Submitted:
20 March 2024
Posted:
22 March 2024
You are already at the latest version
Abstract
Keywords:
Introduction
1.1. Background
1.2. Related Works
2. Proposal Methods
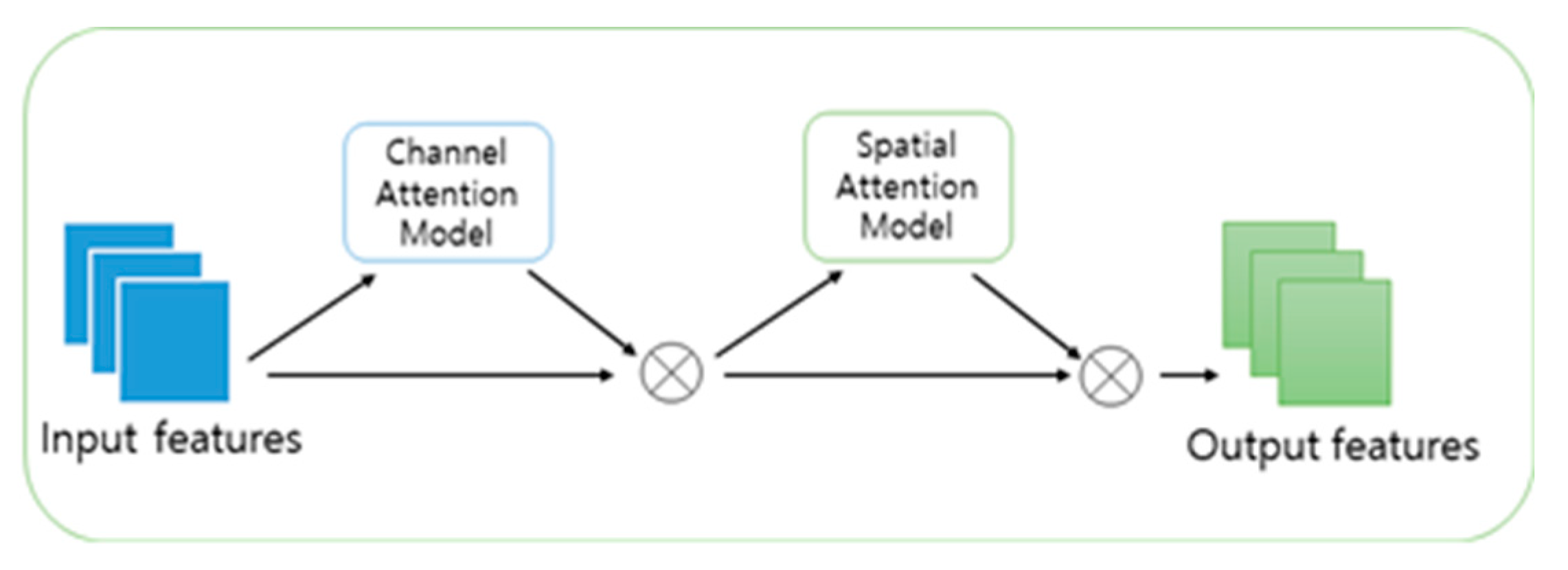
2.1. CBAM(Convolution Block Attention Module)
- The channel attention module learns the weights of each channel through global average pooling and global max pooling of each channel, then weights the channels of the input feature map to create a feature map with better representation ability.
- The spatial attention module learns the weights of each spatial location through max pooling and average pooling in the spatial dimension of the input feature map, then multiplies these weights with the original feature map to enhance useful features and suppress useless background.
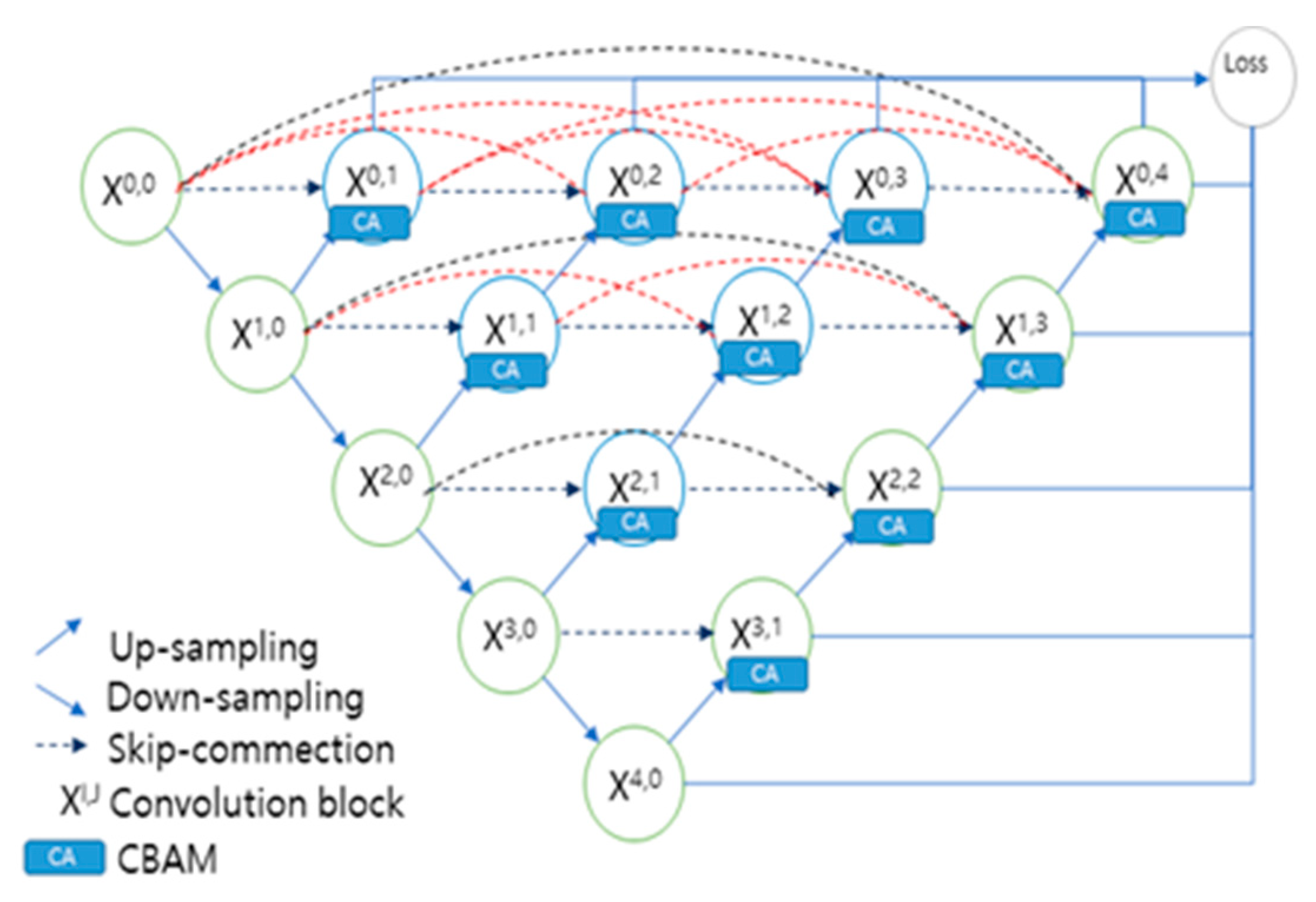
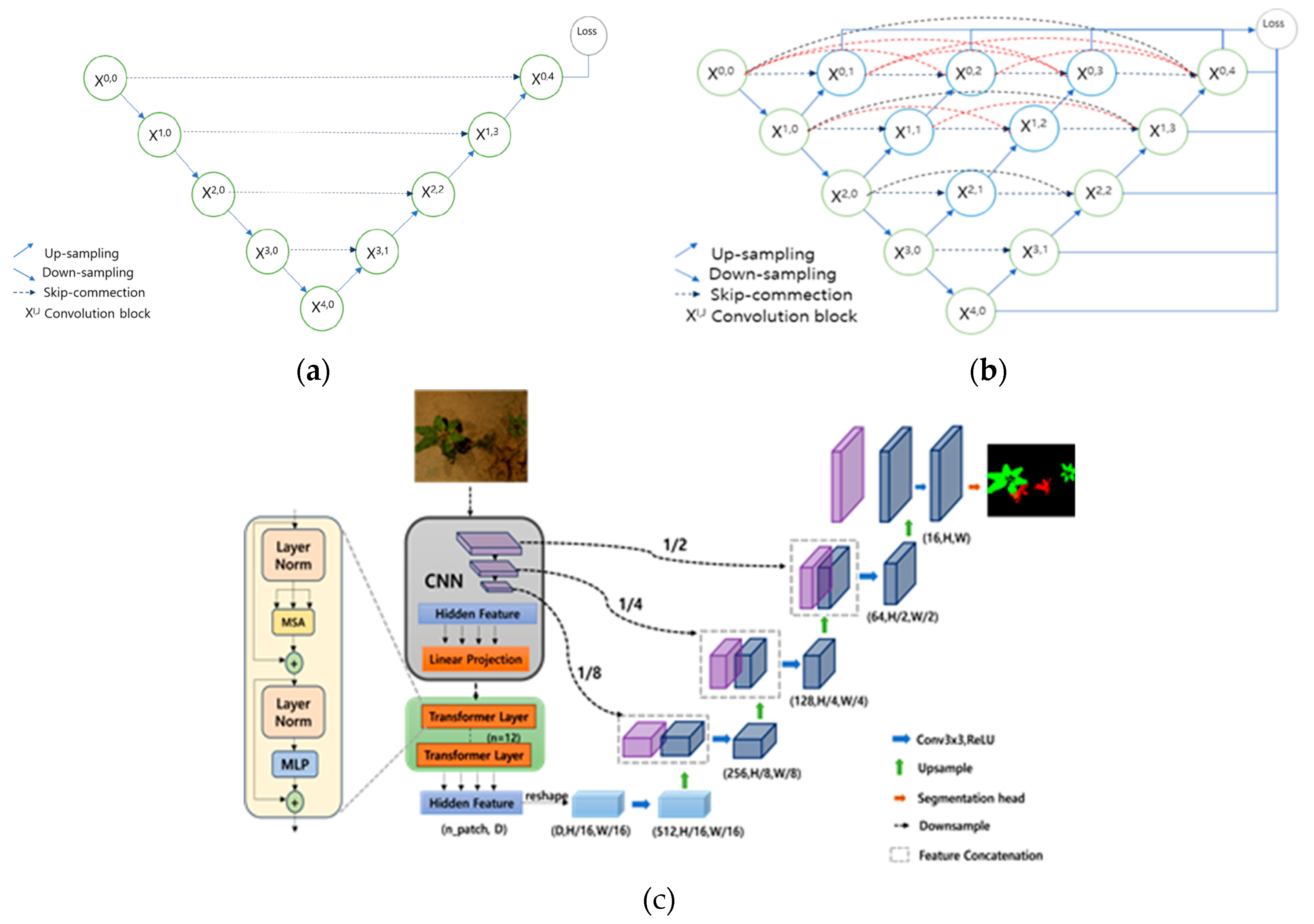
2.2. UNet Overview
- 1)
- Multi-Scale Feature Fusion:
- 2)
- Improved Upsampling Module:
- 3)
- Dense Connection:
- 4)
- Performance Improvement:
- 5)
- Application Areas:
- 6)
- Computational Resource Requirements:
- 7)
-
TransUNet:
- Global Context Detection: TransUNet, based on the Transformer structure, enables the model to capture global context information in images. This helps the model better understand the relationships between different areas of the image and perform tasks more accurately.
- Scalability: The Transformer structure offers good scalability, adapting to images of various sizes and resolutions. This makes TransUNet more flexible in handling different tasks and datasets.
- Self-Attention Mechanism: The model uses the self-attention mechanism to help build long-range dependencies between different areas of the image, useful for handling overall image tasks.
- Fewer Parameters: Compared to traditional CNN-based architectures, Transformers typically require fewer parameters for similar or better performance. This makes the model lighter, resulting in relatively faster training and inference speeds.
- Applicability to Various Tasks: The structure of TransUNet makes it suitable for various computer vision tasks, such as image segmentation and object detection. This diversity allows the model to have applicability in multiple domains.
3. Crop and Weed Identification System
3.1. System Overview
3.2. Extracting Crops from Data Images
3.3. Data Preprocessing
3.4. Loss Function
3.5. Evaluation Metrics
4. Experiments and Results Analysis
4.1. Experimental Environment
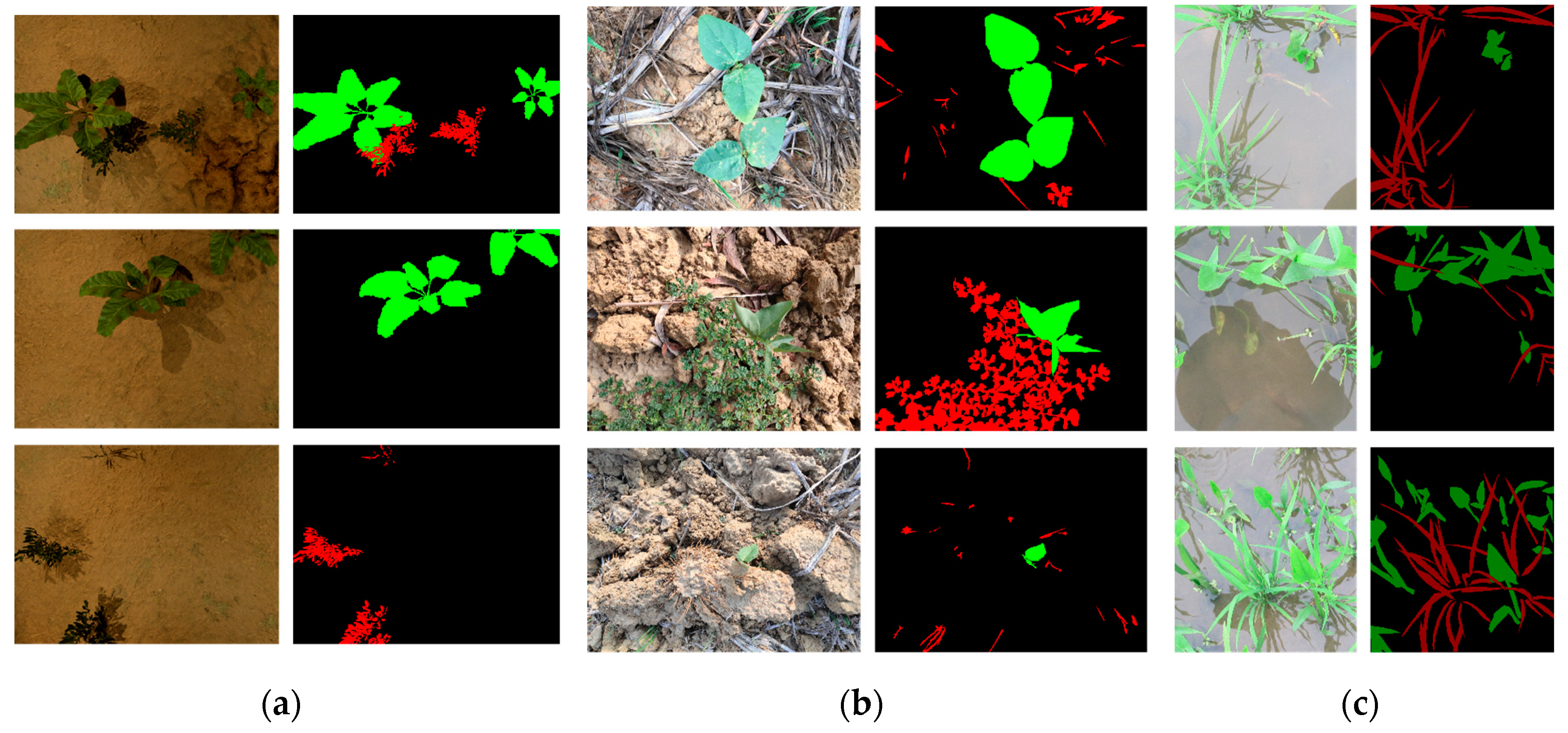
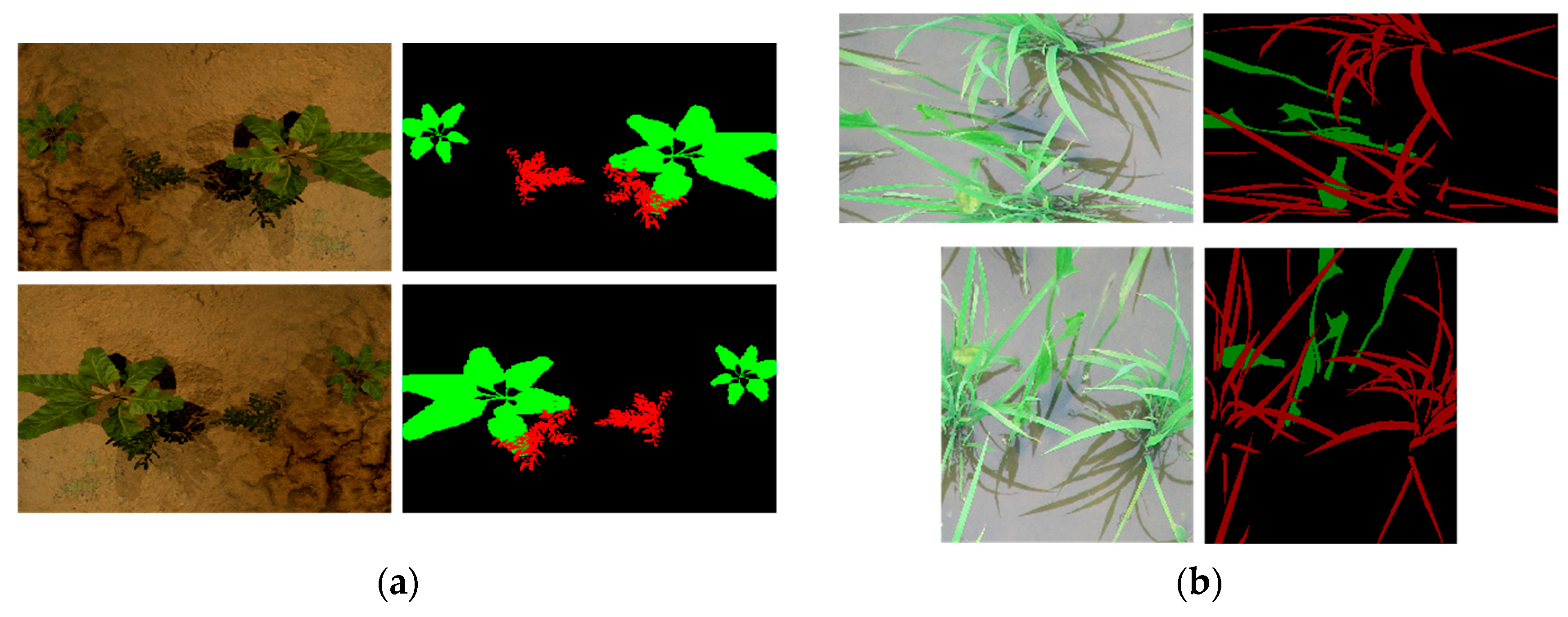
4.2. Experimental Results
| Model | IOU | Background | Weed | Beet |
|---|---|---|---|---|
| UNet | 89.83% | 95.29% | 95.71% | 98.90% |
| UNet++ | 89.75% | 95.30% | 95.71% | 98.91% |
| ATT-UNet | 91.80% | 95.29% | 95.73% | 98.97% |
| TransUNet | 61.11% | 66.94% | 67.22% | 77.75% |
| CNNTransUNet | 68.18% | 66.88% | 56.08% | 80.27% |
| Model | IOU | Background | Weed | Rice |
|---|---|---|---|---|
| UNet | 73.88% | 82.90% | 86.25% | 89.68% |
| UNet++ | 74.90% | 82.89% | 86.26% | 89.37% |
| ATT-UNet | 76.38% | 82.91% | 86.21% | 89.50% |
| TransUNet | 77.09% | 76.32% | 76.44% | 77.75% |
| CNNTransUNet | 76.91% | 75.25% | 76.25% | 77.58% |
| Model | IOU | Background | Weed | Pea |
|---|---|---|---|---|
| UNet | 81.17% | 85.36% | 90.43% | 91.34% |
| UNet++ | 76.52% | 85.32% | 90.40% | 91.38% |
| ATT-UNet | 84.10% | 85.36% | 90.37% | 91.49% |
| TransUNet | 81.56% | 68.77% | 69.87% | 93.26% |
| CNNTransUNet | 81.20% | 68.69% | 69.56% | 92.85% |
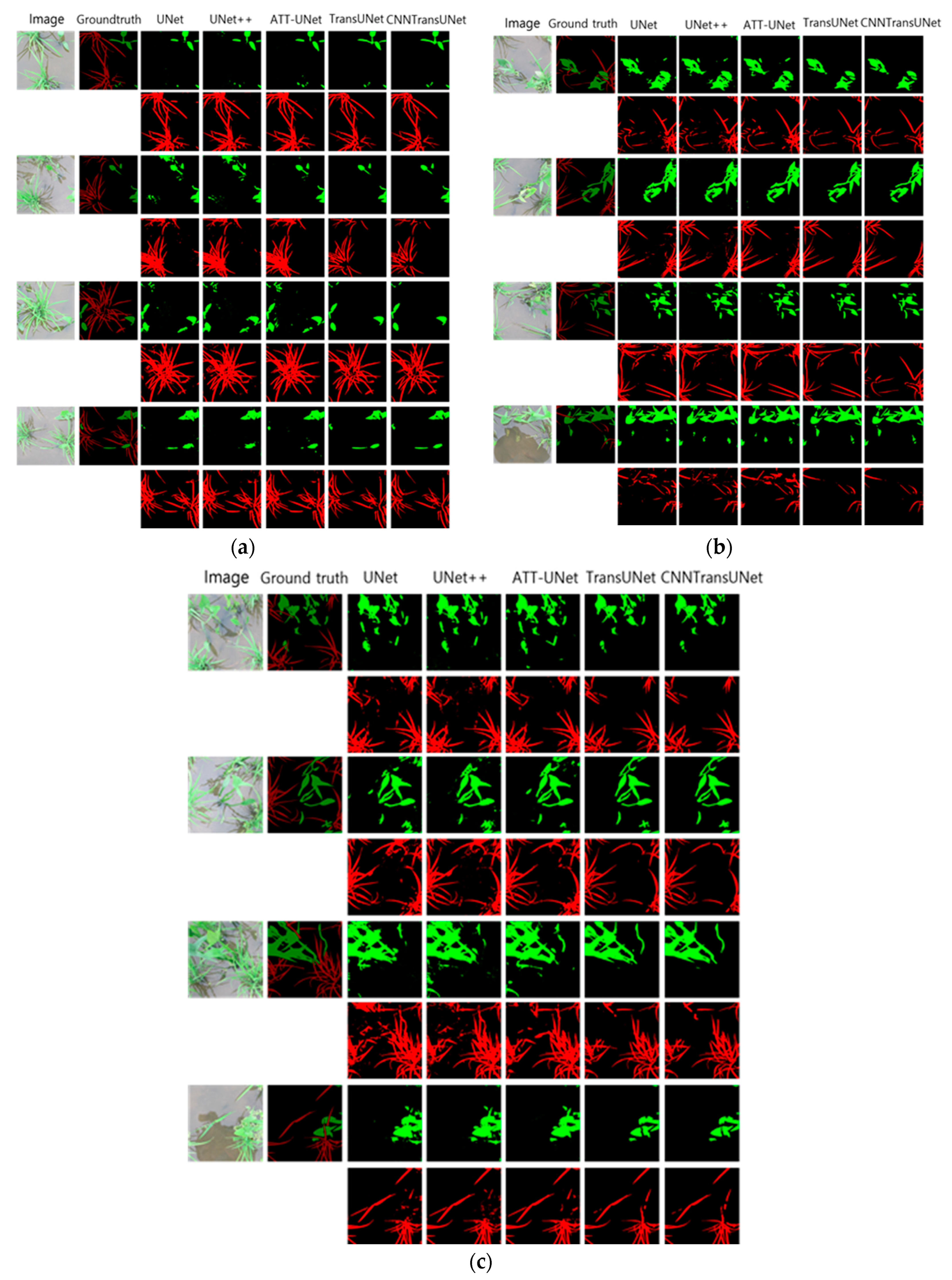
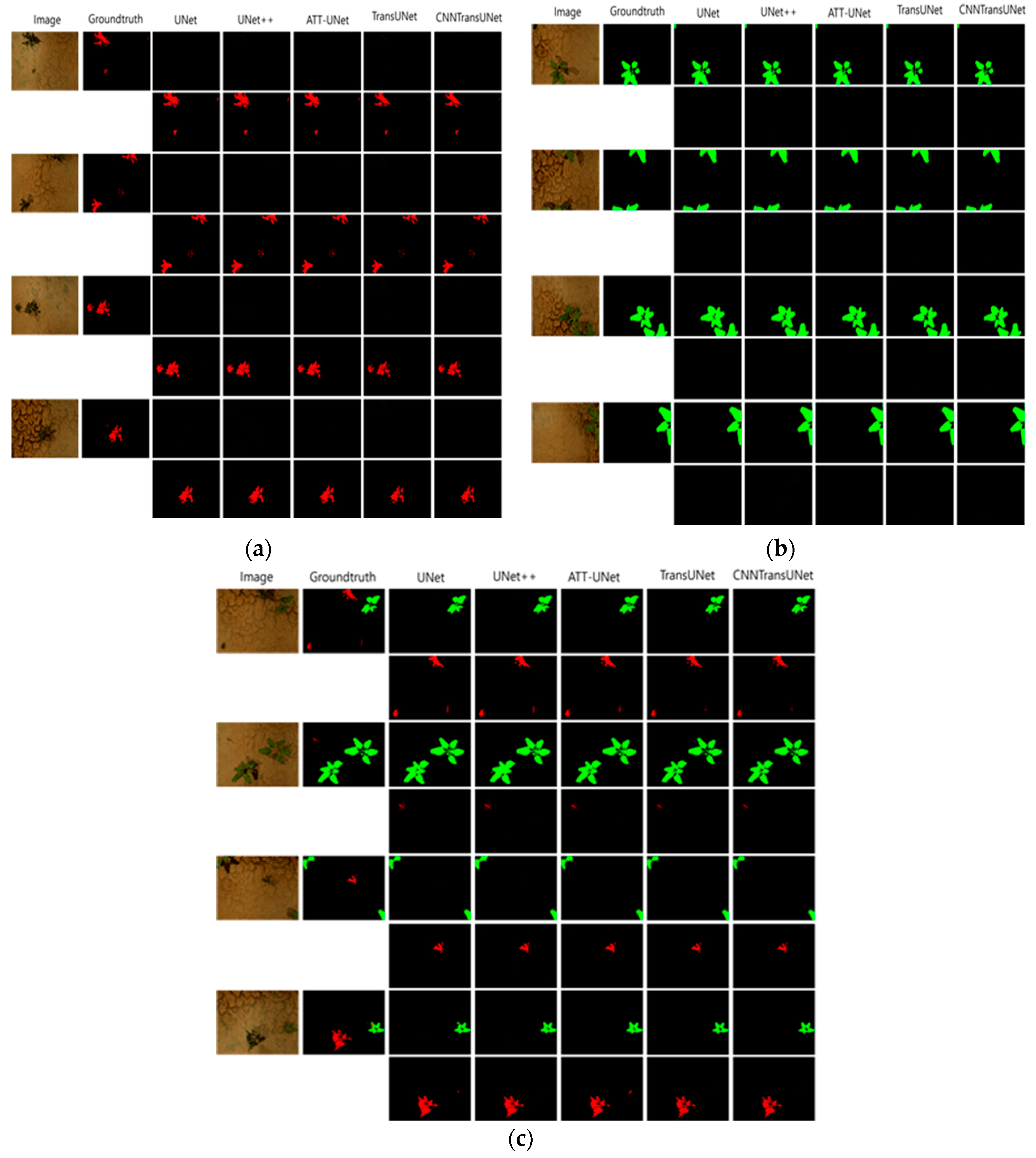
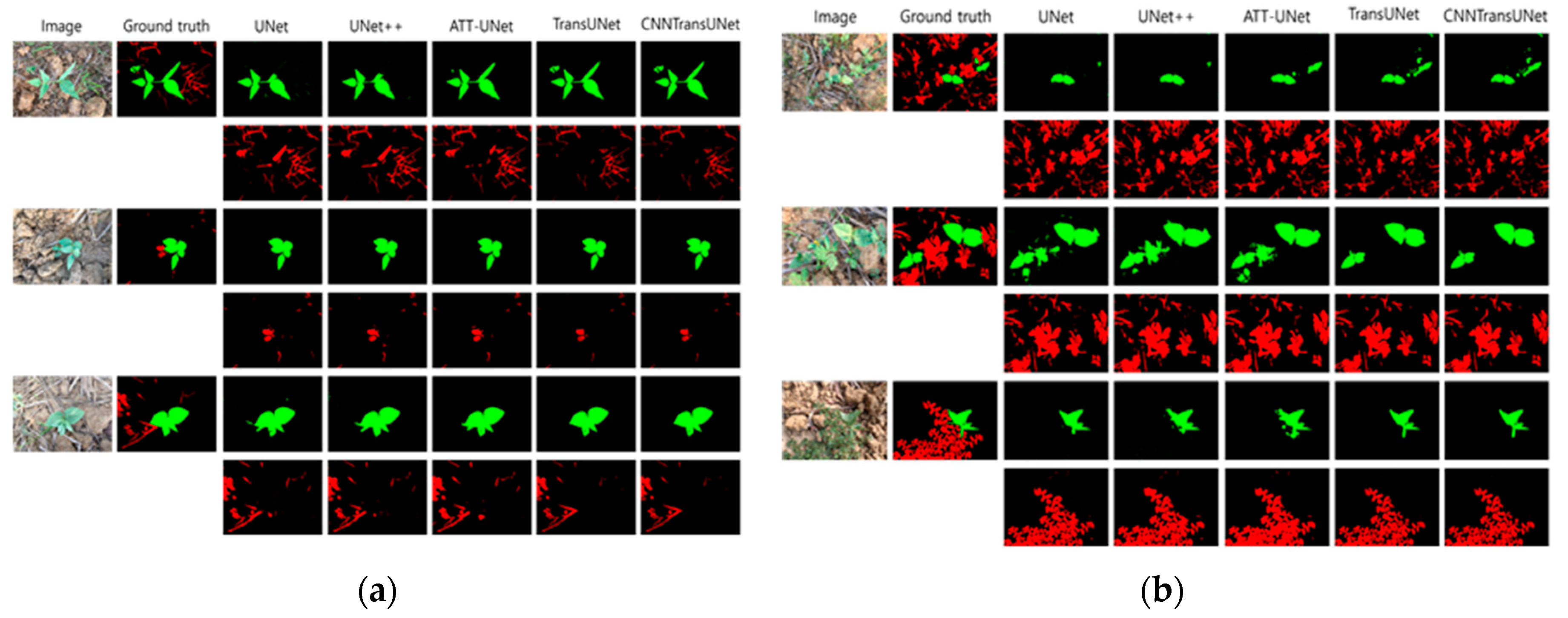
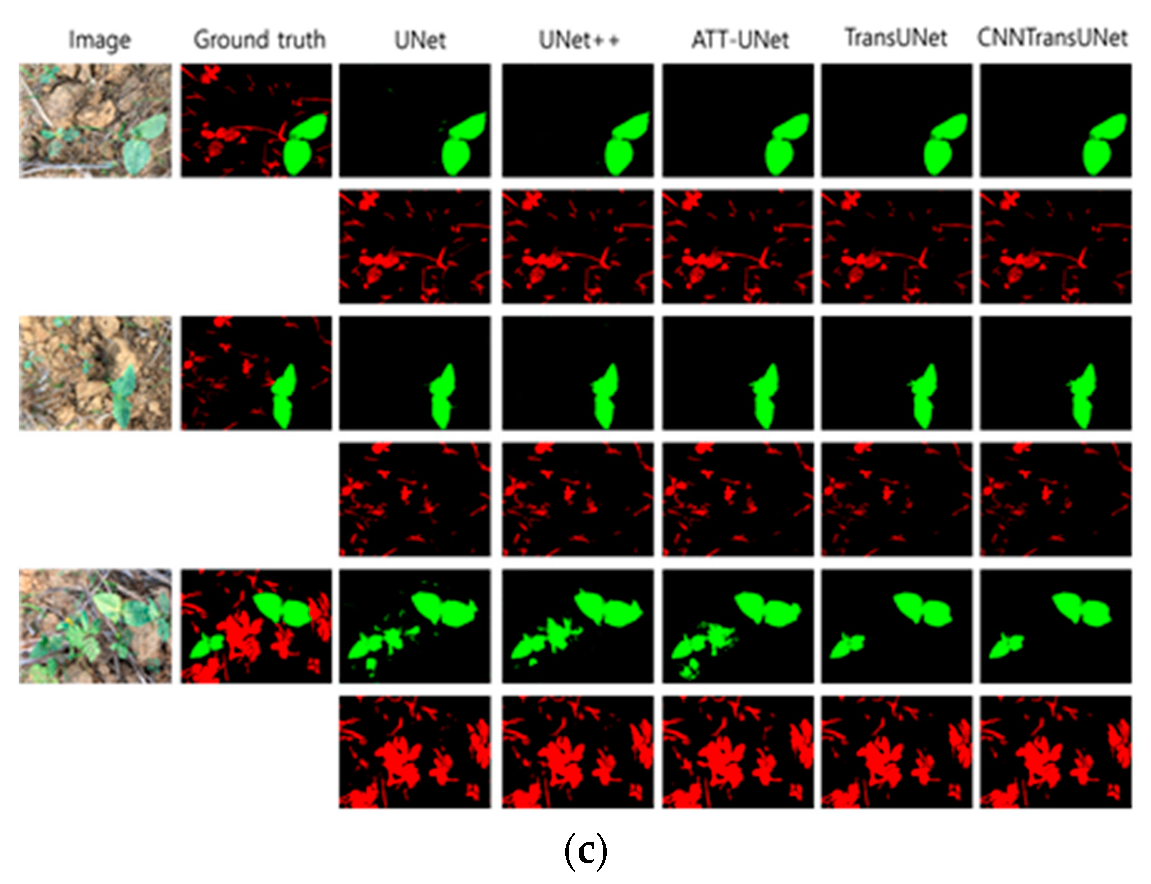
5. Conclusion
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lü Wei, Dong Li, Sun Yuhan, et al. A Brief Discussion on Weed Control Methods at Home and Abroad. Chinese Agricultural Science Bulletin 2018, 34, 34–39.
- Huang Rongxi. Development of a Corn Field Weeding System Based on Computer Vision. Agricultural Mechanization Research 2018, 40, 217–220.
- Wu Lanlan. Identification Research on Weeds in the Corn Seedling Stage in the Field Based on Digital Image Processing. Huazhong Agricultural University, 2010.
- Li, N.、Grift, TE、Yuan, T.、Zhang, C.、Momin, MA、Li, W. (2016). Image processing of crop/weed discrimination in fields with high weed stress.2016 ASABE International Conference, pp. 1.
- Perez AJ、Lopez F、Benlloch, JV. Perez AJ、Lopez F、Benlloch JV. Color and shape analysis techniques for weed detection in grain fields". Agricultural Computer and Electronics 2000, 25, 197–212. [Google Scholar] [CrossRef]
- Ross D. Ram, David C. Slaughter and D. Ken Giles. Cotton precision weed control system. ASAE Transactions 45, 231–238.
- Guijarro M, Pajares G, Riomoros I, et al. Automatic segmentation of relevant textures in agricultural images. Computers and Electronics in Agriculture 2011, 75, 75–83. [CrossRef]
- Cho S I, Lee D S, Jeong J Y. AE—automation and emerging technologies: Weed–plant discrimination by machine vision and artificial neural network. Biosystems engineering 2002, 83, 275–280.
- Ramos P J, Prieto F A, Montoya E C, et al. Automatic fruit count on coffee branches using computer vision. Computers and Electronics in Agriculture 2017, 137, 9–22. [CrossRef]
- Pantazi, Xanthoula Eirini, et al. Wheat yield prediction using machine learning and advanced sensing techniques. Computers and electronics in agriculture 2016, 121, 57–65. [CrossRef]
- Ferentinos, Konstantinos P. Deep learning models for plant disease detection and diagnosis. Computers and electronics in agriculture 2018, 145, 311–318. [CrossRef]
- Liu, Wenjie, et al. DFF-ResNet: An insect pest recognition model based on residual networks. Big Data Mining and Analytics 2020, 3, 300–310. [CrossRef]
- Rustia, Dan Jeric Arcega, et al. Automatic greenhouse insect pest detection and recognition based on a cascaded deep learning classification method. Journal of Applied Entomology 2021, 145, 206–222. [CrossRef]
- Pantazi, Xanthoula-Eirini, Dimitrios Moshou, and Cedric Bravo. Active learning system for weed species recognition based on hyperspectral sensing. Biosystems Engineering 2016, 146, 193–202. [CrossRef]
- Binch, Adam, and C. W. Fox. Controlled comparison of machine vision algorithms for Rumex and Urtica detection in grassland. Computers and Electronics in Agriculture 2017, 140, 123–138. [CrossRef]
- Zhang, Mengyun, Changying Li, and Fuzeng Yang. Classification of foreign matter embedded inside cotton lint using short wave infrared (SWIR) hyperspectral transmittance imaging. Computers and Electronics in Agriculture 2017, 139, 75–90. [CrossRef]
- Maione, Camila, et al. Classification of geographic origin of rice by data mining and inductively coupled plasma mass spectrometry. Computers and Electronics in Agriculture 2016, 121, 101–107. [CrossRef]
- Lin, C. A support vector machine embedded weed identification system. University of Illinois at Urbana-Champaign, 2010.
- Sun Jun, He Xiaofi, Tan Wenjun, Wu Xiaohong, Shen Jifeng, "Deer Tiger. Hollow convolution combined with global convolutional neural networks to identify crop seedlings and weeds. Proceedings of the Chinese Society of Agricultural Engineering 2018, 34, 159–165.
- Zhou, Z.、Rahman Siddiquee, MM、Tajbakhsh, N.、Liang, J. Unet++: A nested u-net architecture for medical image segmentation. Springer International Press, vol. 11042, pp. 3–11.
- Wang Hong, Chen Gongping. Field weed real-time segmentation based on PCAW-UNet. Journal of Xi'an University of Arts and Sciences (Natural Science Edition), 2021, 27–37.
- Shang Jianwei, Jiang Honghai, Yu Gang, "Deep Learning Based Weed Identification System. Software Guide 2020, 19, 127–130.
- Peng Mingxia, Xia Junfang, Peng Hui. Efficient Identification of Cotton Weeds in Complex FPN Context. Journal of Agricultural Engineering 2011, 35, 202–209.
- Chua L O, Roska T. The CNN paradigm. IEEE Transactions on Circuits and Systems I: Fundamental Theory and Applications 1993, 40, 147–156. [CrossRef]
- Ren A, Li Z, Ding C, et al. Sc-dcnn: Highly-scalable deep convolutional neural network using stochastic computing. ACM SIGPLAN Notices 2017, 52, 405–418. [CrossRef]
- Ma B, Li X, Xia Y, et al. Autonomous deep learning: A genetic DCNN designer for image classification. Neurocomputing 2020, 379, 152–161. [CrossRef]
- Woo S, Park J, Lee J Y, et al. Cbam: Convolutional Block Attention Module. Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, pp. 3–19, 2018.
- Chen Ying and Gong Su Ming. Improvement of Human Behavior Recognition Network under Channel Attention Mechanism. Journal of Electronics and Information 2021, 43, 3538–3545.
- Ronneberger, O.、Fischer, P.、Brox, T. U-net: Convolutional network for biomedical image segmentation. Springer International Publishing 2015, 9351, 234–241.
- Guo Lie, Zhang Tuanshan, Sun Weizhen, & Guo Jielong. Image Semantic Description Algorithm of Fusion Spatial Attention Mechanism. Laser & Optoelectronics Progress, vol. 58, no. 12, pp. 1210030, 2021.
- Fan, Xinnan, et al. A Nested Unet with Attention Mechanism for Road Crack Image Segmentation. 2021 IEEE 6th International Conference on Signal and Image Processing (ICSIP), pp. 189–193, 2021.
- Chen J, Lu Y, Yu Q, et al. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint 2021, arXiv:2102.04306.
- Rice/weedy field image dataset for crop/weedy classification. [CrossRef]
- Chebrolu, N. 、Lottes, P. 、Schaefer, A.、Winterhalter、W.、Burgard, W.、Stachniss, C. Agricultural Robot Dataset for Beet Field Plant Classification, Location and Mapping. International Journal of Robotics, 2017, 36, 1045–1052. [Google Scholar]
- Nachiketh RV, Krishnan A, Krishnan KV, Haritha ZA, Sasinas A (2021) Southern pea/weed field image dataset for semantic segmentation and crop/weed classification using an encoder-decoder network. SSRN Electron J. [CrossRef]
- Christopherson, Peter and Chris Jacobs. The importance of the loss function in option valuation. Journal of Financial Economics 2004, 72, 291–318. [CrossRef]
- Barron, Jonathan T. A general and adaptive robust loss function. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4331–4339, 2019.
- Dalianis, Hercules, and Hercules Dalianis. Evaluation metrics and evaluation. Clinical Text Mining: secondary use of electronic patient records, pp. 45–53, 2018.
- Wang, Zhaobin, E. Wang, and Ying Zhu. Image segmentation evaluation: a survey of methods. Artificial Intelligence Review 2020, 53, 5637–5674. [CrossRef]
- Kingma, Diederik P., and Jimmy Ba. Adam: A method for stochastic optimization. arXiv preprint, 2014; arXiv:1412.6980.
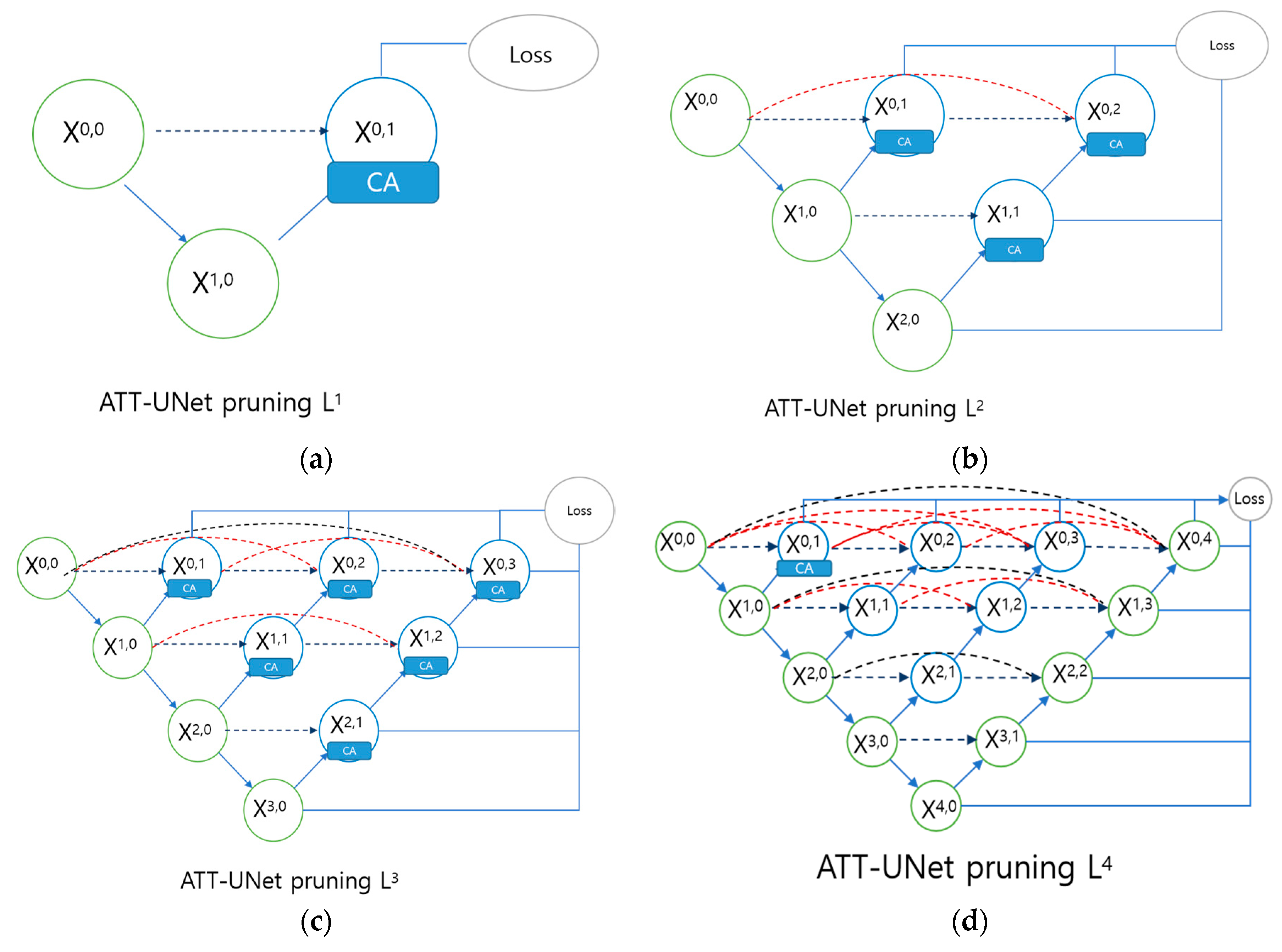

| Model | IOU |
|---|---|
| ATT-UNet L1 | 74.27% |
| ATT-UNet L2 | 73.51% |
| ATT-UNet L3 | 74.28% |
| ATT-UNet L4 | 75.03% |
| Model | IOU |
|---|---|
| ATT-UNet L1 | 89.76% |
| ATT-UNet L2 | 90.24% |
| ATT-UNet L3 | 89.06% |
| ATT-UNet L4 | 89.42% |
| Model | IOU |
|---|---|
| ATT-UNet L1 | 77.96% |
| ATT-UNet L2 | 77.78% |
| ATT-UNet L3 | 77.02% |
| ATT-UNet L4 | 77.60% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





