine Verities Identification?
- (RQ3) What are the main challenges and future development trends in identifying grape varieties using DL-based models?
2.2. Inclusion Criteria
2.3. Search Strategy
2.4. Extraction of Characteristics
3. Results
3.1. Datasets and Benchmarks
3.2. Pre-Processing
3.3. Architecture and Training
3.4. Evaluation
4. Discussion and Future Directions
4.1. Looking into the Grapevine Varieties Identification Problem
- Grapevines are seasonal plants. This means that there are periods when the plants will have leaves, and others when they won’t. This feature has a direct impact on the preparation of the dataset, which ideally should cover different phases of leaf growth. In addition, this feature limits the use of fruits in identification, as they take longer to grow than leaves;
- The presence of some grape varieties (e.g. Syrah, Chardonnay) is more common than others (e.g. Alvarinho), so the datasets are naturally unbalanced and can be treated as a long-tailed data distribution classification (some classes represent the majority of the data, while most classes are under-represented [78]);
- The classification of varieties within a species has a high inter-class similarity and high intra-class variations, placing the task in the fine-grained recognition problems family;
- There will be a high presence of unrelated information in the images acquired in the field, which could contribute to classification errors;
- There is a large amount of publicly available leaf and fruit images that are not annotated for variety identification;
4.2. Datasets
4.3. Pre-Processing
4.4. Architectures and Training
4.5. Evaluation
4.6. Comparison with other Subfields of Precision Viticulture
5. Conclusion
- (RQ1) How Deep-Learning techniques have been used for automatic identification of grapevine varieties? Pre-trained architectures for image classification are the way in which DL has been most widely applied to the identification of grapevine varieties.
- (RQ2) Which are the best DL-based models for Automatic Grapevine Verities Identification? Since the datasets are small, tuning and transfer learning techniques have been used in all image-based classification works. The most commonly used pre-trained architecture was Xception, accompanied by cross entropy loss and static geometric augmentation strategies. In the Evaluation stage, in addition to the popular metrics, Grad-CAM was the most commonly used XAI method.
- (RQ3) What are the main challenges and future development trends in identifying grape varieties using DL-based models? There is still room to evaluate the removal of the complex background; the generation of new samples through GANs can be explored; new architectures can be tested, e.g. Swin Transformers, Capsules Networks, or Bilinear CNNs; different losses still can be explored [78,117,122,123,124]; and other XAI approaches can also be employed, e.g. Guided Integrated Gradients, XRAI, and SmoothGrads.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Acc | Accuracy |
| AUC | Area-Under-The-Curve |
| CapsNet | Capsule Networks |
| CE | Cross Entropy |
| CNN | Convolutional Neural Network |
| DNA | Deoxyribonucleic acid |
| DDR | Douro Demarcated Region |
| DL | Deep Learning |
| ESRGAN | Enhanced Super Resolution Generative Adversarial Networks |
| FL | Focal Loss |
| GAN | Generative Adversarial Network |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| HSI | Hue, Saturation and Intensity |
| ICA | Independent Component Analysis |
| MSC | Multiplicative Scatter Correction |
| MCC | Matthews correlation coefficient |
| LIME | Local Interpretable Model-Agnostic Explanations |
| RGB | Red, Green, Blue |
| RQ | Research Questions |
| SG | Savitzky-Golay |
| SNV | Standard Normal Variant |
| SVM | Support Vector Machines |
| SGD | Stochastic Descent Gradient |
| UAV | Unmanned Aerial Vehicles |
| ViT | Vision Transformer |
| XAI | Explainable Artificial Intelligence |
References
- Eyduran, S.P.; Akin, M.; Ercisli, S.; Eyduran, E.; Maghradze, D. Sugars, organic acids, and phenolic compounds of ancient grape cultivars (Vitis vinifera L.) from Igdir province of Eastern Turkey. Biological Research 2015, 48, 2. [CrossRef]
- Nascimento, R.; Maia, M.; Ferreira, A.E.N.; Silva, A.B.; Freire, A.P.; Cordeiro, C.; Silva, M.S.; Figueiredo, A. Early stage metabolic events associated with the establishment of Vitis vinifera – Plasmopara viticola compatible interaction. Plant Physiology and Biochemistry 2019, 137, 1–13. [CrossRef]
- Cunha, J.; Santos, M.T.; Carneiro, L.C.; Fevereiro, P.; Eiras-Dias, J.E. Portuguese traditional grapevine cultivars and wild vines (Vitis vinifera L.) share morphological and genetic traits. Genetic Resources and Crop Evolution 2009, 56, 975–989. [CrossRef]
- Schneider, A.; Carra, A.; Akkak, A.; This, P.; Laucou, V.; Botta, R. Verifying synonymies between grape cultivars from France and Northwestern Italy using molecular markers. VITIS - Journal of Grapevine Research 2001, 40, 197–197. Number: 4. [CrossRef]
- Lacombe, T. Contribution à l’étude de l’histoire évolutive de la vigne cultivée (<em>Vitis vinifera</em> L.) par l’analyse de la diversité génétique neutre et de gènes d’intérêt. Theses, Institut National d’Etudes Supérieures Agronomiques de Montpellier, 2012. MR AGAP - équipe DAAV - Diversité, adaptation et amélioration de la vigne.
- Distribution of the world’s grapevine varieties. Technical Report 979-10-91799-89-8, International Organisation of Vine and Wine.
- Koklu, M.; Unlersen, M.F.; Ozkan, I.A.; Aslan, M.F.; Sabanci, K. A CNN-SVM study based on selected deep features for grapevine leaves classification. Measurement 2022, 188, 110425. [CrossRef]
- Moncayo, S.; Rosales, J.D.; Izquierdo-Hornillos, R.; Anzano, J.; Caceres, J.O. Classification of red wine based on its protected designation of origin (PDO) using Laser-induced Breakdown Spectroscopy (LIBS). Talanta 2016, 158, 185–191. [CrossRef]
- Giacosa, E. Wine Consumption in a Certain Territory. Which Factors May Have Impact on It? In Production and Management of Beverages; Woodhead Publishing, 2019; pp. 361–380. [CrossRef]
- of Vine, T.I.O.; Wine. STATE OF THE WORLD VITIVINICULTURAL SECTOR IN 2020 2020.
- Jones, G.; Alves, F. Impact of climate change on wine production: a global overview and regional assessment in the Douro Valley of Portugal. Int. J. of Global Warming 2012, 4, 383–406. [CrossRef]
- Chitwood, D.H.; Ranjan, A.; Martinez, C.C.; Headland, L.R.; Thiem, T.; Kumar, R.; Covington, M.F.; Hatcher, T.; Naylor, D.T.; Zimmerman, S.; et al. A Modern Ampelography: A Genetic Basis for Leaf Shape and Venation Patterning in Grape. Plant Physiology 2014, 164, 259–272. [CrossRef]
- Galet, P. Précis d’ampélographie pratique; 1952.
- Garcia-Muñoz, S.; Muñoz-Organero, G.; de Andrés, M.; Cabello, F. Ampelography - An old technique with future uses: the case of minor varieties of Vitis vinifera L. from the Balearic Islands. Journal International des Sciences de la Vigne et du Vin 2011, 45, 125–137. [CrossRef]
- Pavek, D.S.; Lamboy, W.F.; Garvey, E.J. Selecting in situ conservation sites for grape genetic resources in the USA. Genetic Resources and Crop Evolution 2003 50:2 2003, 50, 165–173. [CrossRef]
- Tassie, L. Vine identification – knowing what you have. The Grape and Wine Research and Development Corporation (GWRDC) Innovators network 2010.
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theoretical and Applied Genetics 2004, 109, 1448–1458. [CrossRef]
- Calo, A.; Tomasi, D.; Crespan, M.; Costacurta, A. Relationship between environmental factors and the dynamics of growth and composition of the grapevine. Acta Horticulturae 1996, 427, 217–231. [CrossRef]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A Review on Deep Learning Techniques Applied to Semantic Segmentation 2017.
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M.P. Support Vector Machine and Artificial Neural Network Models for the Classification of Grapevine Varieties Using a Portable NIR Spectrophotometer. PLOS ONE 2015, 10, e0143197. Publisher: Public Library of Science. [CrossRef]
- Karakizi, C.; Oikonomou, M.; Karantzalos, K. Vineyard Detection and Vine Variety Discrimination from Very High Resolution Satellite Data. Remote Sensing 2016, 8, 235. Number: 3 Publisher: Multidisciplinary Digital Publishing Institute. [CrossRef]
- Diago, M.P.; Fernandes, A.M.; Millan, B.; Tardaguila, J.; Melo-Pinto, P. Identification of grapevine varieties using leaf spectroscopy and partial least squares. Computers and Electronics in Agriculture 2013, 99, 7–13. [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Curran Associates, Inc., 2012, Vol. 25.
- Chen, Y.; Huang, Y.; Zhang, Z.; Wang, Z.; Liu, B.; Liu, C.; Huang, C.; Dong, S.; Pu, X.; Wan, F.; et al. Plant image recognition with deep learning: A review. Computers and Electronics in Agriculture 2023, 212, 108072. [CrossRef]
- Mohimont, L.; Alin, F.; Rondeau, M.; Gaveau, N.; Steffenel, L.A. Computer Vision and Deep Learning for Precision Viticulture. Agronomy 2022, 12, 2463. Number: 10 Publisher: Multidisciplinary Digital Publishing Institute. [CrossRef]
- Ferro, M.V.; Catania, P. Technologies and Innovative Methods for Precision Viticulture: A Comprehensive Review. Horticulturae 2023, 9, 399. Number: 3 Publisher: Multidisciplinary Digital Publishing Institute. [CrossRef]
- Chai, J.; Zeng, H.; Li, A.; Ngai, E.W.T. Deep learning in computer vision: A critical review of emerging techniques and application scenarios. Machine Learning with Applications 2021, 6, 100134. [CrossRef]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Computing Surveys 2022, 54, 1–41. arXiv:2101.01169 [cs]. [CrossRef]
- Dhanya, V.G.; Subeesh, A.; Kushwaha, N.L.; Vishwakarma, D.K.; Nagesh Kumar, T.; Ritika, G.; Singh, A.N. Deep learning based computer vision approaches for smart agricultural applications. Artificial Intelligence in Agriculture 2022, 6, 211–229. [CrossRef]
- Doğan, G.; Imak, A.; Ergen, B.; Sengur, A. A new hybrid approach for grapevine leaves recognition based on ESRGAN data augmentation and GASVM feature selection. Neural Computing and Applications 2024. [CrossRef]
- Magalhaes, S.C.; Castro, L.; Rodrigues, L.; Padilha, T.C.; Carvalho, F.D.; Santos, F.N.D.; Pinho, T.; Moreira, G.; Cunha, J.; Cunha, M.; et al. Toward Grapevine Digital Ampelometry Through Vision Deep Learning Models. IEEE Sensors Journal 2023, 23, 10132–10139. [CrossRef]
- Carneiro, G.; Neto, A.; Teixeira, A.; Cunha, A.; Sousa, J. Evaluating Data Augmentation for Grapevine Varieties Identification 2023. pp. 3566–3569. [CrossRef]
- Carneiro, G.A.; Texeira, A.; Morais, R.; Sousa, J.J.; Cunha, A. Can the Segmentation Improve the Grape Varieties’ Identification Through Images Acquired On-Field? Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2023, 14116 LNAI, 351–363. [CrossRef]
- Gupta, R.; Gill, K.S. Grapevine Augmentation and Classification using Enhanced EfficientNetB5 Model. 2023 IEEE Renewable Energy and Sustainable E-Mobility Conference, RESEM 2023 2023. [CrossRef]
- Carneiro, G.A.; Padua, L.; Peres, E.; Morais, R.; Sousa, J.J.; Cunha, A. Segmentation as a Preprocessing Tool for Automatic Grapevine Classification. International Geoscience and Remote Sensing Symposium (IGARSS) 2022, 2022-July, 6053–6056. [CrossRef]
- Carneiro, G.S.; Ferreira, A.; Morais, R.; Sousa, J.J.; Cunha, A. Analyzing the Fine Tuning’s impact in Grapevine Classification. Procedia Computer Science 2022, 196, 364–370. [CrossRef]
- Carneiro, G.A.; Pádua, L.; Peres, E.; Morais, R.; Sousa, J.J.; Cunha, A. Grapevine Varieties Identification Using Vision Transformers. In Proceedings of the IGARSS 2022 - 2022 IEEE International Geoscience and Remote Sensing Symposium, 2022, pp. 5866–5869. [CrossRef]
- Carneiro, G.; Padua, L.; Sousa, J.J.; Peres, E.; Morais, R.; Cunha, A. Grapevine Variety Identification Through Grapevine Leaf Images Acquired in Natural Environment 2021. pp. 7055–7058. [CrossRef]
- Liu, Y.; Su, J.; Shen, L.; Lu, N.; Fang, Y.; Liu, F.; Song, Y.; Su, B. Development of a mobile application for identification of grapevine (Vitis vinifera l.) cultivars via deep learning. International Journal of Agricultural and Biological Engineering 2021, 14, 172–179. [CrossRef]
- Škrabánek, P.; Doležel, P.; Matoušek, R.; Junek, P. RGB Images Driven Recognition of Grapevine Varieties. Springer, 9 2021, Vol. 1268 AISC, Advances in Intelligent Systems and Computing, pp. 216–225. [CrossRef]
- Nasiri, A.; Taheri-Garavand, A.; Fanourakis, D.; Zhang, Y.D.; Nikoloudakis, N. Automated grapevine cultivar identification via leaf imaging and deep convolutional neural networks: A proof-of-concept study employing primary iranian varieties. Plants 2021, 10. [CrossRef]
- Peng, Y.; Zhao, S.; Liu, J.; Peng, Y..; Zhao, S..; Liu, J. Fused Deep Features-Based Grape Varieties Identification Using Support Vector Machine. Agriculture 2021, Vol. 11, Page 869 2021, 11, 869. [CrossRef]
- Franczyk, B.; Hernes, M.; Kozierkiewicz, A.; Kozina, A.; Pietranik, M.; Roemer, I.; Schieck, M. Deep learning for grape variety recognition. Procedia Computer Science 2020, 176, 1211–1220. [CrossRef]
- Fernandes, A.M.; Utkin, A.B.; Eiras-Dias, J.; Cunha, J.; Silvestre, J.; Melo-Pinto, P. Grapevine variety identification using “Big Data” collected with miniaturized spectrometer combined with support vector machines and convolutional neural networks. Computers and Electronics in Agriculture 2019, 163, 104855. [CrossRef]
- Adão, T.; Pinho, T.M.; Ferreira, A.; Sousa, A.; Pádua, L.; Sousa, J.; Sousa, J.J.; Peres, E.; Morais, R. Digital ampelographer: A CNN based preliminary approach, Cham, 2019; Vol. 11804 LNAI, pp. 258–271. [CrossRef]
- Pereira, C.S.; Morais, R.; Reis, M.J.C.S. Deep learning techniques for grape plant species identification in natural images. Sensors (Switzerland) 2019, 19, 4850. [CrossRef]
- Santos, T.; de Souza, L.; Andreza, d.S.; Avila, S. Embrapa Wine Grape Instance Segmentation Dataset – Embrapa WGISD 2019. [CrossRef]
- Al-khazraji, L.R.; Mohammed, M.A.; Abd, D.H.; Khan, W.; Khan, B.; Hussain, A.J. Image dataset of important grape varieties in the commercial and consumer market. Data in Brief 2023, 47, 108906. [CrossRef]
- Vlah, M. Grapevine Leaves, 2021. [CrossRef]
- Sozzi, M.; Cantalamessa, S.; Cogato, A.; Kayad, A.; Marinello, F. wGrapeUNIPD-DL: An open dataset for white grape bunch detection. Data in Brief 2022, 43, 108466. [CrossRef]
- Seng, K.P.; Ang, L.M.; Schmidtke, L.M.; Rogiers, S.Y. Computer vision and machine learning for viticulture technology. IEEE Access 2018, 6, 67494–67510. [CrossRef]
- Rodrigues, A. Um método filométrico de caracterização ampelográfica 1952.
- Hyvärinen, A.; Oja, E. A Fast Fixed-Point Algorithm for Independent Component Analysis. Neural Computation 1997, 9, 1483–1492. [CrossRef]
- Canny, J. A Computational Approach to Edge Detection. IEEE Transactions on Pattern Analysis and Machine Intelligence 1986, PAMI-8, 679–698. [CrossRef]
- Zheng, X.; Wang, X. Leaf Vein Extraction Based on Gray-scale Morphology. International Journal of Image, Graphics and Signal Processing 2010, 2, 25–31. [CrossRef]
- Pereira, C.S.; Morais, R.; Reis, M.J. Pixel-Based Leaf Segmentation from Natural Vineyard Images Using Color Model and Threshold Techniques. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2018, 10882 LNCS, 96–106. [CrossRef]
- Du, Q.; Kopriva, I.; Szu, H. Independent-component analysis for hyperspectral remote sensing imagery classification. Optical Engineering - OPT ENG 2006, 45. [CrossRef]
- Vaseghi, S.; Jetelova, H. Principal and independent component analysis in image processing. Proceeding of the 14th ACM International Conference on Mobile Computing and Networking 2006.
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics 1979, 9, 62–66. [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. CoRR 2015, abs/1505.0.
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 2017, 39, 2481–2495. arXiv: 1511.00561. [CrossRef]
- Wang, X.; Yu, K.; Wu, S.; Gu, J.; Liu, Y.; Dong, C.; Loy, C.C.; Qiao, Y.; Tang, X. ESRGAN: Enhanced Super-Resolution Generative Adversarial Networks, 2018. arXiv:1809.00219 [cs]. [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Communications of the ACM 2014, 63, 139–144. [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings 2014.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2015, 2016-December, 770–778. [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017 2016, 2017-January, 2261–2269. [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. 2017, pp. 1800–1807. [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2018, pp. 4510–4520. [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. Journal of Big Data 2021 8:1 2021, 8, 1–74. [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale 2020.
- Sun, S.Q.; Zeng, S.G.; Liu, Y.; Heng, P.A.; Xia, D.S. A new method of feature fusion and its application in image recognition. Pattern Recognition 2005, 38, 2437–2448. [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization, 2015.
- Lin, T.Y.; Goyal, P.; Girshick, R.B.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. CoRR 2017, abs/1708.0.
- Mukhoti, J.; Kulharia, V.; Sanyal, A.; Golodetz, S.; Torr, P.H.; Dokania, P.K. Calibrating deep neural networks using focal loss. Neural information processing systems foundation, 2 2020, Vol. 2020-Decem.
- Barredo Arrieta, A.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Information Fusion 2020, 58, 82–115. [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. International Journal of Computer Vision 2016, 128, 336–359. [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. Association for Computing Machinery, 2 2016, Vol. 13-17-Augu, pp. 97–101. [CrossRef]
- Cui, Y.; Jia, M.; Lin, T.Y.; Song, Y.; Belongie, S. Class-Balanced Loss Based on Effective Number of Samples. 2019, pp. 9268–9277.
- Barratt, S.; Sharma, R. A Note on the Inception Score 2018. [CrossRef]
- Ravuri, S.; Vinyals, O. Seeing is Not Necessarily Believing: Limitations of BigGANs for Data Augmentation. 2019, pp. 1–5.
- Shmelkov, K.; Schmid, C.; Alahari, K. How good is my GAN? Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2018, 11206 LNCS, 218–234. [CrossRef]
- Gomaa, A.A.; El-Latif, Y.M. Early Prediction of Plant Diseases using CNN and GANs. International Journal of Advanced Computer Science and Applications 2021, 12, 514–519. [CrossRef]
- Nazki, H.; Lee, J.; Yoon, S.; Park, D.S. Image-to-Image Translation with GAN for Synthetic Data Augmentation in Plant Disease Datasets. Smart Media Journal 2019, 8, 46–57. [CrossRef]
- Talukdar, B. Handling of Class Imbalance for Plant Disease Classification with Variants of GANs. 2020, pp. 466–471. [CrossRef]
- Yilma, G.; Belay, S.; Qin, Z.; Gedamu, K.; Ayalew, M. Plant Disease Classification Using Two Pathway Encoder GAN Data Generation. 2020 17th International Computer Conference on Wavelet Active Media Technology and Information Processing, ICCWAMTIP 2020 2020, pp. 67–72. [CrossRef]
- Zeng, Q.; Ma, X.; Cheng, B.; Zhou, E.; Pang, W. GANS-based data augmentation for citrus disease severity detection using deep learning. IEEE Access 2020, 8, 172882–172891. [CrossRef]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le, V.Q. AutoAugment: Learning Augmentation Policies from Data. Cvpr 2019 2018, pp. 113–123. [CrossRef]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond Empirical Risk Minimization, 2018. arXiv:1710.09412 [cs, stat]. [CrossRef]
- DeVries, T.; Taylor, G.W. Dataset Augmentation in Feature Space, 2017, [arXiv:stat.ML/1702.05538].
- Chu, P.; Bian, X.; Liu, S.; Ling, H. Feature Space Augmentation for Long-Tailed Data, 2020. arXiv:2008.03673 [cs]. [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Computers and Electronics in Agriculture 2018, 145, 311–318. [CrossRef]
- Kc, K.; Yin, Z.; Li, D.; Wu, Z. Impacts of Background Removal on Convolutional Neural Networks for Plant Disease Classification In-Situ. Agriculture 2021, 11, 827. Number: 9 Publisher: Multidisciplinary Digital Publishing Institute. [CrossRef]
- Wu, Y.J.; Tsai, C.M.; Shih, F. Improving Leaf Classification Rate via Background Removal and ROI Extraction. Journal of Image and Graphics 2016, 4, 93–98. [CrossRef]
- Howard, A.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.C.; Tan, M.; Chu, G.; Vasudevan, V.; Zhu, Y.; Pang, R.; et al. Searching for mobileNetV3. Institute of Electrical and Electronics Engineers Inc., 5 2019, Vol. 2019-Octob, pp. 1314–1324. [CrossRef]
- Tan, M.; Le, V.Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. 36th International Conference on Machine Learning, ICML 2019 2019, 2019-June, 10691–10700.
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training, 2021. arXiv:2104.00298 [cs]. [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s, 2022. arXiv:2201.03545 [cs]. [CrossRef]
- Woo, S.; Debnath, S.; Hu, R.; Chen, X.; Liu, Z.; Kweon, I.S.; Xie, S. ConvNeXt V2: Co-designing and Scaling ConvNets with Masked Autoencoders, 2023. arXiv:2301.00808 [cs]. [CrossRef]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic Routing Between Capsules. Advances in Neural Information Processing Systems 2017, 2017-December, 3857–3867.
- Kwabena Patrick, M.; Felix Adekoya, A.; Abra Mighty, A.; Edward, B.Y. Capsule Networks – A survey. Journal of King Saud University - Computer and Information Sciences 2022, 34, 1295–1310. [CrossRef]
- Andrushia, A.D.; Neebha, T.M.; Patricia, A.T.; Sagayam, K.M.; Pramanik, S. Capsule network-based disease classification for Vitis Vinifera leaves. Neural Computing and Applications 2024, 36, 757–772. [CrossRef]
- Raghu, M.; Unterthiner, T.; Kornblith, S.; Zhang, C.; Dosovitskiy, A. Do Vision Transformers See Like Convolutional Neural Networks? 2021.
- Steiner, A.; Kolesnikov, A.; Zhai, X.; Wightman, R.; Uszkoreit, J.; Beyer, L. How to train your ViT? Data, Augmentation, and Regularization in Vision Transformers 2021.
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Computing Surveys 2021. [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows, 2021. arXiv:2103.14030 [cs]. [CrossRef]
- Guo, Y.; Lan, Y.; Chen, X. CST: Convolutional Swin Transformer for detecting the degree and types of plant diseases. Computers and Electronics in Agriculture 2022, 202, 107407. [CrossRef]
- Chang, B.; Wang, Y.; Zhao, X.; Li, G.; Yuan, P. A general-purpose edge-feature guidance module to enhance vision transformers for plant disease identification. Expert Systems with Applications 2024, 237, 121638. [CrossRef]
- Wang, F.; Rao, Y.; Luo, Q.; Jin, X.; Jiang, Z.; Zhang, W.; Li, S. Practical cucumber leaf disease recognition using improved Swin Transformer and small sample size. Computers and Electronics in Agriculture 2022, 199, 107163. [CrossRef]
- El-Nouby, A.; Izacard, G.; Touvron, H.; Laptev, I.; Jegou, H.; Grave, E. Are Large-scale Datasets Necessary for Self-Supervised Pre-training?, 2021. arXiv:2112.10740 [cs]. [CrossRef]
- Doersch, C.; Gupta, A.; Zisserman, A. CrossTransformers: spatially-aware few-shot transfer, 2021. arXiv:2007.11498 [cs]. [CrossRef]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Computers and Electronics in Agriculture 2020, 178, 105760. [CrossRef]
- Nazki, H.; Yoon, S.; Fuentes, A.; Park, D.S. Unsupervised image translation using adversarial networks for improved plant disease recognition. Computers and Electronics in Agriculture 2020, 168, 105117. [CrossRef]
- Homan, D.; du Preez, J.A. Automated feature-specific tree species identification from natural images using deep semi-supervised learning. Ecological Informatics 2021, 66, 101475. [CrossRef]
- Güldenring, R.; Nalpantidis, L. Self-supervised contrastive learning on agricultural images. Computers and Electronics in Agriculture 2021, 191. Type: Article. [CrossRef]
- Buda, M.; Maki, A.; Mazurowski, M.A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Networks 2018, 106, 249–259. arXiv:1710.05381 [cs, stat]. [CrossRef]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-Shot Learning approach for plant disease classification using images taken in the field. Computers and Electronics in Agriculture 2020, 175, 105542. [CrossRef]
- Park, S.; Lim, J.; Jeon, Y.; Choi, J.Y. Influence-Balanced Loss for Imbalanced Visual Classification. 2021, pp. 735–744.
- Wei, X.S.; Song, Y.Z.; Mac Aodha, O.; Wu, J.; Peng, Y.; Tang, J.; Yang, J.; Belongie, S. Fine-Grained Image Analysis with Deep Learning: A Survey, 2021. arXiv:2111.06119 [cs]. [CrossRef]
- Lin, T.Y.; RoyChowdhury, A.; Maji, S. Bilinear CNNs for Fine-grained Visual Recognition, 2017. arXiv:1504.07889 [cs]. [CrossRef]
- Gao, Y.; Beijbom, O.; Zhang, N.; Darrell, T. Compact Bilinear Pooling. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 317–326. ISSN: 1063-6919. [CrossRef]
- Min, S.; Yao, H.; Xie, H.; Zha, Z.J.; Zhang, Y. Multi-Objective Matrix Normalization for Fine-grained Visual Recognition. IEEE Transactions on Image Processing 2020, 29, 4996–5009. arXiv:2003.13272 [cs]. [CrossRef]
- Dubey, A.; Gupta, O.; Guo, P.; Raskar, R.; Farrell, R.; Naik, N. Pairwise Confusion for Fine-Grained Visual Classification, 2018. arXiv:1705.08016 [cs]. [CrossRef]
- Sun, G.; Cholakkal, H.; Khan, S.; Khan, F.S.; Shao, L. Fine-grained Recognition: Accounting for Subtle Differences between Similar Classes, 2019. arXiv:1912.06842 [cs]. [CrossRef]
- Chang, D.; Ding, Y.; Xie, J.; Bhunia, A.K.; Li, X.; Ma, Z.; Wu, M.; Guo, J.; Song, Y.Z. The Devil is in the Channels: Mutual-Channel Loss for Fine-Grained Image Classification. IEEE Transactions on Image Processing 2020, 29, 4683–4695. arXiv:2002.04264 [cs]. [CrossRef]
- Subramanya, A.; Pillai, V.; Pirsiavash, H. Fooling network interpretation in image classification. Proceedings of the IEEE International Conference on Computer Vision 2019, 2019-Octob, 2020–2029. [CrossRef]
- Alvarez-Melis, D.; Jaakkola, T.S. On the Robustness of Interpretability Methods 2018.
- Garreau, D.; von Luxburg, U. Explaining the Explainer: A First Theoretical Analysis of LIME 2020.
- Stiffler, M.; Hudler, A.; Lee, E.; Braines, D.; Mott, D.; Harborne, D. An Analysis of Reliability Using LIME with Deep Learning Models 2018.
- Kapishnikov, A.; Venugopalan, S.; Avci, B.; Wedin, B.; Terry, M.; Bolukbasi, T. Guided Integrated Gradients: An Adaptive Path Method for Removing Noise. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2021, abs/2106.0, 5048–5056. [CrossRef]
- Kapishnikov, A.; Bolukbasi, T.; Viegas, F.; Terry, M. XRAI: Better Attributions Through Regions. Proceedings of the IEEE International Conference on Computer Vision 2019, 2019-October, 4947–4956. [CrossRef]
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. SmoothGrad: removing noise by adding noise 2017. [CrossRef]
- Jin, H.; Chu, X.; Qi, J.; Zhang, X.; Mu, W. CWAN: Self-supervised learning for deep grape disease image composition. Engineering Applications of Artificial Intelligence 2023, 123, 106458. [CrossRef]
- Jin, H.; Li, Y.; Qi, J.; Feng, J.; Tian, D.; Mu, W. GrapeGAN: Unsupervised image enhancement for improved grape leaf disease recognition. Computers and Electronics in Agriculture 2022, 198, 107055. [CrossRef]
- Kerkech, M.; Hafiane, A.; Canals, R. Vine disease detection in UAV multispectral images using optimized image registration and deep learning segmentation approach. Computers and Electronics in Agriculture 2020, 174, 105446. [CrossRef]
- Jin, H.; Chu, X.; Qi, J.; Feng, J.; Mu, W. Learning multiple attention transformer super-resolution method for grape disease recognition. Expert Systems with Applications 2024, 241, 122717. [CrossRef]
| 1 |

| Study | Year | Dataset Location | Dataset Description | Focus | Architecture | Results |
|---|---|---|---|---|---|---|
| Doğan et al. [30] | 2024 | Turkey | 7000 RGB images distributed between 5 classes acquired in controlled environment |
Leaves | Fused Deep Features + SVM |
1.00 (F1) |
| Magalhães et al. [31] | 2023 | Portugal | 40428 RGB images distributed between 26 classes acquired in a controlled environment |
Leaves | MobileNetV2, ResNet-34 and VGG-11 |
94.75 (F1) |
| Carneiro et al. [32] | 2023 | Portugal | 6216 RGB images distributed between 14 classes acquired in field |
Leaves | EfficientNetV2S | 0.89 (F1) |
| Carneiro et al. [33] | 2023 | Portugal | 675 RGB images distributed between 12 classes; 4354 RGB images distributed between 14 classes; both acquired in field |
Leaves | EfficientNetV2S | 0.88 (F1) |
| Gupta and Gill [34] | 2023 | Turkey | 2500 RGB images distributed between 5 classes acquired in controlled environment |
Leaves | EfficientNetB5 | 0.86 (Acc) |
| Carneiro et al. [35] | 2022 | Portugal | 28427 RGB images distributed between 6 classes acquired in field |
Leaves | Xception | 0.92 (F1) |
| Carneiro et al. [36] | 2022 | Portugal | 6922 RGB images distributed between 12 classes acquired in-field |
Leaves | Xception | 0.92 (F1) |
| Carneiro et al. [37] | 2022 | Portugal | 6922 RGB images distributed between 12 classes acquired in-field |
Leaves | Vision Transformer (ViT_B) |
0.96 (F1) |
| Koklu et al. [7] | 2022 | Turkey | 2500 RGB images distributed between 5 classes acquired in controlled environment |
Leaves | MobileNetV2 + SVM |
97.60 (Acc) |
| Carneiro et al. [38] | 2021 | Portugal | 6922 RGB images distributed between 12 classes acquired in-field |
Leaves | Xception | 0.93 (F1) |
| Liu et al. [39] | 2021 | China | 5091 RGB images distributed between 21 classes acquired in-field |
Leaves | GoogLeNet | 99.91 (Acc) |
| Škrabánek et al. [40] | 2021 | Czech Republic | 7200 RGB images distributed between 7 classes acquired in-field |
Fruits | DenseNet | 98.00 (Acc) |
| Nasiri et al. [41] | 2021 | Iran | 300 RGB images distributed between 6 classes acquired in a controlled environment |
Leaves | VGG-16 | 99.00 (Acc) |
| Peng et al.[42] | 2021 | Brazil | 300 RGB images distributed between 6 classes acquired in-field |
Fruits | Fused Deep Features |
96.80 (F1) |
| Franczyk et al. [43] | 2020 | Brazil | 3957 RGB images distributed between 5 classes acquired in-field |
Fruits | ResNet | 99.00 (Acc) |
| Fernandes et al. [44] | 2019 | Portugal | 35933 Spectra distributed between 64 classes in-field |
Leaves | Handcraft | 0.68 (AUC) 0.98 (AUC) |
| Adão et al. [45] | 2019 | Portugal | 3120 RGB images distributed between 6 classes acquired in controlled environment |
Leaves | Xception | 100.00 (Acc) |
| Pereira et al. [46] | 2019 | Portugal | 224 RGB images distributed between 6 classes acquired in controlled environment |
Leaves | AlexNet | 77.30 (Acc) |
| Study | Acquisition Device | Publicly Available | Acquisition Period | D. Augmentation | Acquisition Environment |
|---|---|---|---|---|---|
| Doğan et al. [30] | Camera - Prosilica GT2000C | Yes | - | Static augmentations and artificially generated images | Special illumination box |
| Magalhães et al. [31] | Kyocera TASKalfa 2552ci | No | 1 day in June 2021 | Blur, rotations, variations in brightness, horizontal flips, and gaussian noise | Special illumination box |
| Carneiro et al. [32] | Smartphones | No | Seasons 2021 and 2020 | Static augmentations, CutMix and RandAugment | In-field |
| Carneiro et al. [33] | Camera and Smarthphone | No | 1 Season and 2 Seasons | Rotations, Flips and Zoom | In-field |
| Gupta and Gill [34] | Camera - Prosilica GT2000C | Yes | - | Angle, scaling factor, translation | Special illumination box |
| Carneiro et al. [35] | Mixed Camera and Smartphone | No | Seasons 2017 and 2020 | Rotation, shift, flip, and brightness changes | In-field |
| Carneiro et al. [36] | Camera - Canon EOS 600D | No | - | Rotations, shifts, variations in brightness and flips | In-field |
| Carneiro et al. [37] | Camera - Canon EOS 600D | No | 1 Season | Rotations, shifts, variations in brightness and flips | In-field |
| Koklu et al. [7] | Camera - Prosilica GT2000C | Yes | - | Angle, scaling factor, translation | Special illumination box |
| Carneiro et al. [38] | Camera - Canon EOS 600D | No | 1 Season | Rotations, shifts, variations in brightness and flips | In-field |
| Liu et al. [39] | Camera - Canon EOS 70D | No | - | Scaling, transposing, rotation and flips | In-field |
| Škrabánek et al. [40] | Camera – Canon EOS 100D and Canon EOS 1100D | No | 2 days in August 2015 | - | In-field |
| Nasiri et al. [41] | Camera – Canon SX260 HS | On request | 1 day in July 2018 | Rotation, height and width shift | Capture station with artificial light |
| Peng et al.[42] | Mixed Camera and Smartphone | Yes | 1 day in April 2017 and 1 day in April 2018 | - | In-field |
| Franczyk et al. [43] | Mixed Camera and Smartphone | Yes | 1 day in April 2017 and 1 day in April 2018 | - | In-field |
| Fernandes et al. [44] | Spectrometer – OceanOptics Flame-S | No | 4 days in July 2017 | - | In-field |
| Adão et al. [45] | Camera - Canon EOS 600D | No | Season 2017 | Rotations, contrasts/brightness, vertical/horizontal mirroring, and scale variations | Controlled environment with white background |
| Pereira et al. [46] | Mixed Camera and Smartphone | No | Seasons 2016 and 2019 | Translation, reflection, rotation | In-field |
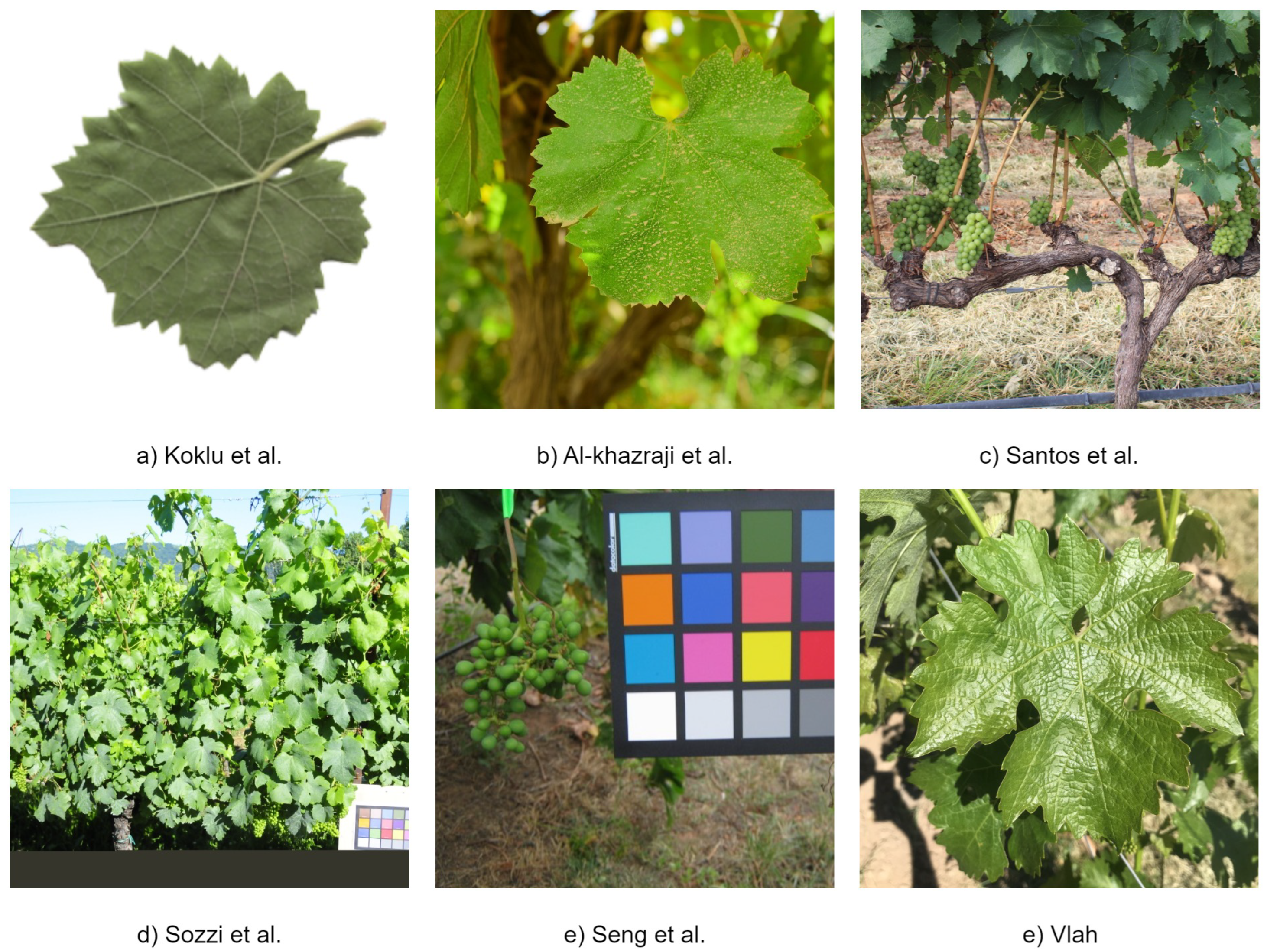
| Dataset | Number of Classes | Number of images | Classes | Balanced |
|---|---|---|---|---|
| Koklu et al. [7] | 5 | 500 | Ak, Ala Idris, Buzgulu, Dimnit, Nazli | Yes, 100 images per class |
| Al-khazraji et al. [48] | 8 | 8000 | deas al-annz, kamali, halawani, thompson seedless, aswud balad, riasi, frinsi, shdah | Yes, 1000 images per class |
| Santos et al. [47] | 6 | 300 | Chardonnay, Cabernet Franc, Cabernet Sauvignon, Sauvignon Blanc, Syrah | No |
| Sozzi et al. [50] | 3 | 312 | Glera, Chardonnay, Trebbiano | No |
| Seng et al. [51] | 15 | 2078 | Merlot, Cabernet Sauvignon, Saint Macaire, Flame Seedless, Viognier, Ruby Seedless, Riesling, Muscat Hamburg, Purple Cornichon, Sultana, Sauvignon Blanc, Chardonnay | No |
| Vlah [49] | 11 | 1009 | Auxerrois, Cabernet Franc, Cabernet Sauvignon, Chardonnay, Merlot, Müller Thurgau, Pinot Noir, Riesling, Sauvignon Blanc, Syrah, Tempranillo | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





