Submitted:
13 March 2024
Posted:
14 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Group Dynamics Models
2.1.1. Group Dynamics Model with Coupling Forces
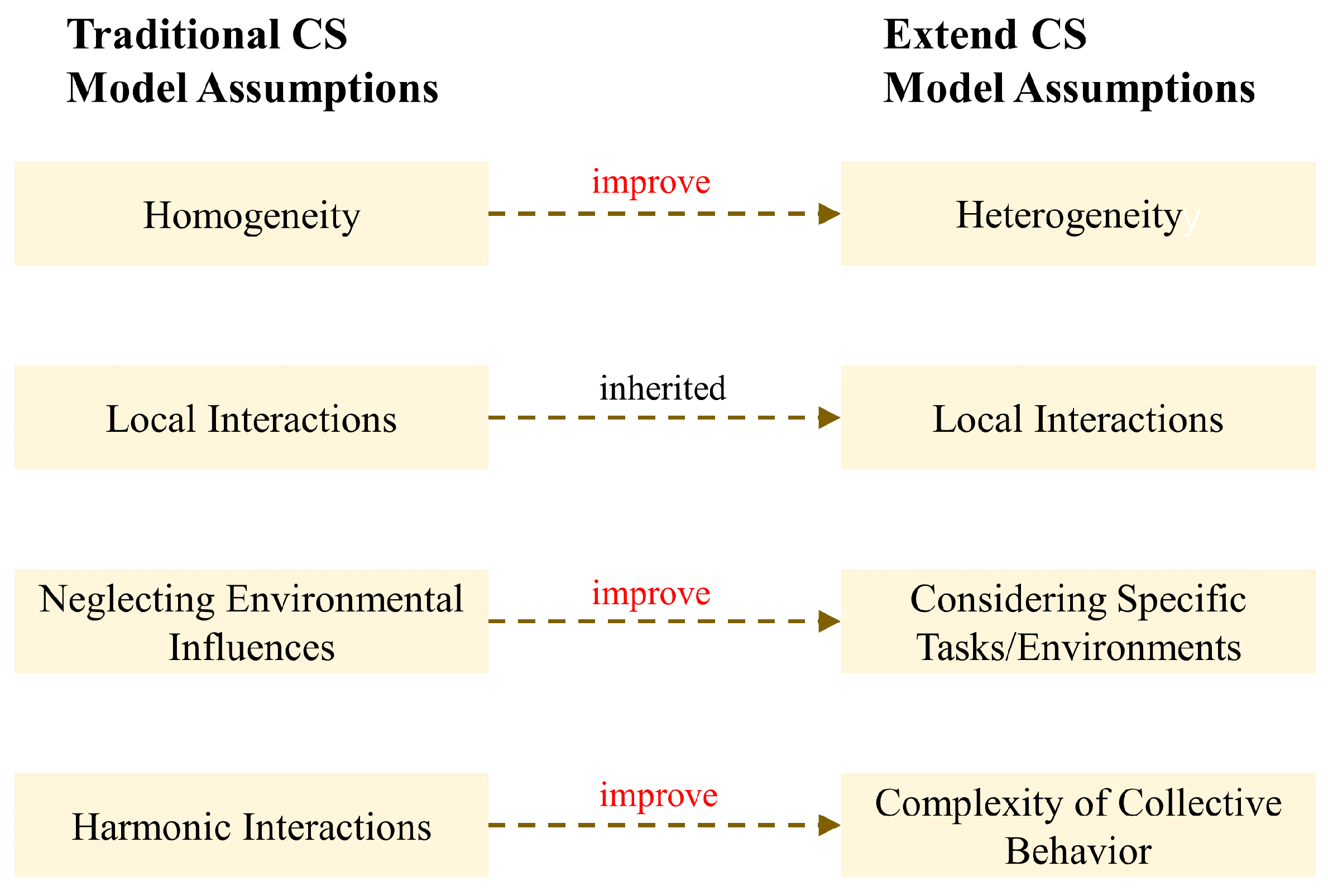
- (1)Homogeneity: The model presupposes that all individuals within the system exhibit uniform behavior, adhering to a common set of rules for updating their states. Specifically, each individual adjusts its velocity by calculating the weighted average of velocity differences with other individuals.
- (2)Local Interactions: The update of an individual’s motion state is contingent upon the states of its immediate neighbors. This mechanism is consistent with the phenomenon of information transmission through visual, auditory, or other sensory means among natural groups such as flocks of birds and schools of fish.
- (3)Neglecting Environmental Influences: The model does not directly incorporate the impact of environmental factors on the motion states of individuals.
- (4)Harmonic Interactions: The interaction force between individuals diminishes with increasing distance, with this force designed to facilitate cohesive and coordinated behavior within the group.
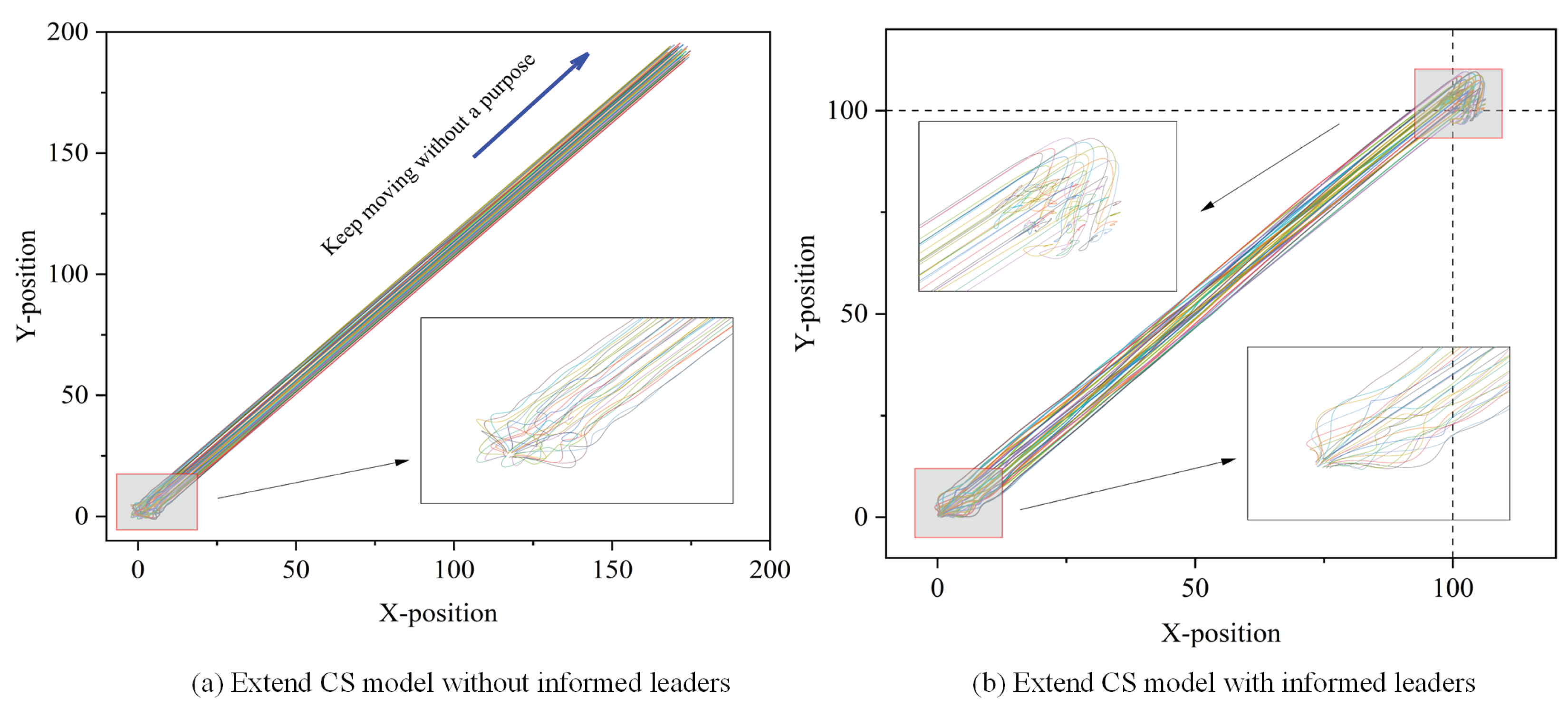
2.1.2. Biological Group Model with Informed Leaders
2.2. Individual Interaction Mode
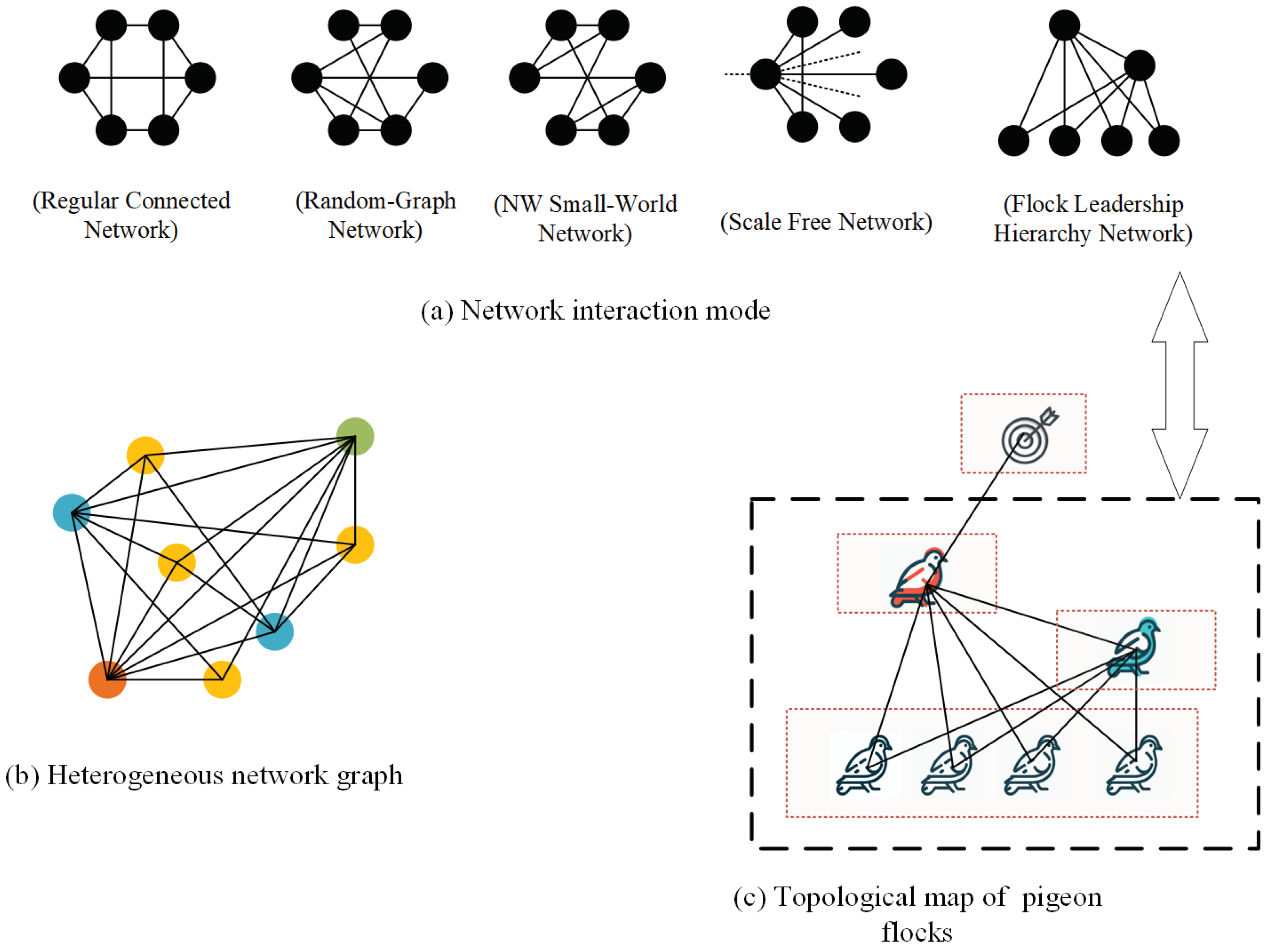
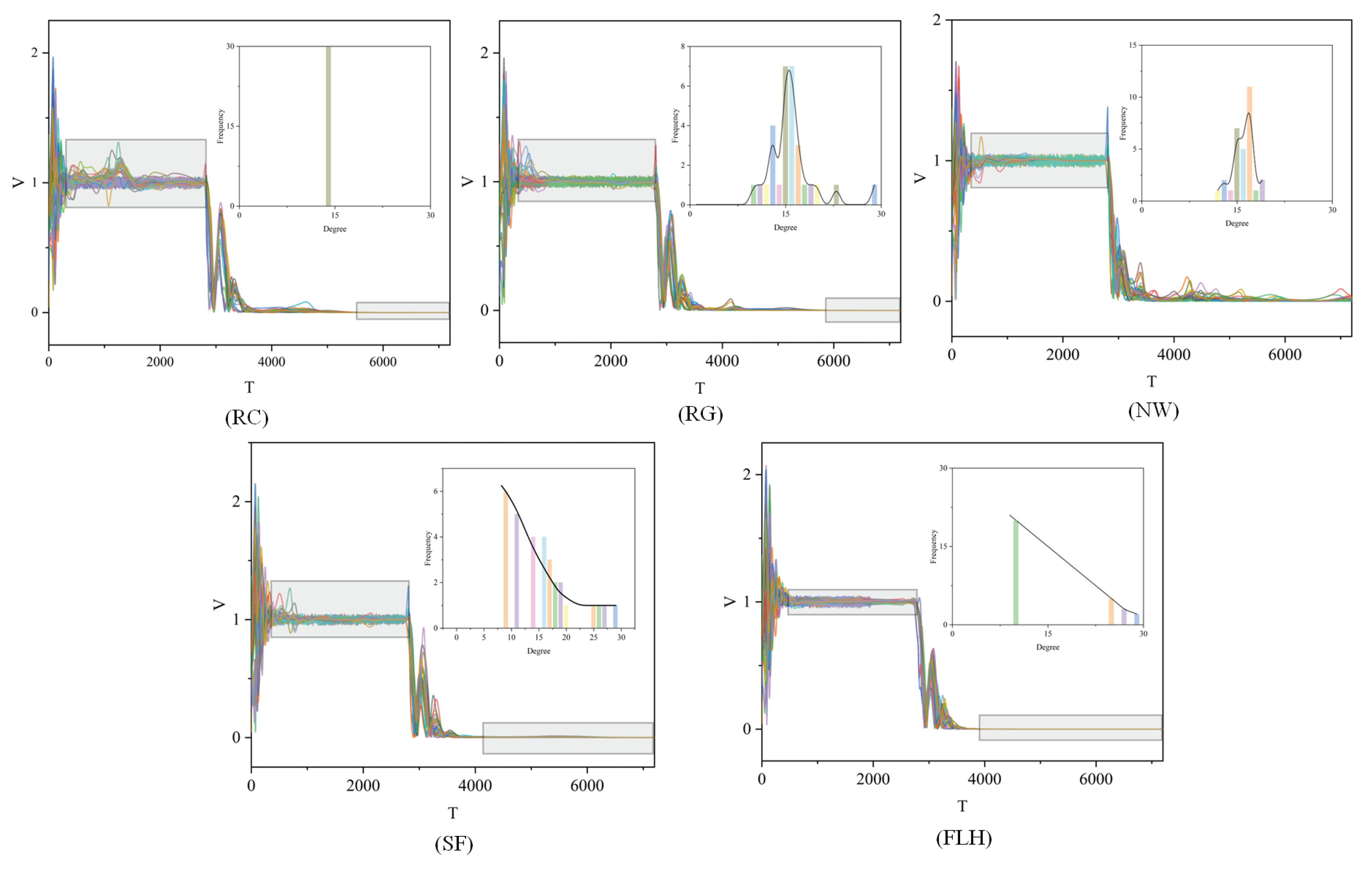
2.2.1. Regular Connected Network(RC)
2.2.2. Random-Graph Network(RG)
2.2.3. Newman-Watts-Strogatz Small-World Network (NW)
2.2.4. Scale Free Network(SF)
2.2.5. Flock Leadership Hierarchy Network(FLH)
| Algorithm 1 FLH network generation algorithm |
|
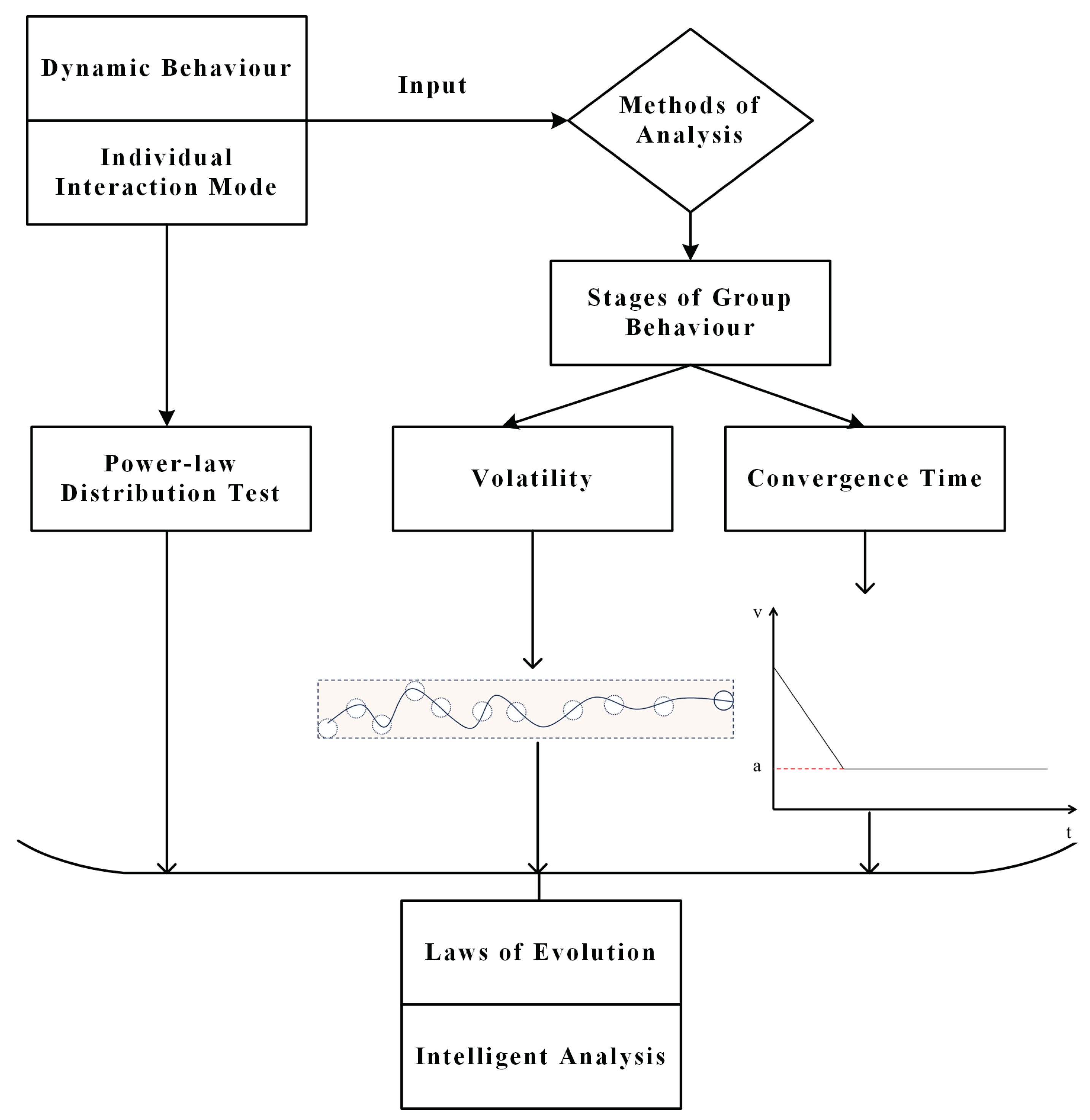
3. Model Building
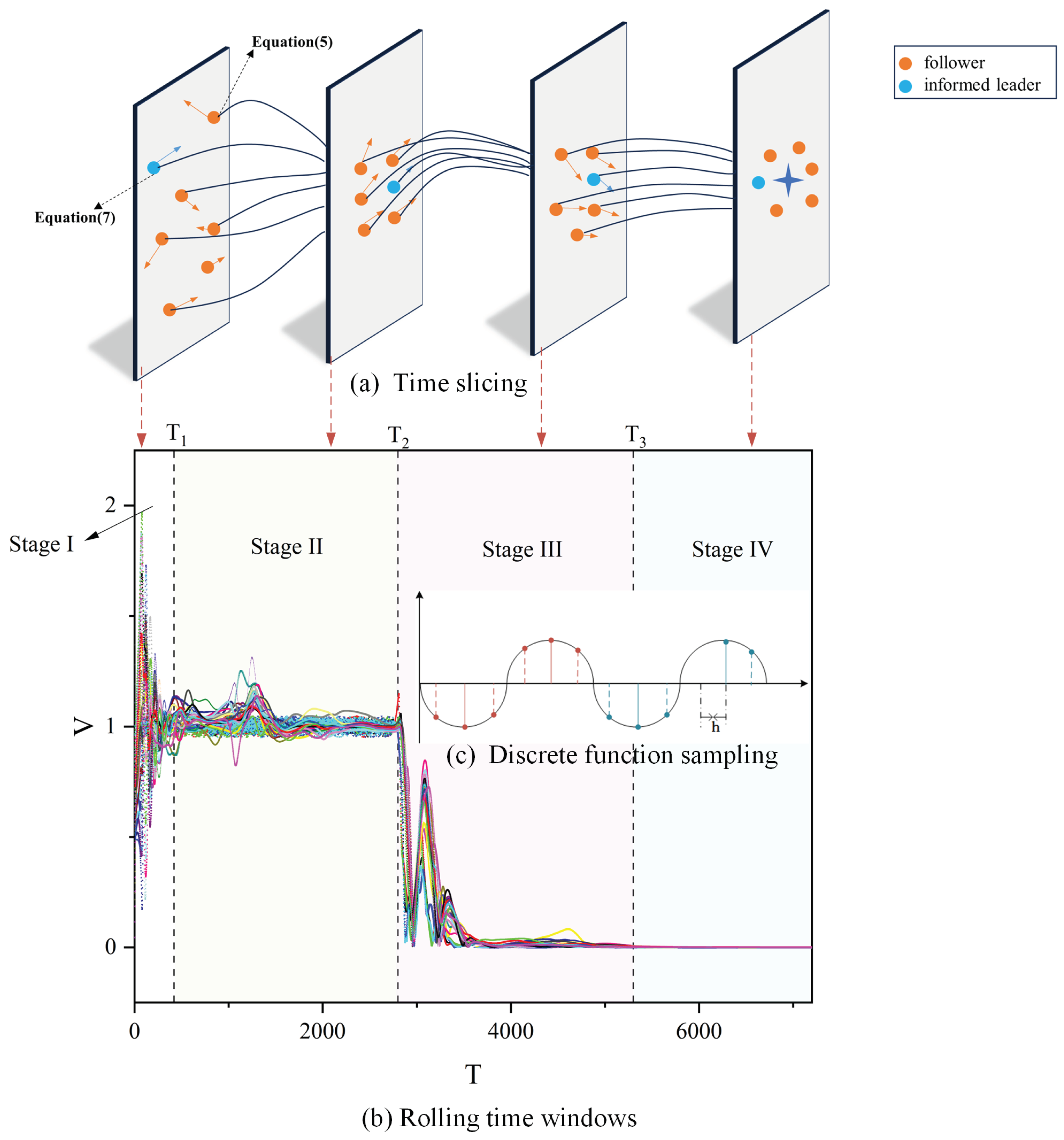
3.1. Stages of Group Behaviour and Demonstration
3.2. Metric Performance
3.2.1. Volatility
3.2.2. Convergence Time
3.3. Power-Law Distribution Test
4. Numerical Simulation
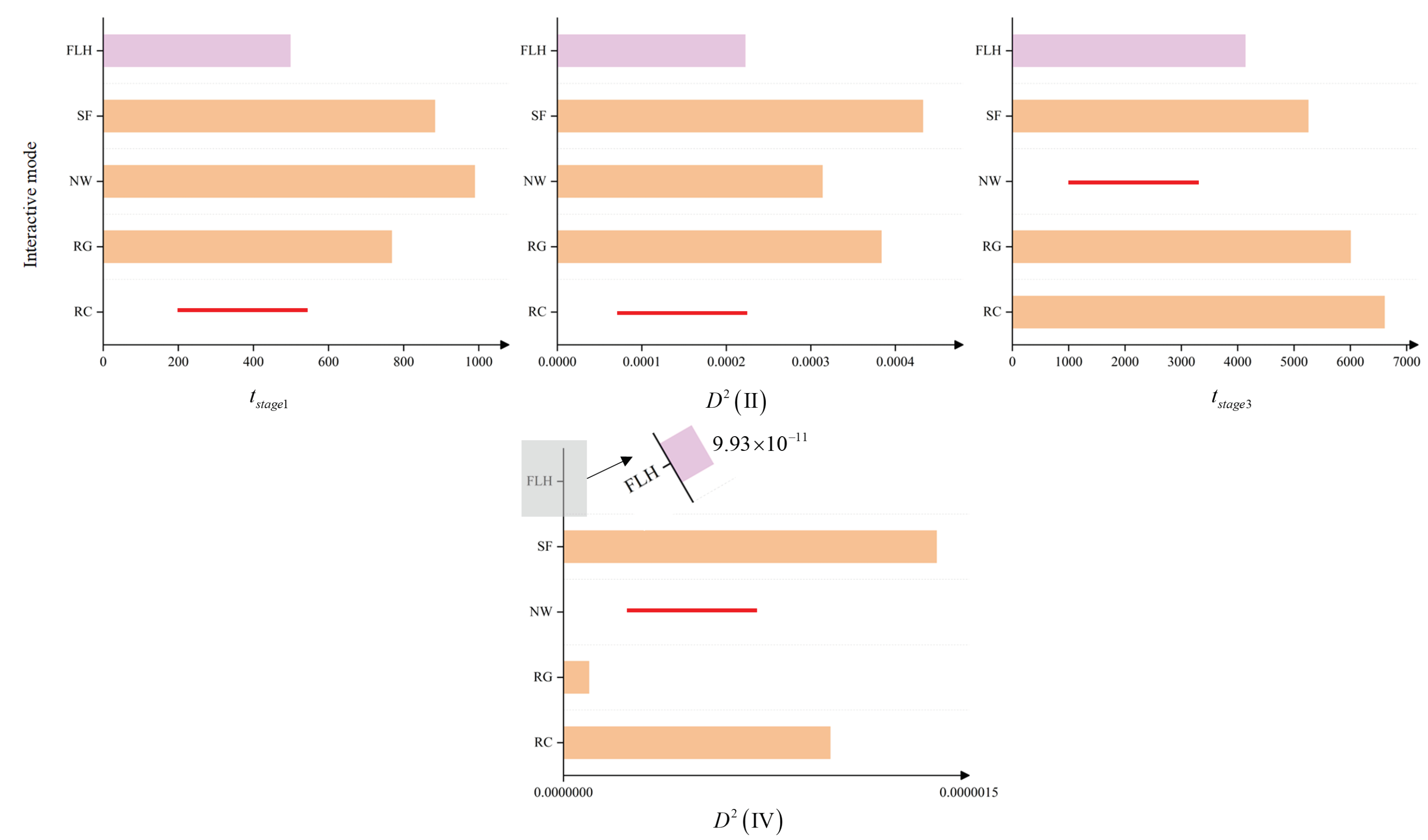
4.1. Experimental Design
4.2. Discussion
5. Conclusions
6. Future Research
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CS | Cucker-Smale model |
| RC | Regular Connected Network |
| RG | Random-Graph Network |
| NW | Newman-Watts-Strogatz Small-World Network |
| SF | Scale-Free Network |
| FLH | Flock Leadership Hierarchy Network |
| hd | high-degree nodes |
| ld | low-degree nodes |
References
- Reynolds, C.W. Flocks, Herds and Schools : A Distributed Behavior Model. Proc. SIGGRAPH ’87 1987, 25. [Google Scholar]
- Marras, S.; Killen, S.S.; Lindström, J.; Mckenzie, D.J.; Steffensen, J.F.; Domenici, P. Fish swimming in schools save energy regardless of their spatial position. Behavioral Ecology&Sociobiology 2015. [Google Scholar]
- AOKI; Ichiro. A Simulation Study on the Schooling Mechanism in Fish. Nihon-suisan-gakkai-shi 1982, 48, 1081–1088. [Google Scholar] [CrossRef]
- Hubbard, S.; Babak, P.; Sigurdsson, S.T.; Magnússon, K.G. A model of the formation of fish schools and migrations of fish. Ecological Modelling 2004, 174, 359–374. [Google Scholar] [CrossRef]
- Zhong, C.; Wengao Lou, Y.L. A Projection Pursuit Dynamic Cluster Model for Tourism Safety Early Warning and Its Implications for Sustainable Tourism. Mathematics 2023, 11, 4919. [Google Scholar] [CrossRef]
- Alan, P.; Scott, T.J. Can we identify general architectural principles that impact the collective behaviour of both human and animal systems? Philosophical Transactions of the Royal Society B Biological Sciences 2018, 373. [Google Scholar]
- Huang, S.; Brangwynne, C.P.; Parker, K.K.; Ingber, D.E. Symmetry-breaking in mammalian cell cohort migration during tissue pattern formation: Role of random-walk persistence. Cell Motility & the Cytoskeleton 2010, 61, 201–213. [Google Scholar]
- Yaxley, K.J.; Joiner, K.F.; Abbass, H.A. Drone approach parameters leading to lower stress sheep flocking and movement: sky shepherding. Scientific Reports 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Hakim, V.; Silberzan, P. Collective cell migration: a physics perspective. Reports on progress in physics. Physical Society (Great Britain) 2017, 80, 076601. [Google Scholar] [CrossRef]
- Belmonte, J.M.; Thomas, G.L.; Brunnet, L.G.; Almeida, R.M.C.D.; Chate, H. Self-propelled particle model for cell-sorting phenomena. Physical Review Letters 2008, 100, 248702. [Google Scholar] [CrossRef]
- R?Rth, P. Collective guidance of collective cell migration. Trends in Cell Biology 2007, 17, 575–579. [Google Scholar] [CrossRef]
- Wei, X.; Liu, J.C.; Bi, S. Uncertainty quantification and propagation of crowd behaviour effects on pedestrian-induced vibrations of footbridges. Mechanical Systems and Signal Processing 2022. [Google Scholar] [CrossRef]
- Smith, J.; Gavrilets, S.; Mulder, M.; Hooper, P.; Mouden, C.; Nettle, D.; Hauert, C.; Hill, K.; Perry, S.; Pusey, A. Leadership in Mammalian Societies: Emergence, Distribution, Power, and Payoff. Trends in Ecology & Evolution 2015, 31, 54–66. [Google Scholar]
- Dell'Ariccia, G.; Dell'Omo, G.; Wolfer, D.P.; Lipp, H.P. Flock flying improves pigeons’ homing: GPS track analysis of individual flyers versus small groups. Animal Behaviour London Bailliere Tindall Then Academic Press 2008. [Google Scholar]
- Nagy, M.; Akos, Z.; Biro, D.; Vicsek, T. Hierarchical group dynamics in pigeon flocks. Nature 2010, 464, 890. [Google Scholar] [CrossRef] [PubMed]
- Nagy, M.; Vasarhelyi, G.; Pettit, B.; Roberts-Mariani, I.; Vicsek, T.; Biro, D. Context-dependent hierarchies in pigeons. Proceedings of the National Academy of Sciences of the United States of America 2013, 110. [Google Scholar] [CrossRef] [PubMed]
- Flack, A.; Ákos, Z.; Nagy, M.; Vicsek, T.; Biro, D. Robustness of flight leadership relations in pigeons. Animal Behaviour 2013. [Google Scholar] [CrossRef]
- Watts, I.; Nagy, M.; Theresa, B.D.P.; Biro, D. Misinformed leaders lose influence over pigeon flocks. Biol Lett 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Garland, J.; Berdahl, A.M.; Sun, J.; Bollt, E.M. Anatomy of leadership in collective behaviour. Chaos 2018, 7, 5308. [Google Scholar]
- Strandburg-Peshkin, A.; Twomey, C.R.; Bode, N.W.F.; Kao, A.B.; Katz, Y.; Ioannou, C.C.; Rosenthal, S.B.; Torney, C.J.; Wu, H.S.; Levin, S.A.a. Visual sensory networks and effective information transfer in animal groups. Current biology: CB 2013, 23, 709–11. [Google Scholar] [CrossRef]
- Stephan, G.; Reebs. Can a minority of informed leaders determine the foraging movements of a fish shoal? Animal Behaviour 2000, 59, 403–409. [Google Scholar]
- Flocks, R.C.W. A distributed behavioral model. ACM SIGGRAPH Computer Graphics 1987, 21, 25–34. [Google Scholar]
- Vicsek, T.; Czirók, A.; Ben-Jacob, E.; Cohen, I.; Shochet, O. Novel Type of Phase Transition in a System of Self-Driven Particles. Physical Review Letters 1995, 75. [Google Scholar] [CrossRef]
- Jadbabaie, A.; Lin, J.; Morse, A.S. Coordination of groups of mobile autonomous agents using nearest neighbor rules. Proceedings of the 41st IEEE Conference on Decision and Control, 2002. 2002, 3, 2953–2958. [Google Scholar]
- Cucker F, S.S. Emergent behavior in flocks. IEEE Transactions on Automatic Control 2007, 52, 852–862. [Google Scholar] [CrossRef]
- Tunstrøm, K. Determining interaction rules in animal swarms. Behavioral Ecology 2010, 21, 1106–1111. [Google Scholar]
- Lukeman, R.; Li, Y.X.; Edelstein-Keshet, L. Inferring individual rules from collective behavior. Proceedings of the National Academy of Sciences 2010, 107, 12576–12580. [Google Scholar] [CrossRef]
- Zhdankin, V.; Sprott, J.C. Simple predator-prey swarming model. Physical Review E Statistical Nonlinear & Soft Matter Physics 2010, 82, 056209. [Google Scholar]
- Couzin, I.; Krause, J.; Franks, N.; Levin, S. Effective leadership and decision-making in animal groups on the move. Nature 2005, 433, 513–516. [Google Scholar] [CrossRef]
- Guo, H.; Han, J.; Zhang, G. Hopf Bifurcation and Control for the Bioeconomic Predator–Prey Model with Square Root Functional Response and Nonlinear Prey Harvesting. Mathematics 2023, 11, 4958. [Google Scholar] [CrossRef]
- Cucker F, S.S. On the mathematics of emergence. Japanese Journal of Mathematics 2007, 2, 197–227. [Google Scholar] [CrossRef]
- Ha, S.; Liu, J. A simple proof of the Cucker-Smale flocking dynamics and mean-field limit. Communications in Mathematical Sciences 2009, 7, 297–325. [Google Scholar] [CrossRef]
- Park, J.; Kim, H.; Ha, S. Cucker-Smale Flocking With Inter-Particle Bonding Forces. IEEE Transactions on Automatic Control 2010, 55, 2617–2623. [Google Scholar] [CrossRef]
- Liang, J.; Qi, M.; Gu, K.; Liang, Y.; Zhang, Z.; Duan, X.J. The structure inference of flocking systems based on the trajectories. Chaos 2022, 32 10, 101103. [Google Scholar] [CrossRef]
- Chen, G.; Lou, Y.; Wang, L. A Comparative Study on Controllability Robustness of Complex Networks. IEEE Transactions on Circuits and Systems II: Express Briefs 2019, 66, 828–832. [Google Scholar] [CrossRef]
- Bürgi, H.B. Crystal structures. Acta crystallographica Section B, Structural science, crystal engineering and materials 2022, 78 Pt 3, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Huang, X.; Scarpa, F.; Wen, G.; Chen, Y.; Zhang, C. In-plane crashworthiness of bio-inspired hierarchical honeycombs. Composite Structures 2018, 192, 516–527. [Google Scholar] [CrossRef]
- Lee, S.; Cha, Y.; Han, S.; taek Hyun, C. Application of Association Rule Mining and Social Network Analysis for Understanding Causality of Construction Defects. Sustainability 2019. [Google Scholar] [CrossRef]
- Dragicevic, A.Z.; Gurtoo, A. Stochastic control of ecological networks. Journal of Mathematical Biology 2022, 85. [Google Scholar] [CrossRef] [PubMed]
- Brinkley, C. The Small World of the Alternative Food Network. Sustainability 2018, 10, 2921. [Google Scholar] [CrossRef]
- Glazier, V.E.; Krysan, D.J. Transcription factor network efficiency in the regulation of Candida albicans biofilms: it is a small world. Current Genetics 2018, 64, 883–888. [Google Scholar] [CrossRef]
- Aparicio, S.; Villazón-Terrazas, J.; Álvarez, G. A Model for Scale-Free Networks: Application to Twitter. Entropy 2015, 17, 5848–5867. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lim, W. Cluster burst synchronization in a scale-free network of inhibitory bursting neurons. Cognitive Neurodynamics 2018, 14, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Watts, I.; Pettit, B.; Nagy, M.; de Perera, T.B.; Biro, D. Lack of experience-based stratification in homing pigeon leadership hierarchies. Royal Society Open Science 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Huang, J. SpreadRank: A Novel Approach for Identifying Influential Spreaders in Complex Networks. Entropy 2023, 25. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Small, M.; Pérez-Barbería, F.J. Uncovering interaction patterns of multi-agent collective motion via complex network analysis. In Proceedings of the 2014 IEEE International Symposium on Circuits and Systems (ISCAS); 2014; pp. 2213–2216. [Google Scholar]
- Zhang, Y.; Chen, F.; Rohe, K. Correction to: Social Media Public Opinion as Flocks in a Murmuration: Conceptualizing and Measuring Opinion Expression on Social Media. J. Comput. Mediat. Commun. 2022, 27. [Google Scholar]
- Wang, X.; Lu, Y.; Shi, C.; Wang, R.; Cui, P.; Mou, S. Dynamic Heterogeneous Information Network Embedding With Meta-Path Based Proximity. IEEE Transactions on Knowledge and Data Engineering 2022, 34, 1117–1132. [Google Scholar] [CrossRef]
- Ouellette, N. A physics perspective on collective animal behavior. Physical Biology 2022, 19. [Google Scholar] [CrossRef]
- Jeon, J.G. Valuation of Commodity-Linked Bond with Stochastic Convenience Yield, Stochastic Volatility, and Credit Risk in an Intensity-Based Model. Mathematics 2023, 11, 4969. [Google Scholar] [CrossRef]
- Blythe, D.A.J.; Nikulin, V.V.; Müller, K.R. Robust Statistical Detection of Power-Law Cross-Correlation. Scientific Reports 2016, 6. [Google Scholar] [CrossRef]

| Symbol | Meaning | Value | Symbol | Meaning | Value |
|---|---|---|---|---|---|
| N | Number of individuals | 30 | Corrdination parameter | 0.2/0.5/1 | |
| Coupling strength | 10 | Collision coefficient | 10 | ||
| Anti-collision upper boundary | 100 | Anti-collision lower boundary | 3 | ||
| Velocity upper bound | 5 | Acceleration upper bound | 1 | ||
| R | Maximum radius of communication | 50 | t | Time | 360 |
| D | Target position | (100,100) | Initial position | near (0,0) | |
| Number of leaders | 5 | h | Discretized step size | 0.05 | |
| Velocity threshold | c | Acceptable range of volatility | 0.2 / | ||
| Volatility threshold |
| Network Topology | Power Law Distribution | ||||
|---|---|---|---|---|---|
| RC | 1,370 | 5,271 | —— | ||
| RG | 871 | 5,720 | —— | ||
| NW | 640 | —— | —— | —— | |
| SF | 802 | 3,821 | Obey | ||
| FLH | 467 | 3,672 | Obey |
| Network Topology | ||||
|---|---|---|---|---|
| RC | 2,154 | 7,061 | ||
| RG | 726 | 5,160 | ||
| NW | 881 | —— | —— | |
| SF | 292 | 4,261 | ||
| FLH | 253 | 4,157 |
| Network Topology | ||||
|---|---|---|---|---|
| RC | 1,387 | 6,206 | ||
| RG | 226 | 6,250 | ||
| NW | 241 | —— | —— | |
| SF | 230 | 4,541 | ||
| FLH | 177 | 3,999 |
| Network Topology | ||||
|---|---|---|---|---|
| RC | —— | —— | 6,613 | |
| RG | 769 | 6,010 | ||
| NW | 990 | —— | —— | |
| SF | 884 | 5,258 | ||
| FLH | 499 | 4,140 |
| Network Topology | ||||
|---|---|---|---|---|
| RC | 1,671 | 5,036 | ||
| RG | 1,098 | 4,084 | ||
| NW | 986 | —— | —— | |
| SF | 861 | 3,984 | ||
| FLH | 489 | 3,658 |
| Network Topology | ||||
|---|---|---|---|---|
| RC | 1,532 | 4,494 | ||
| RG | 1,674 | 5,216 | ||
| NW | —— | —— | 5,284 | |
| SF | 1,144 | 4,819 | ||
| FLH | 857 | 4,574 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





