Submitted:
13 March 2024
Posted:
14 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. HHpred Search
2.2. File Parsing
2.3. Molecular Similarity
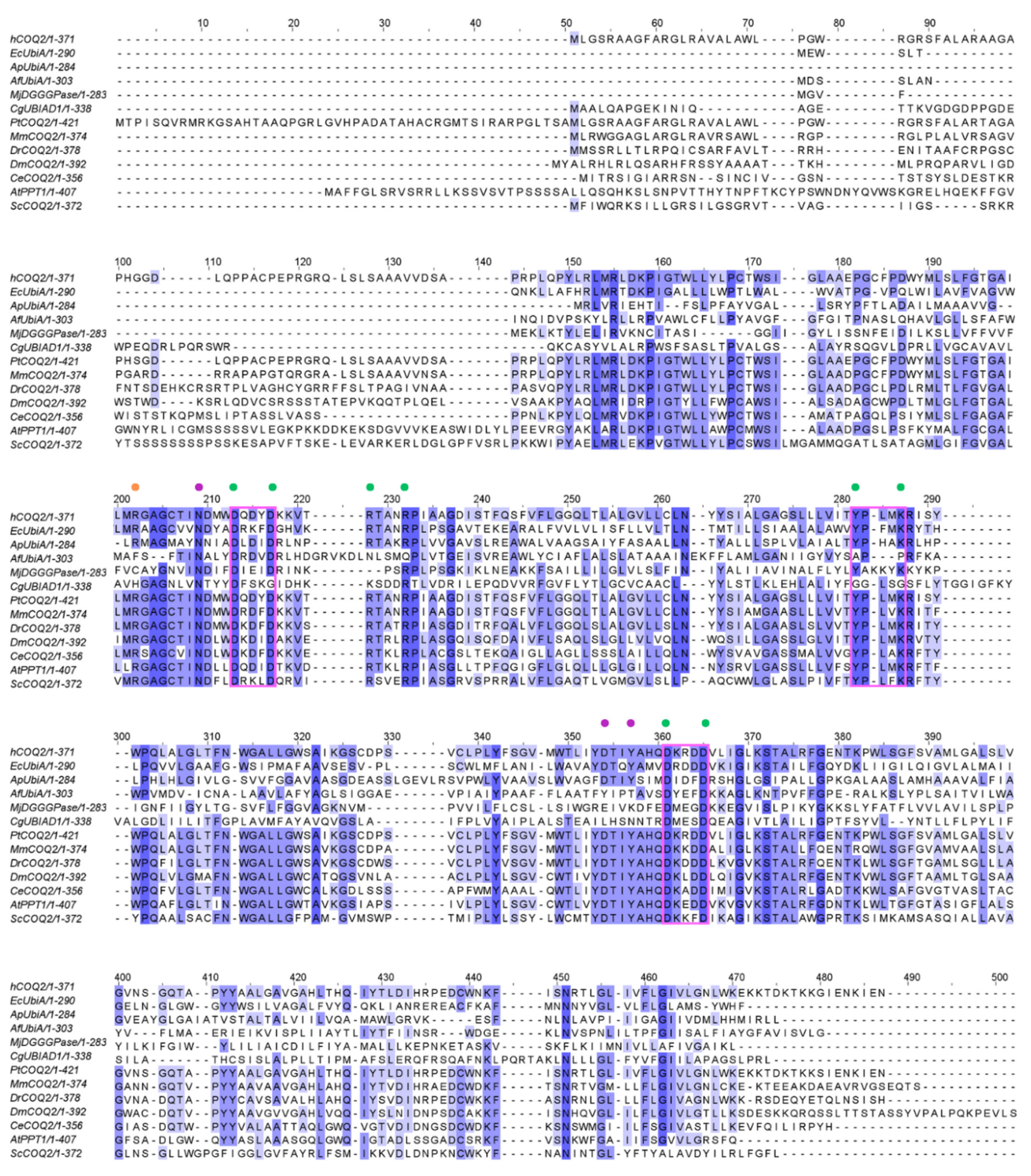
2.4. Multiple Sequence Alignment (MSA)
2.6. AlphaFold2 Prediction Model
2.7. Variant Structure Prediction
2.8. Structure Superposition
3. Results
3.1. Homology Search
3.2. Ligand Similarity
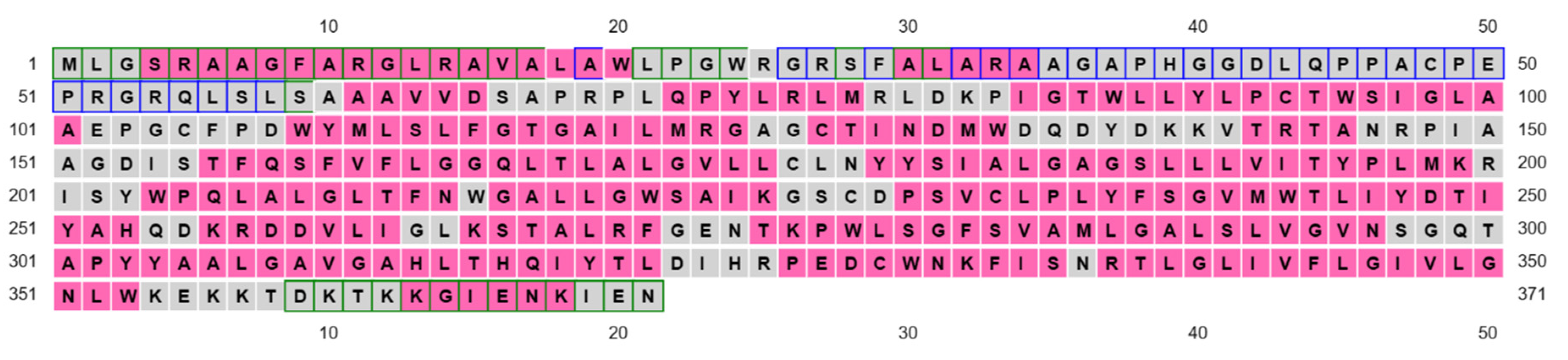
3.3. Residues Potentially Involved in Substrate Binding
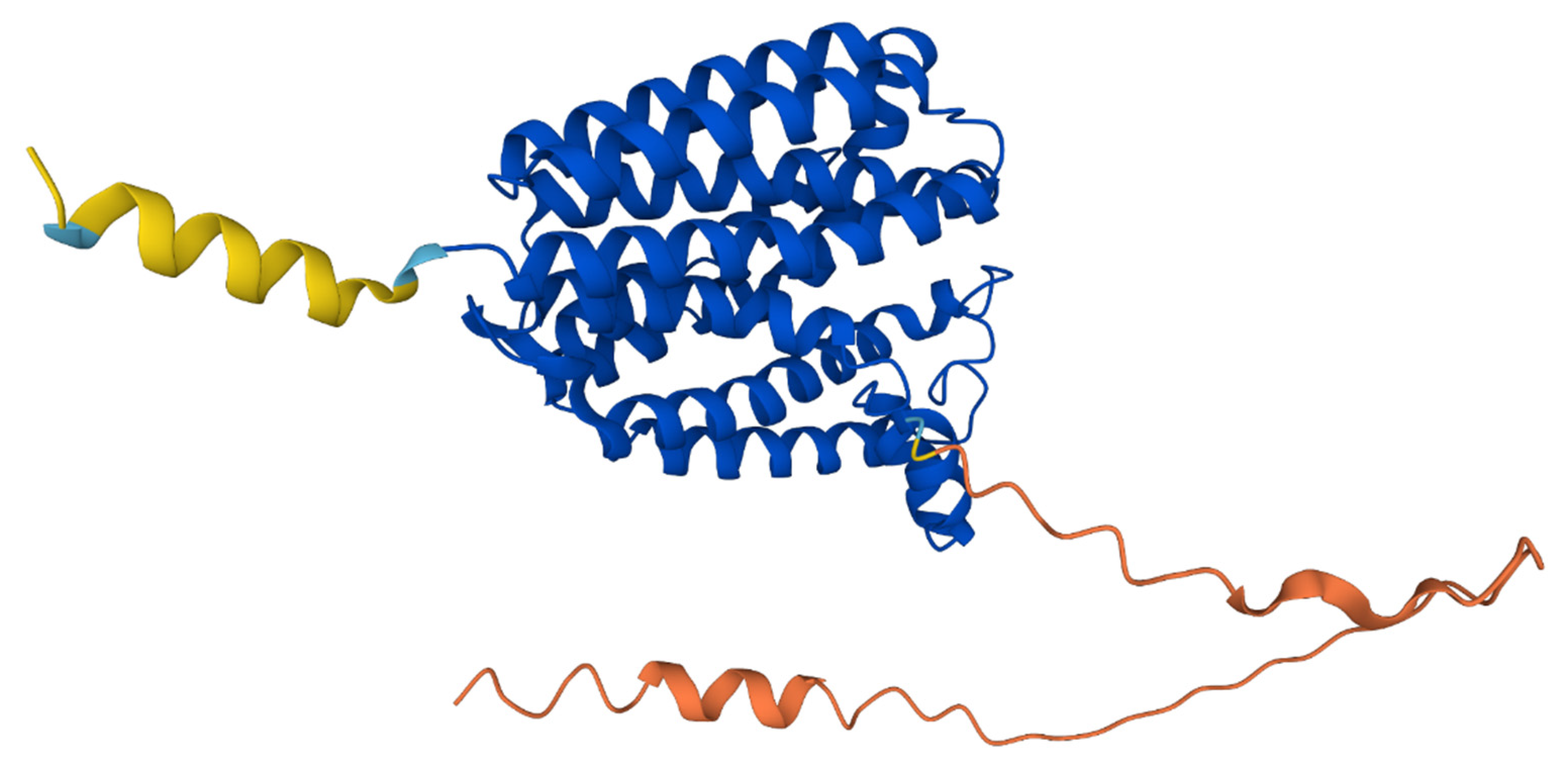
3.4. COQ2 Structural Model
3.5. Structural Superposition of Homologous Structures to COQ2
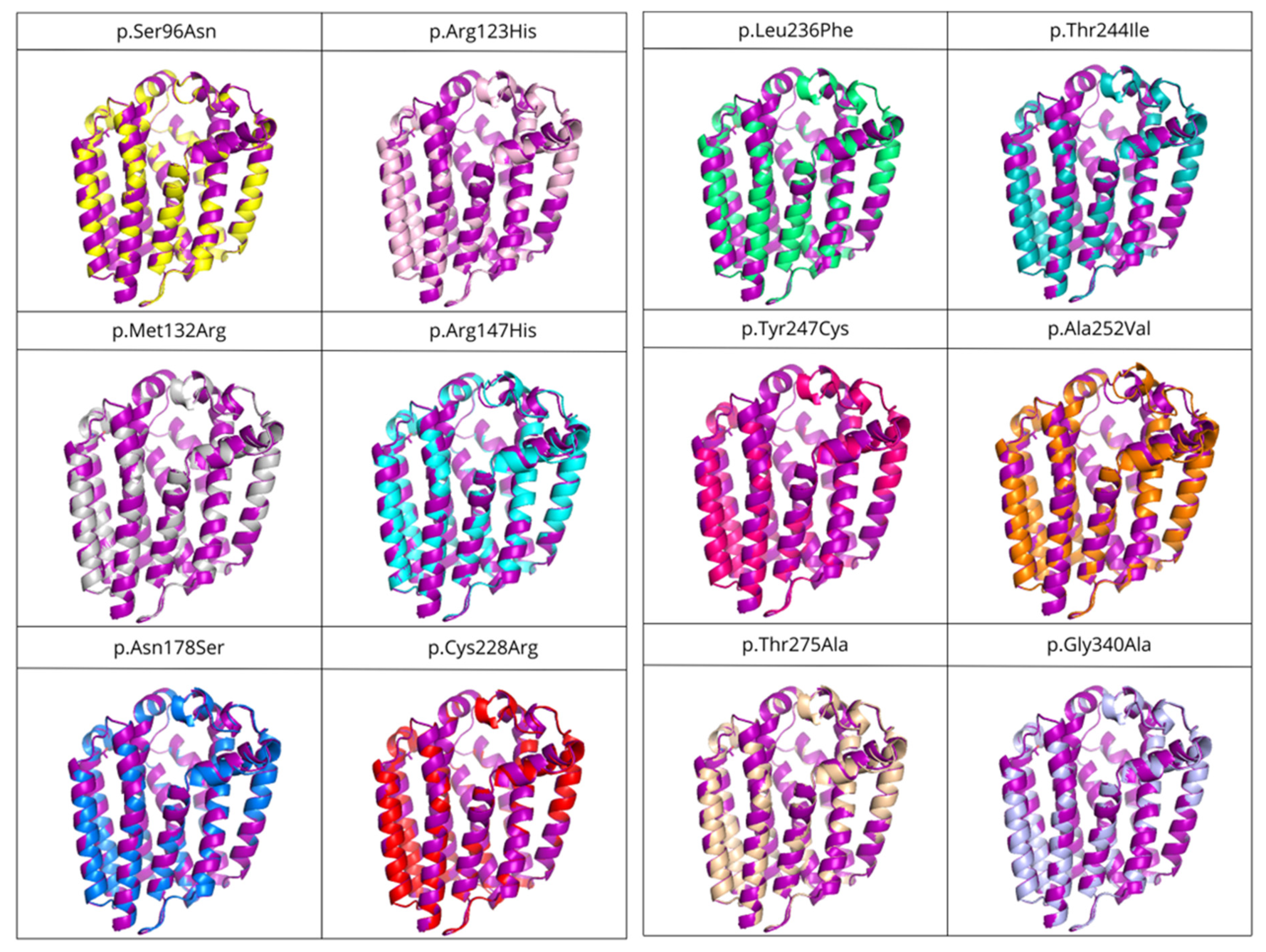
3.6. Effects of Mutations in COQ2 Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alcazar-Fabra, M.; Rodriguez-Sanchez, F.; Trevisson, E.; Brea-Calvo, G. Primary Coenzyme Q deficiencies: A literature review and online platform of clinical features to uncover genotype-phenotype correlations. Free Radic Biol Med 2021, 167, 141-180. [CrossRef]
- Hernandez-Camacho, J.D.; Garcia-Corzo, L.; Fernandez-Ayala, D.J.M.; Navas, P.; Lopez-Lluch, G. Coenzyme Q at the Hinge of Health and Metabolic Diseases. Antioxidants (Basel) 2021, 10. [CrossRef]
- Stefely, J.A.; Pagliarini, D.J. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem Sci 2017, 42, 824-843. [CrossRef]
- Wang, S.; Jain, A.; Novales, N.A.; Nashner, A.N.; Tran, F.; Clarke, C.F. Predicting and Understanding the Pathology of Single Nucleotide Variants in Human COQ Genes. Antioxidants (Basel) 2022, 11. [CrossRef]
- Aussel, L.; Pierrel, F.; Loiseau, L.; Lombard, M.; Fontecave, M.; Barras, F. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 2014, 1837, 1004-1011. [CrossRef]
- Kawamukai, M. Biosynthesis of coenzyme Q in eukaryotes. Biosci Biotechnol Biochem 2016, 80, 23-33. [CrossRef]
- Crane, F.L. Discovery of ubiquinone (coenzyme Q) and an overview of function. Mitochondrion 2007, 7 Suppl, S2-7. [CrossRef]
- Barroso, M.P.; Gomez-Diaz, C.; Lopez-Lluch, G.; Malagon, M.M.; Crane, F.L.; Navas, P. Ascorbate and alpha-tocopherol prevent apoptosis induced by serum removal independent of Bcl-2. Arch Biochem Biophys 1997, 343, 243-248. [CrossRef]
- Barroso, M.P.; Gomez-Diaz, C.; Villalba, J.M.; Buron, M.I.; Lopez-Lluch, G.; Navas, P. Plasma membrane ubiquinone controls ceramide production and prevents cell death induced by serum withdrawal. J Bioenerg Biomembr 1997, 29, 259-267. [CrossRef]
- Hadian, K. Ferroptosis Suppressor Protein 1 (FSP1) and Coenzyme Q(10) Cooperatively Suppress Ferroptosis. Biochemistry 2020, 59, 637-638. [CrossRef]
- Kagan, V.E.; Straub, A.C.; Tyurina, Y.Y.; Kapralov, A.A.; Hall, R.; Wenzel, S.E.; Mallampalli, R.K.; Bayir, H. Vitamin E/Coenzyme Q-Dependent "Free Radical Reductases": Redox Regulators in Ferroptosis. Antioxid Redox Signal 2023. [CrossRef]
- Fernandez-Del-Rio, L.; Clarke, C.F. Coenzyme Q Biosynthesis: An Update on the Origins of the Benzenoid Ring and Discovery of New Ring Precursors. Metabolites 2021, 11. [CrossRef]
- Crane, F.L.; Hatefi, Y.; Lester, R.L.; Widmer, C. Isolation of a quinone from beef heart mitochondria. Biochim Biophys Acta 1957, 25, 220-221. [CrossRef]
- Morton, R.A. Ubiquinone. Nature 1958, 182, 1764-1767. [CrossRef]
- Awad, A.M.; Bradley, M.C.; Fernandez-Del-Rio, L.; Nag, A.; Tsui, H.S.; Clarke, C.F. Coenzyme Q(10) deficiencies: Pathways in yeast and humans. Essays Biochem 2018, 62, 361-376. [CrossRef]
- Desbats, M.A.; Lunardi, G.; Doimo, M.; Trevisson, E.; Salviati, L. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency. J Inherit Metab Dis 2015, 38, 145-156. [CrossRef]
- Ren, S.; de Kok, N.A.W.; Gu, Y.; Yan, W.; Sun, Q.; Chen, Y.; He, J.; Tian, L.; Andringa, R.L.H.; Zhu, X.; et al. Structural and Functional Insights into an Archaeal Lipid Synthase. Cell Rep 2020, 33, 108294. [CrossRef]
- Pelosi, L.; Morbiato, L.; Burgardt, A.; Tonello, F.; Bartlett, A.K.; Guerra, R.M.; Ferizhendi, K.K.; Desbats, M.A.; Rascalou, B.; Marchi, M.; et al. COQ4 is required for the oxidative decarboxylation of the C1 carbon of coenzyme Q in eukaryotic cells. Molecular Cell 2024. [CrossRef]
- Tran, U.C.; Clarke, C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 2007, 7 Suppl, S62-71. [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 2022, 50, D439-D444. [CrossRef]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023, 51, D523-D531. [CrossRef]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Soding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 2020, 72, e108. [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 2018, 430, 2237-2243. [CrossRef]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 2005, 33, W244-248. [CrossRef]
- Sydow, D.; Morger, A.; Driller, M.; Volkamer, A. TeachOpenCADD: A teaching platform for computer-aided drug design using open source packages and data. J Cheminform 2019, 11, 29. [CrossRef]
- Kuwahara, H.; Gao, X. Analysis of the effects of related fingerprints on molecular similarity using an eigenvalue entropy approach. J Cheminform 2021, 13, 27. [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J Mol Biol 1990, 215, 403-410. [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 2010, 27, 221-224. [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004, 5, 113. [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189-1191. [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 1999, 292, 195-202. [CrossRef]
- Jones, D.T.; Cozzetto, D. DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015, 31, 857-863. [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res 2019, 47, W402-W407. [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 2021, 30, 70-82. [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat Methods 2022, 19, 679-682. [CrossRef]
- Align. In PyMolWiki. Available online: (accessed on September 2023).
- Super. In PyMolWiki. Available online: (accessed on September 2023).
- Cheng, W.; Li, W. Structural insights into ubiquinone biosynthesis in membranes. Science 2014, 343, 878-881. [CrossRef]
- Bertoline, L.M.F.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An overview of protein structure prediction. Front Bioinform 2023, 3, 1120370. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583-589. [CrossRef]
- Chen, H.; Qi, X.; Faulkner, R.A.; Schumacher, M.M.; Donnelly, L.M.; DeBose-Boyd, R.A.; Li, X. Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain. Nat Commun 2022, 13, 4273. [CrossRef]
- Huang, H.; Levin, E.J.; Liu, S.; Bai, Y.; Lockless, S.W.; Zhou, M. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol 2014, 12, e1001911. [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat Rev Mol Cell Biol 2019, 20, 681-697. [CrossRef]
- Li, W. Bringing Bioactive Compounds into Membranes: The UbiA Superfamily of Intramembrane Aromatic Prenyltransferases. Trends Biochem Sci 2016, 41, 356-370. [CrossRef]
- Mugoni, V.; Postel, R.; Catanzaro, V.; De Luca, E.; Turco, E.; Digilio, G.; Silengo, L.; Murphy, M.P.; Medana, C.; Stainier, D.Y.; et al. Ubiad1 is an antioxidant enzyme that regulates eNOS activity by CoQ10 synthesis. Cell 2013, 152, 504-518. [CrossRef]
- Desbats, M.A.; Morbidoni, V.; Silic-Benussi, M.; Doimo, M.; Ciminale, V.; Cassina, M.; Sacconi, S.; Hirano, M.; Basso, G.; Pierrel, F.; et al. The COQ2 genotype predicts the severity of coenzyme Q10 deficiency. Hum Mol Genet 2016, 25, 4256-4265. [CrossRef]
- Yeh, V.; Goode, A.; Bonev, B.B. Membrane Protein Structure Determination and Characterisation by Solution and Solid-State NMR. Biology (Basel) 2020, 9. [CrossRef]
- Yang, Y.; Ke, N.; Liu, S.; Li, W. Methods for Structural and Functional Analyses of Intramembrane Prenyltransferases in the UbiA Superfamily. Methods Enzymol 2017, 584, 309-347. [CrossRef]
- Melzer, M.; Heide, L. Characterization of polyprenyldiphosphate: 4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim Biophys Acta 1994, 1212, 93-102. [CrossRef]
- Herebian, D.; Seibt, A.; Smits, S.H.J.; Bunning, G.; Freyer, C.; Prokisch, H.; Karall, D.; Wredenberg, A.; Wedell, A.; Lopez, L.C.; et al. Detection of 6-demethoxyubiquinone in CoQ(10) deficiency disorders: Insights into enzyme interactions and identification of potential therapeutics. Mol Genet Metab 2017, 121, 216-223. [CrossRef]
- Diomedi-Camassei, F.; Di Giandomenico, S.; Santorelli, F.M.; Caridi, G.; Piemonte, F.; Montini, G.; Ghiggeri, G.M.; Murer, L.; Barisoni, L.; Pastore, A.; et al. COQ2 nephropathy: A newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol 2007, 18, 2773-2780. [CrossRef]
- Jakobs, B.S.; van den Heuvel, L.P.; Smeets, R.J.; de Vries, M.C.; Hien, S.; Schaible, T.; Smeitink, J.A.; Wevers, R.A.; Wortmann, S.B.; Rodenburg, R.J. A novel mutation in COQ2 leading to fatal infantile multisystem disease. J Neurol Sci 2013, 326, 24-28. [CrossRef]
- Santos-Ocana, C.; Cascajo, M.V.; Alcazar-Fabra, M.; Staiano, C.; Lopez-Lluch, G.; Brea-Calvo, G.; Navas, P. Cellular Models for Primary CoQ Deficiency Pathogenesis Study. Int J Mol Sci 2021, 22. [CrossRef]
- Perrakis, A.; Sixma, T.K. AI revolutions in biology: The joys and perils of AlphaFold. EMBO Rep 2021, 22, e54046. [CrossRef]
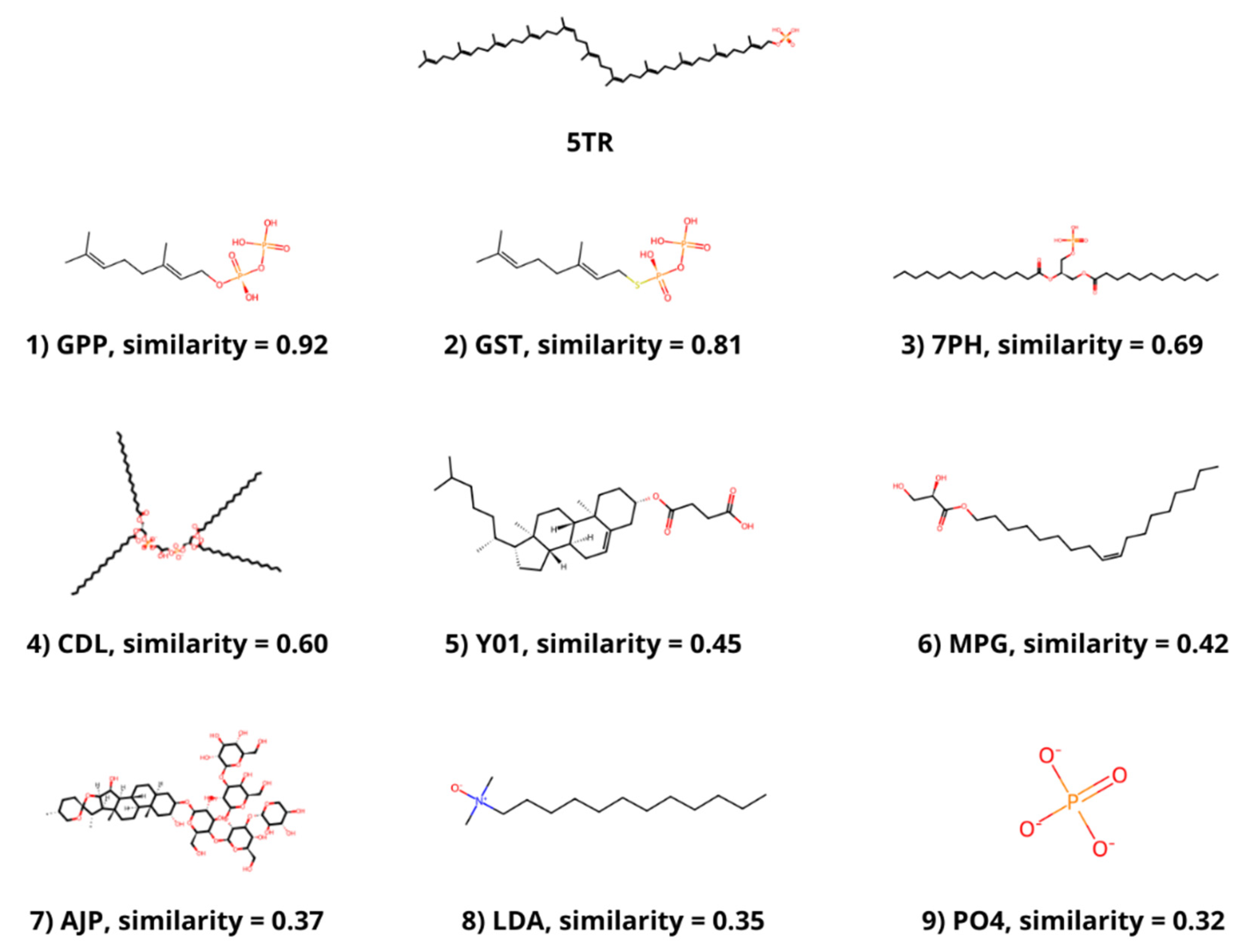

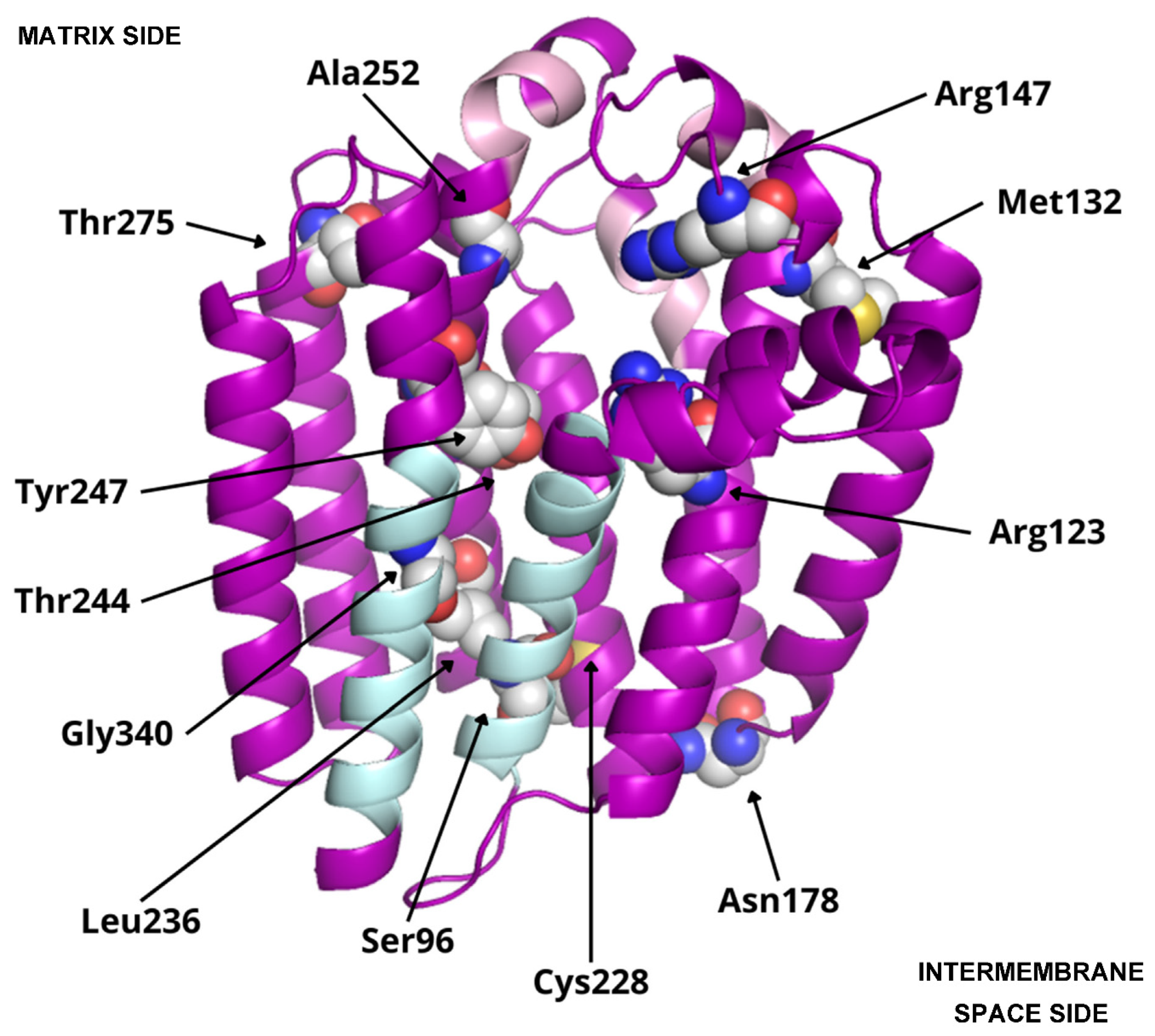
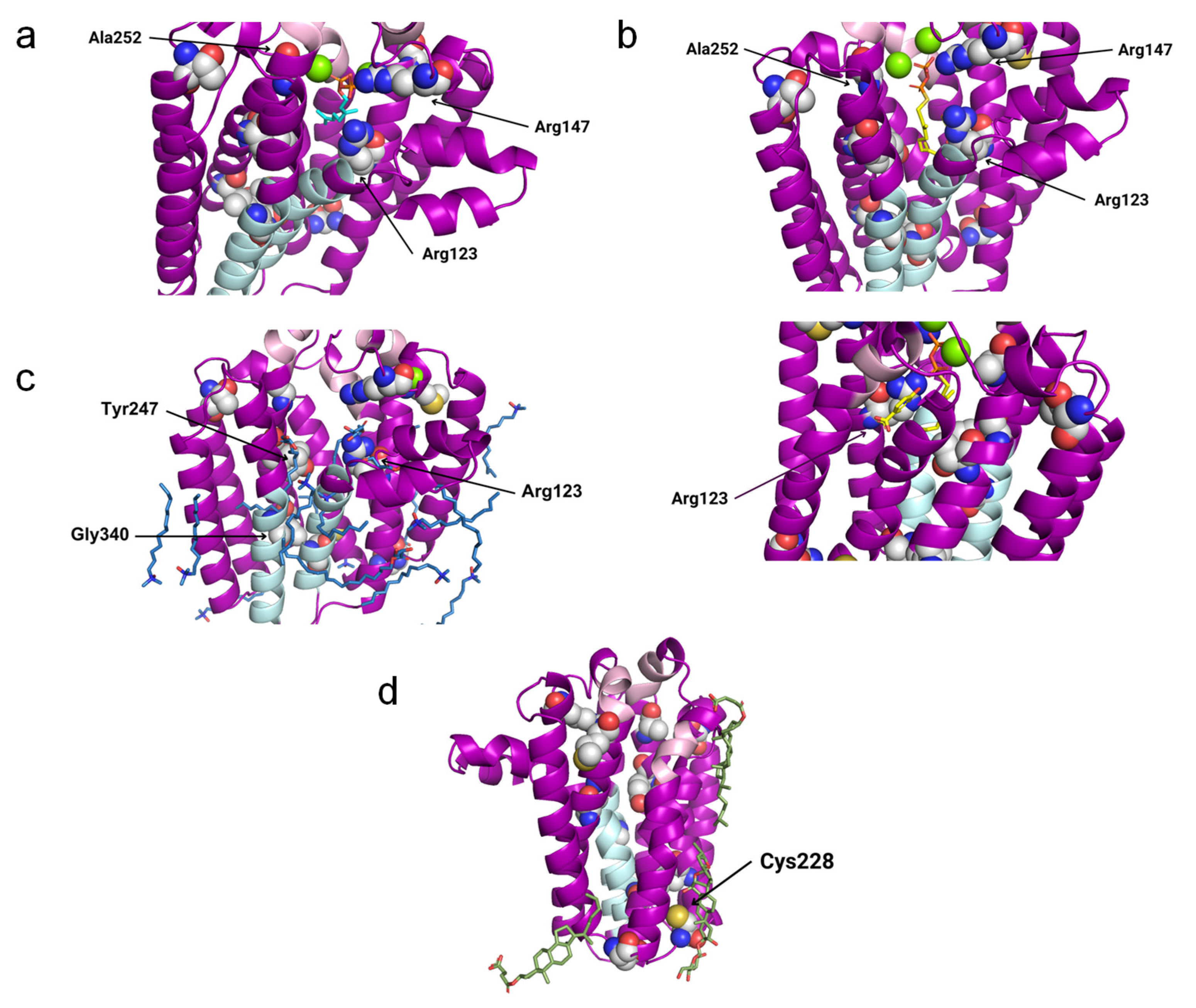
| PDB ID | Protein | Ligands |
|---|---|---|
| 4OD5_A | UbiA homolog from Aeropyrum pernix K1 |
p-hydroxybenzoic acid (PHB) |
| Geranyl S-thiolodiphosphate (GST) | ||
| Magnesium ion (MG) | ||
| 6M31_B | Digeranylgeranylglyceryl phosphate synthase from Methanocaldococcus jannaschii DSM 2661 | [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate (MPG) |
| Lauryl dimethylamine-n-oxide (LDA) | ||
| Phosphate ion (PO4) | ||
| Magnesium ion (MG) | ||
| 8DJM_B | UbiA prenyltransferase domain-containing protein from Cricetulus griseus | Cholesterol hemisuccinate (Y01) |
| Digitonin (AJP) | ||
| 4TQ3_B | UbiA homolog from Archaeoglobus fulgidus DSM 4304 | Geranyl diphosphate (GPP) |
| Magnesium ion (MG) | ||
| 7Q21_f | Cytochrome c oxidase polypeptide 4 from Corynebacterium glutamicum ATCC 13032 | Phosphatidic acid (7PH) |
| Cardiolipin (CDL) | ||
| Tridecane (TRD) |
| Amino acid modification | Location | SIFT prediction |
|---|---|---|
| Ser96Asn | Transmembrane helix S1 | Pathogenic |
| Arg123His | Transmembrane helix S2 | Pathogenic |
| Met132Arg | Loop between S2-S3 (matrix side) | Pathogenic |
| Arg147His | Loop between S2-S3 (matrix side) | Pathogenic |
| Asn178Ser | Loop between S3-S4 (intermembrane space side) |
Tolerated |
| Cys228Arg | Loop between S5-S6 (intermembrane space side) |
Pathogenic |
| Leu236Phe | Transmembrane helix S6 | Pathogenic |
| Thr244Ile | Transmembrane helix S6 | Tolerated |
| Tyr247Cys | Loop between S6-S7 (matrix side) | Pathogenic |
| Ala252Val | Loop between S6-S7 (matrix side) | Pathogenic |
| Thr275Ala | Loop between S6-S7 (matrix side) | Tolerated |
| Gly340Ala | Transmembrane helix S9 | Pathogenic |
| Variant | SIFT prediction | RMSD (Å) |
|---|---|---|
| p.Ser96Asn | Pathogenic | 0.183 |
| p.Met132Arg | Pathogenic | 0.199 |
| p.Leu236Phe | Pathogenic | 0.207 |
| p.Arg147His | Pathogenic | 0.210 |
| p.Asn178Ser | Tolerated | 0.225 |
| p.Gly340Ala | Pathogenic | 0.237 |
| p.Tyr247Cys | Pathogenic | 0.246 |
| p.Cys228Arg | Pathogenic | 0.247 |
| p.Thr244Ile | Tolerated | 0.247 |
| p.Arg123His | Pathogenic | 0.256 |
| p.Thr275Ala | Tolerated | 0.257 |
| p.Ala252Val | Pathogenic | 0.268 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





