Submitted:
19 March 2024
Posted:
21 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Design and Sampling
2.2. Extraction and RNA Amplification
2.3. Statistical Analysis
2.4. Phylogenetic Analysis
2.5. Mutational Analysis
3. Results
3.1. Study design and sampling
3.2. Real-Time PCR results
3.3. Statistical Analysis
3.4. Phylogenetic Analysis
3.5. Mutational Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3, . [CrossRef]
- Postler, T.S.; Beer, M.; Blitvich, B.J.; Bukh, J.; de Lamballerie, X.; Drexler, J.F.; Imrie, A.; Kapoor, A.; Karganova, G.G.; Lemey, P.; et al. Renaming of the genus Flavivirus to Orthoflavivirus and extension of binomial species names within the family Flaviviridae. Arch. Virol. 2023, 168, 1–7, . [CrossRef]
- Burbelo, P.D.; Dubovi, E.J.; Simmonds, P.; Medina, J.L.; Henriquez, J.A.; Mishra, N.; Wagner, J.; Tokarz, R.; Cullen, J.M.; Iadarola, M.J.; et al. Serology-Enabled Discovery of Genetically Diverse Hepaciviruses in a New Host. J. Virol. 2012, 86, 6171–6178, . [CrossRef]
- Smith, D.B.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, A.S.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J. Gen. Virol. 2016, 97, 2894–2907, . [CrossRef]
- Ramsay, J.D.; Evanoff, R.; Wilkinson, T.E., Jr.; Divers, T.J.; Knowles, D.P.; Mealey, R.H. Experimental transmission of equine hepacivirus in horses as a model for hepatitis C virus. Hepatology 2015, 61, 1533–1546, . [CrossRef]
- Scheel, T.K.H.; Kapoor, A.; Nishiuchi, E.; Brock, K.V.; Yu, Y.; Andrus, L.; Gu, M.; Renshaw, R.W.; Dubovi, E.J.; McDonough, S.P.; et al. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc. Natl. Acad. Sci. 2015, 112, 2192–2197, . [CrossRef]
- Pfaender, S.; Walter, S.; Grabski, E.; Todt, D.; Bruening, J.; Romero-Brey, I.; Gather, T.; Brown, R.J.P.; Hahn, K.; Puff, C.; et al. Immune protection against reinfection with nonprimate hepacivirus. Proc. Natl. Acad. Sci. 2017, 114, E2430–E2439, . [CrossRef]
- Tomlinson, J.E.; Wolfisberg, R.; Fahnøe, U.; Patel, R.S.; Trivedi, S.; Kumar, A.; Sharma, H.; Nielsen, L.; McDonough, S.P.; Bukh, J.; et al. Pathogenesis, MicroRNA-122 Gene-Regulation, and Protective Immune Responses After Acute Equine Hepacivirus Infection. J. Hepatol. 2021, 74, 1148–1163, . [CrossRef]
- Gather, T.; Walter, S.; Todt, D.; Pfaender, S.; Brown, R.J.P.; Postel, A.; Becher, P.; Moritz, A.; Hansmann, F.; Baumgaertner, W.; et al. Vertical transmission of hepatitis C virus-like non-primate hepacivirus in horses. J. Gen. Virol. 2016, 97, 2540–2551, . [CrossRef]
- Pronost, S.; Fortier, C.; Marcillaud-Pitel, C.; Tapprest, J.; Foursin, M.; Saunier, B.; Pitel, P.-H.; Paillot, R.; Hue, E.S. Further Evidence for in Utero Transmission of Equine Hepacivirus to Foals. Viruses 2019, 11, 1124, . [CrossRef]
- Badenhorst, M.; de Heus, P.; Auer, A.; Rümenapf, T.; Tegtmeyer, B.; Kolodziejek, J.; Nowotny, N.; Steinmann, E.; Cavalleri, J.-M. No Evidence of Mosquito Involvement in the Transmission of Equine Hepacivirus (Flaviviridae) in an Epidemiological Survey of Austrian Horses. Viruses 2019, 11, 1014, . [CrossRef]
- S. Pronost, et al. Hépacivirus, pégivirus, TDAV: Une nouvelle triade de virus hépatiques chez le ceval?. Pratique vétérinaire équine 197, 24-31 (2018).
- Altan, E.; Li, Y.; Jr, G.S.-S.; Sawaswong, V.; Barnum, S.; Pusterla, N.; Deng, X.; Delwart, E. Viruses in Horses with Neurologic and Respiratory Diseases. Viruses 2019, 11, 942, . [CrossRef]
- Yoon, J.; Park, T.; Kim, A.; Song, H.; Park, B.; Ahn, H.; Go, H.; Kim, D.; Lee, J.; Park, S.; et al. First report of equine parvovirus-hepatitis and equine hepacivirus coinfection in horses in Korea. Transbound. Emerg. Dis. 2021, 69, 2735–2746, . [CrossRef]
- Postel, A.; Cavalleri, J.-M.V.; Pfaender, S.; Walter, S.; Steinmann, E.; Fischer, N.; Feige, K.; Haas, L.; Becher, P. Frequent presence of hepaci and pegiviruses in commercial equine serum pools. Veter- Microbiol. 2015, 182, 8–14, . [CrossRef]
- Paim, W.; Weber, M.; Cibulski, S.; da Silva, M.; Puhl, D.; Budaszewski, R.; Varela, A.; Mayer, F.; Canal, C. Characterization of the viral genomes present in commercial batches of horse serum obtained by high-throughput sequencing. Biologicals 2019, 61, 1–7, . [CrossRef]
- Lu, G.; Huang, J.; Yang, Q.; Xu, H.; Wu, P.; Fu, C.; Li, S. Identification and genetic characterization of hepacivirus and pegivirus in commercial equine serum products in China. PLOS ONE 2017, 12, e0189208–e0189208, . [CrossRef]
- Meister, T.L.; Tegtmeyer, B.; Postel, A.; Cavalleri, J.-M.; Todt, D.; Stang, A.; Steinmann, E. Equine Parvovirus-Hepatitis Frequently Detectable in Commercial Equine Serum Pools. Viruses 2019, 11, 461, . [CrossRef]
- Kopper, J.J.; Schott, H.C.; Divers, T.J.; Mullaney, T.; Huang, L.; Noland, E.; Smedley, R. Theiler's disease associated with administration of tetanus antitoxin contaminated with nonprimate (equine) hepacivirus and equine parvovirus-hepatitis virus. Equine Veter- Educ. 2018, 32, E5–E9, . [CrossRef]
- Tomlinson, J.E.; Kapoor, A.; Kumar, A.; Tennant, B.C.; Laverack, M.A.; Beard, L.; Delph, K.; Davis, E.; Ii, H.S.; Lascola, K.; et al. Viral testing of 18 consecutive cases of equine serum hepatitis: A prospective study (2014-2018). J. Veter- Intern. Med. 2018, 33, 251–257, . [CrossRef]
- Pfaender, S.; Cavalleri, J.M.; Walter, S.; Doerrbecker, J.; Campana, B.; Brown, R.J.; Burbelo, P.D.; Postel, A.; Hahn, K.; Anggakusuma; et al. Clinical course of infection and viral tissue tropism of hepatitis C virus–like nonprimate hepaciviruses in horses. J. Hepatol. 2014, 61, 447–459, . [CrossRef]
- Cardone, R.; Buonavoglia, A.; Lanave, G.; Vasinioti, V.I.; Mininni, V.; Lorusso, E.; Decaro, N.; Martella, V.; Elia, G.; Diakoudi, G. Description of an Equine Hepacivirus Cluster in a Horse Stable in Italy. Transbound. Emerg. Dis. 2023, 2023, 1–7, . [CrossRef]
- Cavalleri, J.V.; Korbacska-Kutasi, O.; Leblond, A.; Paillot, R.; Pusterla, N.; Steinmann, E.; Tomlinson, J. European College of Equine Internal Medicine consensus statement on equine flaviviridae infections in Europe. J. Veter- Intern. Med. 2022, 36, 1858–1871, . [CrossRef]
- Tegtmeyer, B.; Echelmeyer, J.; Pfankuche, V.M.; Puff, C.; Todt, D.; Fischer, N.; Durham, A.; Feige, K.; Baumgärtner, W.; Steinmann, E.; et al. Chronic equine hepacivirus infection in an adult gelding with severe hepatopathy. Veter- Med. Sci. 2019, 5, 372–378, . [CrossRef]
- Pronost, S.; Hue, E.; Fortier, C.; Foursin, M.; Fortier, G.; Desbrosse, F.; Rey, F.; Pitel, P.-H.; Saunier, B. Identification of equine hepacivirus infections in France: Facts and Physiopathological insights. J. Equine Veter- Sci. 2016, 39, S22, . [CrossRef]
- Date, T.; Sugiyama, M.; Lkhagvasuren, D.; Wakita, T.; Oyunsuren, T.; Mizokami, M. Prevalence of equine hepacivirus infection in Mongolia. Virus Res. 2020, 282, 197940, . [CrossRef]
- Elia, G.; Lanave, G.; Lorusso, E.; Parisi, A.; Trotta, A.; Buono, R.; Martella, V.; Decaro, N.; Buonavoglia, C. Equine hepacivirus persistent infection in a horse with chronic wasting. Transbound. Emerg. Dis. 2017, 64, 1354–1358, . [CrossRef]
- Reuter, G.; Maza, N.; Pankovics, P.; Boros, . Non-primate hepacivirus infection with apparent hepatitis in a horse — Short communication. Acta Veter- Hung. 2014, 62, 422–427, . [CrossRef]
- Lyons, S.; Kapoor, A.; Sharp, C.; Schneider, B.S.; Wolfe, N.D.; Culshaw, G.; Corcoran, B.; McGorum, B.C.; Simmonds, P. Nonprimate Hepaciviruses in Domestic Horses, United Kingdom. Emerg. Infect. Dis. 2012, 18, 1976–1982, . [CrossRef]
- Figueiredo, A.S.; Lampe, E.; Espírito-Santo, M.P.D.; Mello, F.C.D.A.; de Almeida, F.Q.; de Lemos, E.R.S.; Godoi, T.L.O.S.; Dimache, L.A.G.; dos Santos, D.R.L.; Villar, L.M. Identification of two phylogenetic lineages of equine hepacivirus and high prevalence in Brazil. Veter- J. 2015, 206, 414–416, . [CrossRef]
- Matsuu, A.; Hobo, S.; Ando, K.; Sanekata, T.; Sato, F.; Endo, Y.; Amaya, T.; Osaki, T.; Horie, M.; Masatani, T.; et al. Genetic and serological surveillance for non-primate hepacivirus in horses in Japan. Veter- Microbiol. 2015, 179, 219–227, . [CrossRef]
- Kim, H.-S.; Moon, H.-W.; Sung, H.W.; Kwon, H.M. First identification and phylogenetic analysis of equine hepacivirus in Korea. Infect. Genet. Evol. 2017, 49, 268–272, . [CrossRef]
- Reichert, C.; Campe, A.; Walter, S.; Pfaender, S.; Welsch, K.; Ruddat, I.; Sieme, H.; Feige, K.; Steinmann, E.; Cavalleri, J.M. Frequent occurrence of nonprimate hepacivirus infections in Thoroughbred breeding horses – A cross-sectional study for the occurrence of infections and potential risk factors. Veter- Microbiol. 2017, 203, 315–322, . [CrossRef]
- Badenhorst, M.; Tegtmeyer, B.; Todt, D.; Guthrie, A.; Feige, K.; Campe, A.; Steinmann, E.; Cavalleri, J.M. First detection and frequent occurrence of Equine Hepacivirus in horses on the African continent. Veter- Microbiol. 2018, 223, 51–58, . [CrossRef]
- Figueiredo, A.S.; Lampe, E.; de Albuquerque, P.P.L.F.; Chalhoub, F.L.L.; de Filippis, A.M.B.; Villar, L.M.; Cruz, O.G.; Pinto, M.A.; de Oliveira, J.M. Epidemiological investigation and analysis of the NS5B gene and protein variability of non-primate hepacivirus in several horse cohorts in Rio de Janeiro state, Brazil. Infect. Genet. Evol. 2018, 59, 38–47, . [CrossRef]
- Abbadi, I.; Lkhider, M.; Kitab, B.; Jabboua, K.; Zaidane, I.; Haddaji, A.; Nacer, S.; Matsuu, A.; Pineau, P.; Tsukiyama-Kohara, K.; et al. Non-primate hepacivirus transmission and prevalence: Novel findings of virus circulation in horses and dogs in Morocco. Infect. Genet. Evol. 2021, 93, 104975, . [CrossRef]
- Wu, et al., First identification and genomic characterization of equine hepacivirus sub-type 3 strain in China. Virus Genes 56, 777–780 (2020).
- Pronost, S.; Hue, E.; Fortier, C.; Foursin, M.; Fortier, G.; Desbrosse, F.; Rey, F.A.; Pitel, P.-H.; Richard, E.; Saunier, B. Prevalence of Equine Hepacivirus Infections in France and Evidence for Two Viral Subtypes Circulating Worldwide. Transbound. Emerg. Dis. 2016, 64, 1884–1897, . [CrossRef]
- B. S. Gemaque, et al., Hepacivirus infection in domestic horses, Brazil, 2011-2013. Emerg. Infect. Dis. 20, 2011–2013 (2014).
- G. Lu, et al., First description of hepacivirus and pegivirus infection in domestic Horses in China: A study in guangdong province, heilongjiang province and Hong Kong district. PLoS One 11, 1–12 (2016).
- Lu, G.; Ou, J.; Sun, Y.; Wu, L.; Xu, H.; Zhang, G.; Li, S. Natural recombination of equine hepacivirus subtype 1 within the NS5A and NS5B genes. Virology 2019, 533, 93–98, . [CrossRef]
- Tanaka, T.; Kasai, H.; Yamashita, A.; Okuyama-Dobashi, K.; Yasumoto, J.; Maekawa, S.; Enomoto, N.; Okamoto, T.; Matsuura, Y.; Morimatsu, M.; et al. Hallmarks of Hepatitis C Virus in Equine Hepacivirus. J. Virol. 2014, 88, 13352–13366, . [CrossRef]
- Hayashi, S.; Tanaka, T.; Moriishi, K.; Hirayama, K.; Yamada, A.; Hotta, K. Seroepidemiology of non-primate hepacivirus (NPHV) in Japanese native horses. J. Veter- Med Sci. 2018, 80, 186–189, . [CrossRef]
- Chen, Y.; Cai, S.; Zhang, Y.; Lai, Z.; Zhong, L.; Sun, X.; Li, S.; Lu, G. First identification and genomic characterization of equine hepacivirus subtype 2 in China. Arch. Virol. 2021, 166, 3221–3224, . [CrossRef]
- C. Fortier, et al., Hepatitis viruses: prevalence of equine parvovirus-hepatitis virus and equine hepacivirus in France and Australia. Equine Vet. J. 53, 68–68 (2021).
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Lukashev, A.N.; Gmyl, A.; Coutard, B.; Adam, A.; Ritz, D.; Leijten, L.M.; van Riel, D.; et al. Evidence for Novel Hepaciviruses in Rodents. PLOS Pathog. 2013, 9, e1003438, . [CrossRef]
- Schlottau, K.; Fereidouni, S.; Beer, M.; Hoffmann, B. Molecular identification and characterization of nonprimate hepaciviruses in equines. Arch. Virol. 2018, 164, 391–400, . [CrossRef]
- Elia, G.; Lanave, G.; Lorusso, E.; Parisi, A.; Cavaliere, N.; Patruno, G.; Terregino, C.; Decaro, N.; Martella, V.; Buonavoglia, C. Identification and genetic characterization of equine hepaciviruses in Italy. Veter- Microbiol. 2017, 207, 239–247, . [CrossRef]
- Thrusfield, M., Christley, R., Brown, H., Diggle, P. J., French, N., Howe, K., Kelly, L., O'Connor, A., Sargeant, J., & Wood, H. (2017). Veterinary Epidemiology: Fourth Edition. (4th ed.) Wiley-Blackwell. [CrossRef]
- K. Tamura, G. Stecher, S. Kumar, MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol.38, 3022–3027 (2021).
- B. Q. Minh, et al., IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol.37, 1530–1534 (2020).
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589, . [CrossRef]
- T.A. Hall, BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98 (1999).
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 2015, 12, 7–8, . [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309, . [CrossRef]
- Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Huang, C.C.; E Ferrin, T. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform. 2006, 7, 339–339, . [CrossRef]
- Lyons, S.; Kapoor, A.; Schneider, B.S.; Wolfe, N.D.; Culshaw, G.; Corcoran, B.; Durham, A.E.; Burden, F.; McGorum, B.C.; Simmonds, P. Viraemic frequencies and seroprevalence of non-primate hepacivirus and equine pegiviruses in horses and other mammalian species. J. Gen. Virol. 2014, 95, 1701–1711, . [CrossRef]
- Badenhorst, M.; de Heus, P.; Auer, A.; Tegtmeyer, B.; Stang, A.; Dimmel, K.; Tichy, A.; Kubacki, J.; Bachofen, C.; Steinmann, E.; et al. Active equine parvovirus-hepatitis infection is most frequently detected in Austrian horses of advanced age. Equine Veter- J. 2021, 54, 379–389, . [CrossRef]
- Gather, T.; Walter, S.; Pfaender, S.; Todt, D.; Feige, K.; Steinmann, E.; Cavalleri, J.M.V. Acute and chronic infections with nonprimate hepacivirus in young horses. Veter- Res. 2016, 47, 1–5, . [CrossRef]
- Pacchiarotti, G.; Nardini, R.; Scicluna, M.T. Equine Hepacivirus: A Systematic Review and a Meta-Analysis of Serological and Biomolecular Prevalence and a Phylogenetic Update. Animals 2022, 12, 2486, . [CrossRef]
- Simmonds, P.; Bukh, J.; Combet, C.; Deléage, G.; Enomoto, N.; Feinstone, S.; Halfon, P.; Inchauspé, G.; Kuiken, C.; Maertens, G.; et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. J. Hepatol. 2005, 42, 962–973, . [CrossRef]
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. J. Hepatol. 2013, 59, 318–327, . [CrossRef]
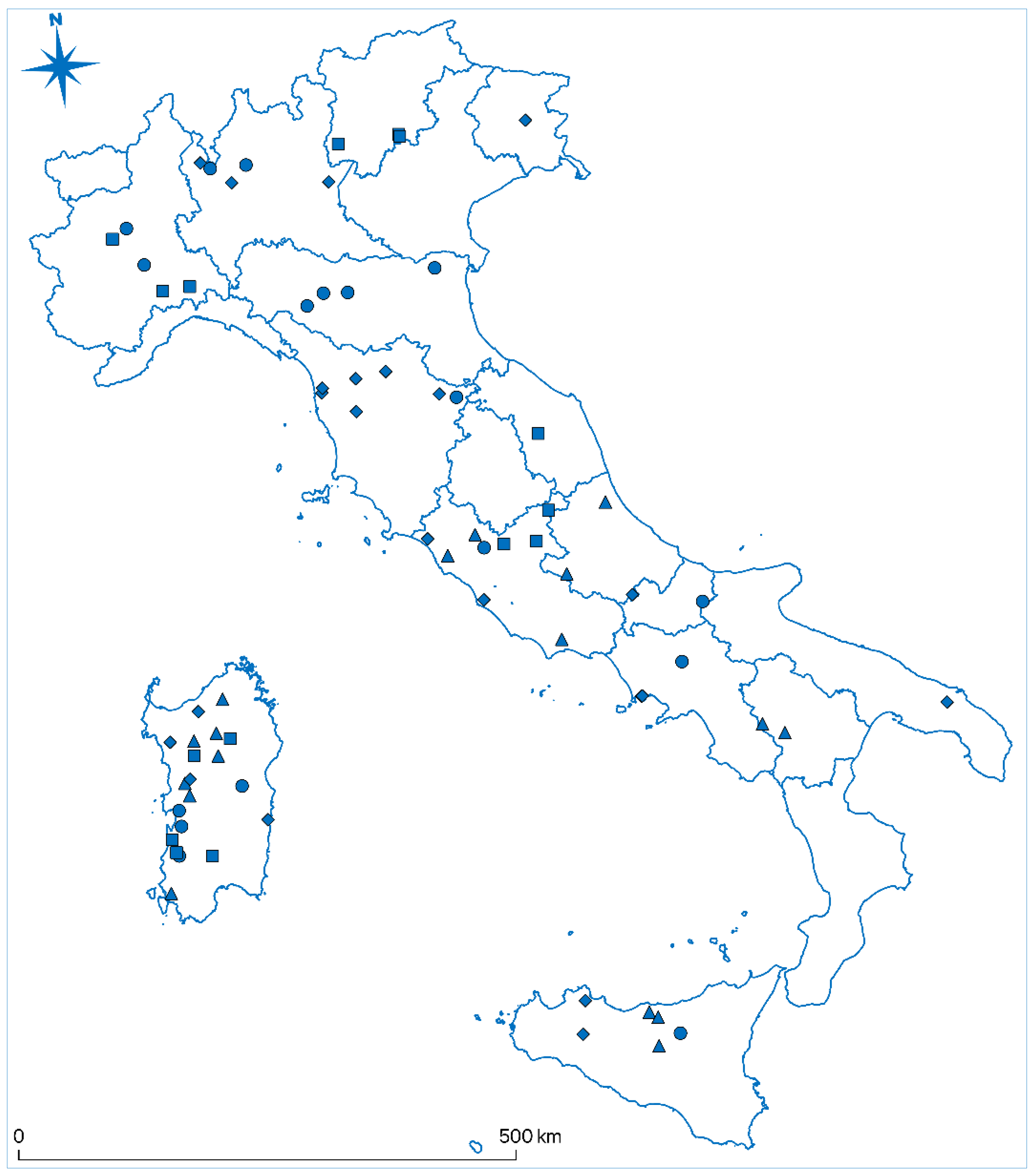
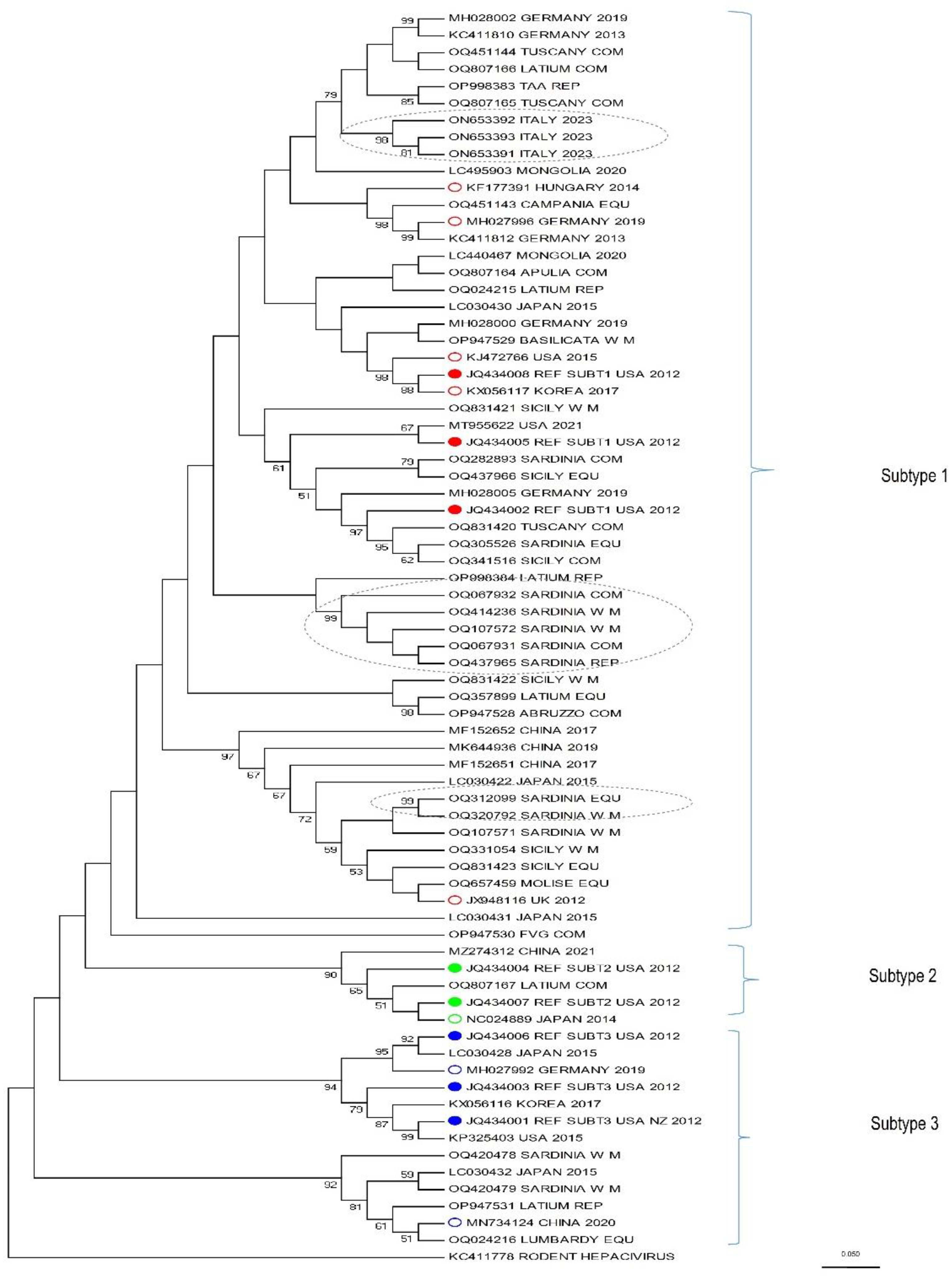
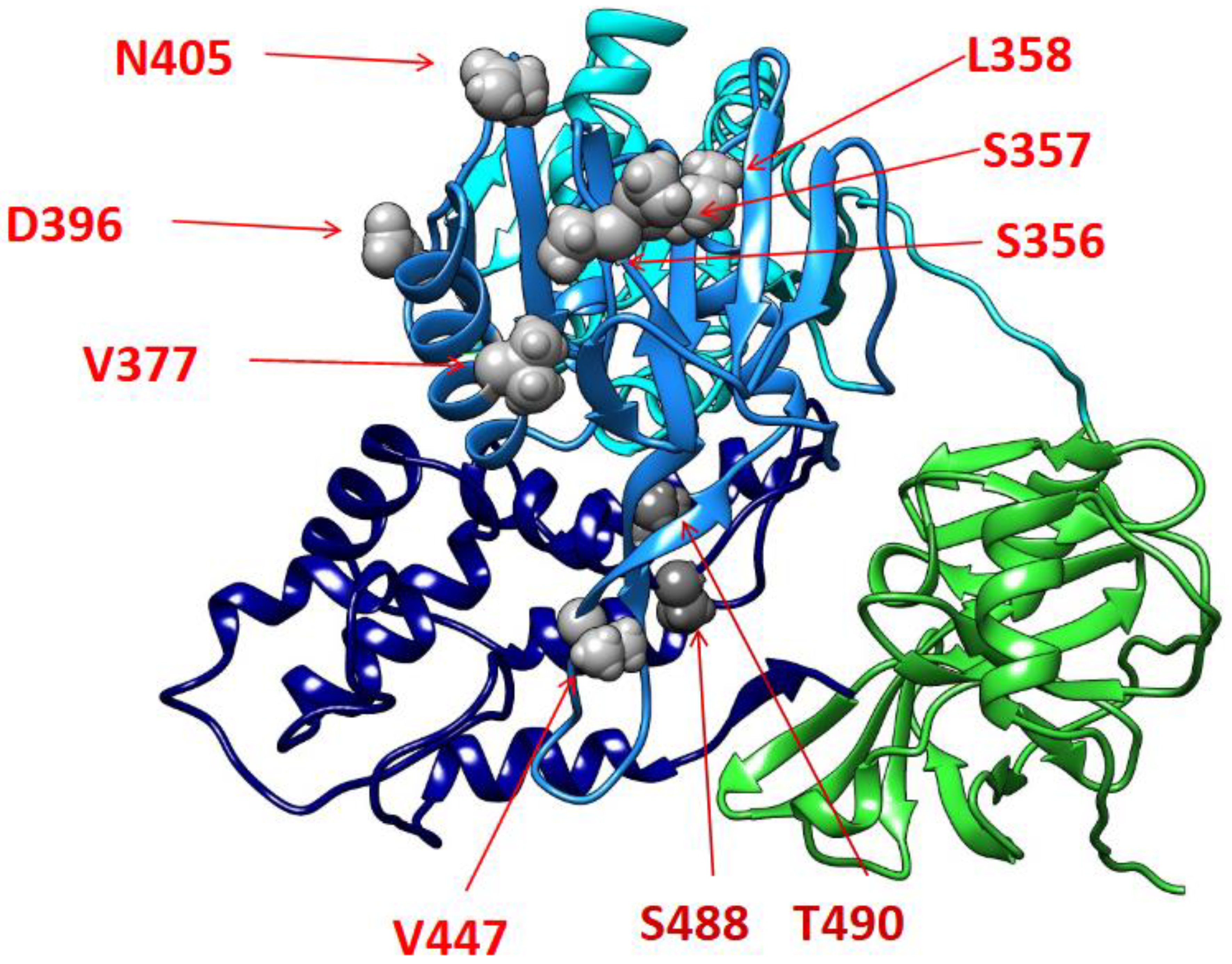
| Analysed samples | Positive samples | P (%) | Lower IC (%) | Upper IC (%) | SE (%) | |
|---|---|---|---|---|---|---|
| Production categories | ||||||
| Equestrian | 497 | 17 | 3.42 | 0 | 7.82 | ± 4.40 |
| Competition | 464 | 23 | 4.96 | 0.41 | 9.51 | ± 4.55 |
| Work/Meat | 442 | 17 | 3.85 | 0 | 8.51 | ± 4.66 |
| Reproduction | 398 | 20 | 5.03 | 0.11 | 9.94 | ± 4.91 |
| Italy macroregions | ||||||
| North | 393 | 18 | 4.58 | 0 | 9.52 | ± 4.94 |
| Center | 315 | 17 | 5.40 | 0 | 10.92 | ± 5.52 |
| South | 435 | 13 | 2.99 | 0 | 7.96 | ± 4.70 |
| Sardinia | 376 | 20 | 5.32 | 0.26 | 10.37 | ± 5.05 |
| Sicily | 282 | 9 | 3.19 | 0 | 9.03 | ± 5.84 |
| Chi Square P | Reproduction | Work/Meat | Competition |
|---|---|---|---|
| Equestrian | 0.23 | 0.72 | 0.23 |
| Competition | 0.96 | 0.41 | |
| Work/Meat | 0.40 |
| Chi Square P | Sicily | Sardinia | South | Center |
|---|---|---|---|---|
| North | 0.43 | 0.740 | 0.272 | 0.73 |
| Center | 0.23 | 1.00 | 0.130 | |
| South | 1.00 | 0.11 | ||
| Sardinia | 0.25 |
| NS3 mutation | Overall, N (%) | Competition, N (%) T=12 | Equestrian, N (%) T=8 | Reproduction, N (%), T=5 | Work meat, N (%) T=10 |
|---|---|---|---|---|---|
| T490V | 27 (77.1) | 9 (75) | 6 (75) | 4 (80) | 8 (80) |
| L358I | 7 (20.0) | 1 (8.3) | 2 (25) | 1 (20) | 3 (30) |
| S488T | 5 (14.3) | 2 (16.7) | 0 (0) | 1 (20) | 2 (20) |
| V377I | 3 (8.6) | 0 (0) | 0 (0) | 0 (0) | 3 (30) |
| N405K | 2 (5.7) | 1 (8.3) | 0 (0) | 1 (20) | 0 (0) |
| V447I | 2 (5.7) | 1 (8.3) | 1 (12.5) | 0 (0) | 0 (0) |
| D396E | 1 (2.9) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) |
| S356A | 1 (2.9) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) |
| S357A | 1 (2.9) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) |
| T358L | 1 (2.9) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) |
| NS3 Mutations | Overall, N (%) N=35 | Sardinia, N (%) (N=12) | Sicily, N (%) (N=6) | Central Italya, N (%) (N=9) | Southern Italyb, N (%) (N=5) |
|---|---|---|---|---|---|
| T490V | 27 (77.1) | 10 (83.3) | 4 (66.7) | 6 (66.7) | 5 (100) |
| L358I | 7 (20.0) | 4 (33.3) | 1 (16.7) | 1 (11.1) | 0 (0) |
| S488T | 5 (14.3) | 5 (41.7) | 0 (0) | 0 (0) | 0 (0) |
| V377I | 3 (8.6) | 2 (16.7) | 1 (16.7) | 0 (0) | 0 (0) |
| N405K | 2 (5.7) | 2 (16.7) | 0 (0) | 0 (0) | 0 (0) |
| V447I | 2 (5.7) | 0 (0) | 1 (16.7) | 1 (11.1) | 0 (0) |
| D396E | 1 (2.9) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) |
| S356A | 1 (2.9) | 0 (0) | 0 (0) | 1 (11.1) | 0 (0) |
| S357A | 1 (2.9) | 0 (0) | 0 (0) | 1 (11.1) | 0 (0) |
| T358L | 1 (2.9) | 0 (0) | 0 (0) | 1 (11.1) | 0 (0) |
| NS3 Mutations | Localization as secondary structure | NS3 Functional Domain |
|---|---|---|
| S356A | Helix 356-358 | Helicase Subdomain II |
| S357A | Helix 356-358 | Helicase Subdomain II |
| T358L | Helix 356-358 | Helicase Subdomain II |
| L358I | Helix 356-358 | Helicase Subdomain II |
| V377I | Helix 371-384 | Helicase Subdomain II |
| D396E | Coil 392-405 | Helicase Subdomain II |
| N405K | Strand 405-410 | Helicase Subdomain II |
| V447I | Helix 447-502 | Helicase Subdomain II |
| S488T | Helix 488-500 | Helicase Subdomain III |
| T490V | Helix 488-500 | Helicase Subdomain III |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





