Submitted:
22 March 2024
Posted:
25 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
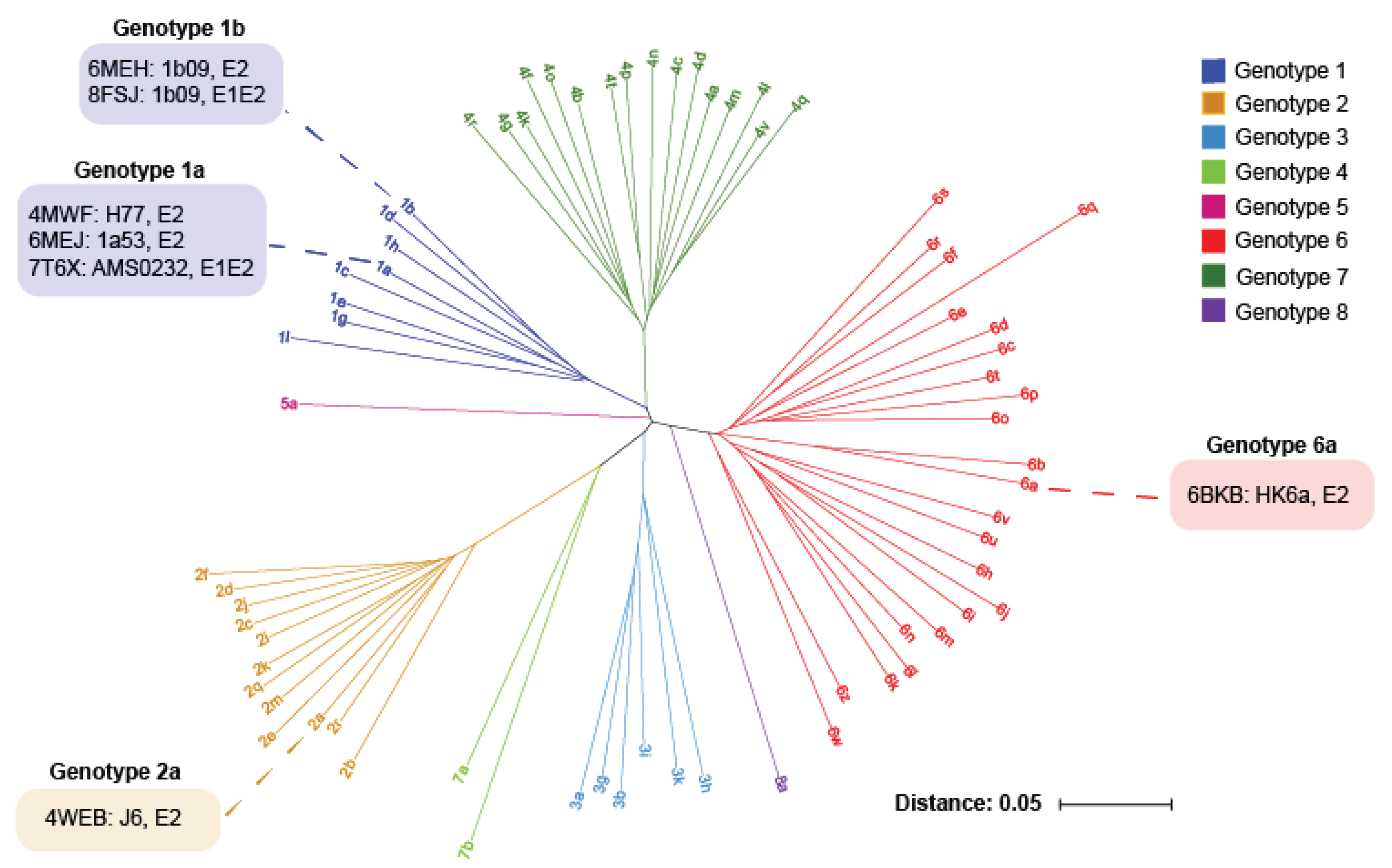
2. HCV E1E2 Diversity
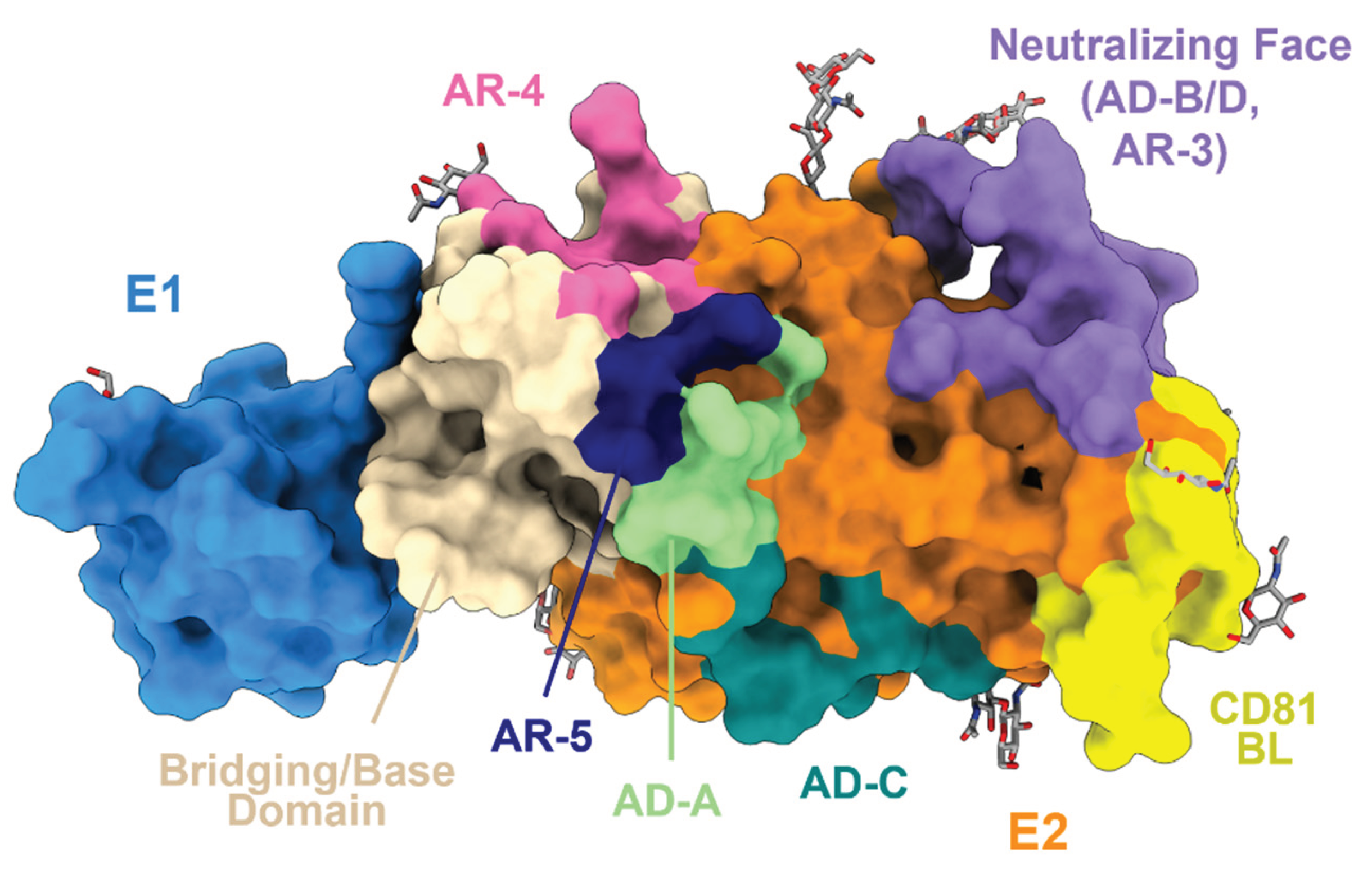
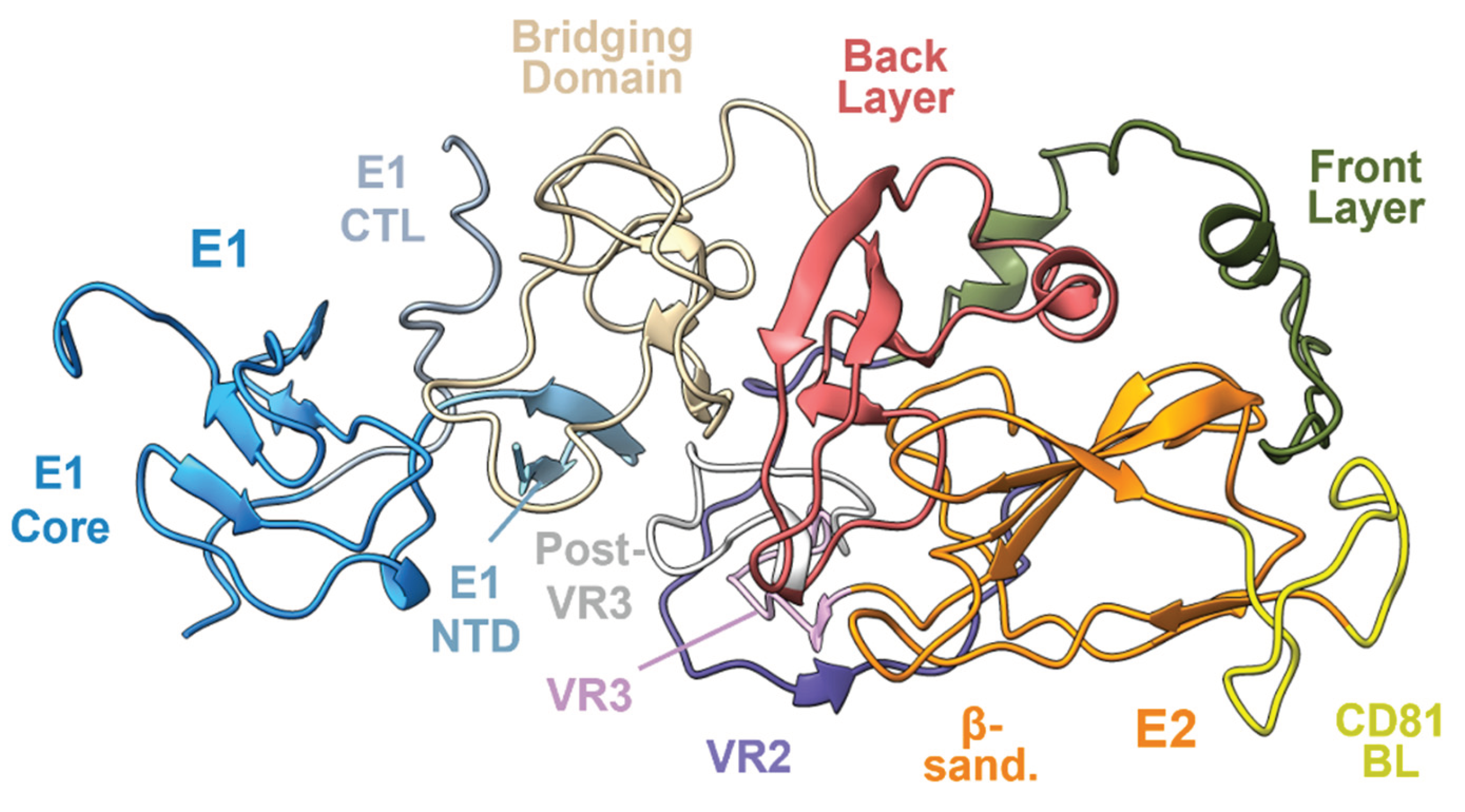
3. HCV E2 and E1E2 Structure
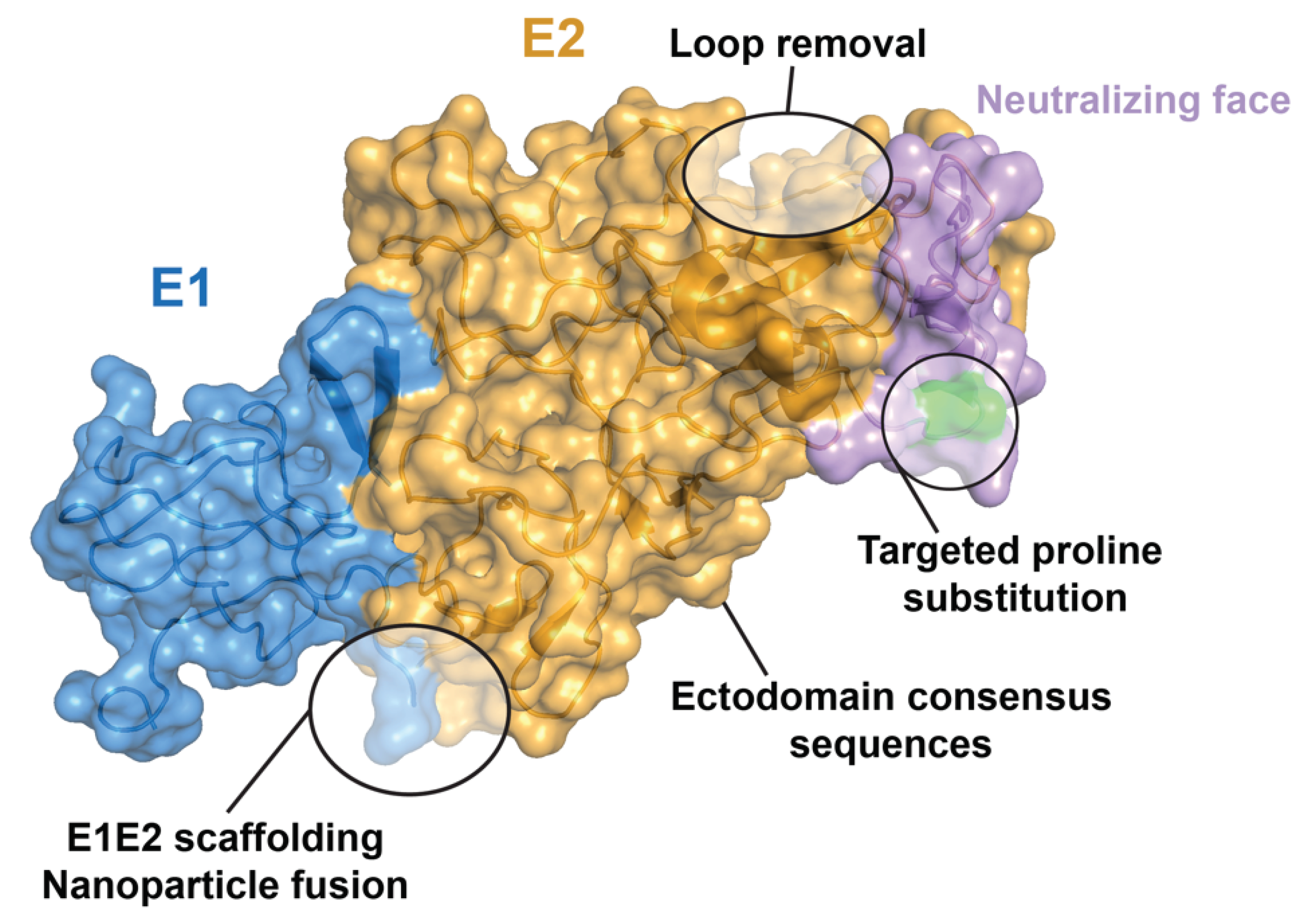
4. Structure-Based Vaccine Design
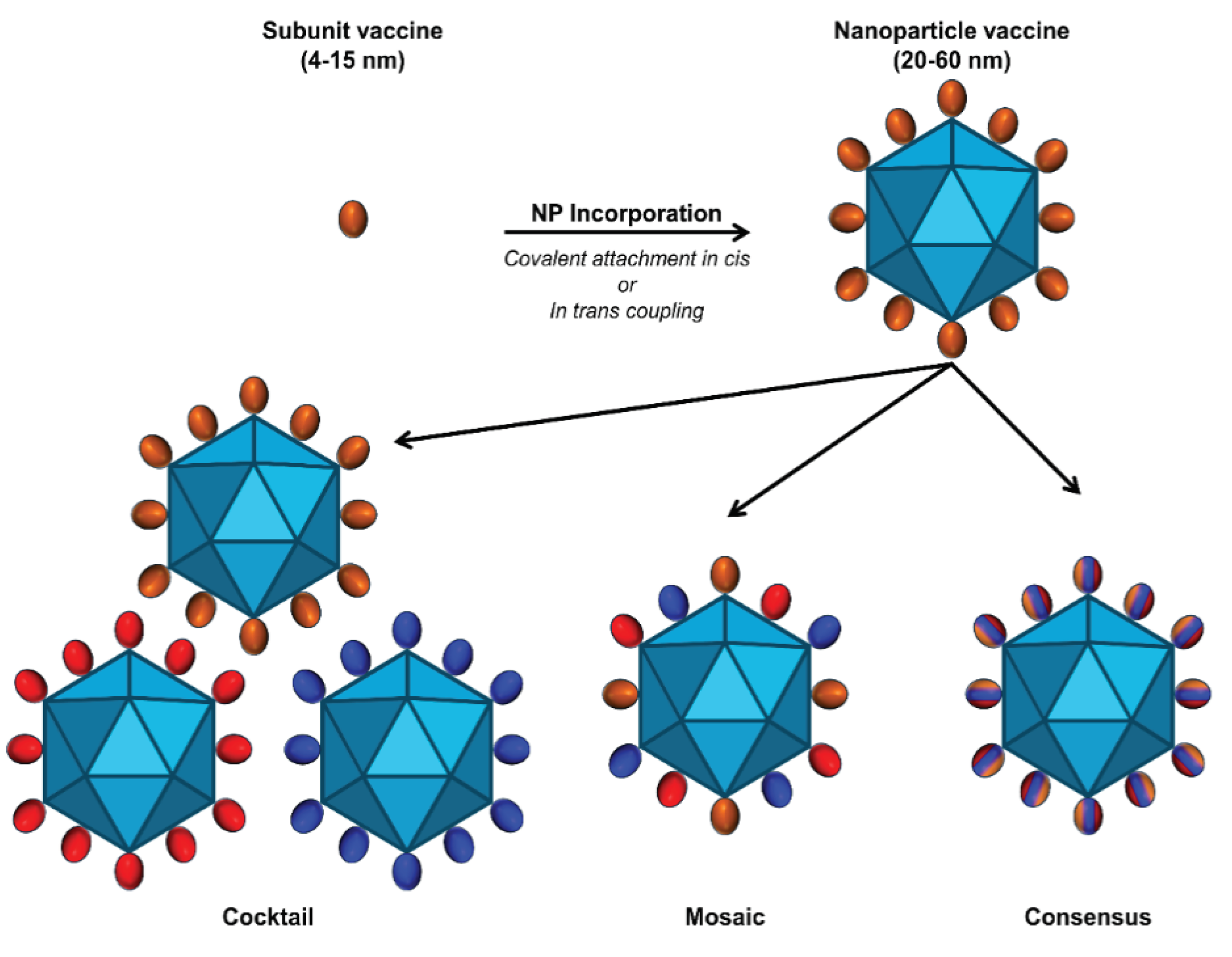
5. Multivalent Delivery Platforms and Considerations
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest Statement
References
- Metcalf, M.C.; Janus, B.M.; Yin, R.; Wang, R.; Guest, J.D.; Pozharski, E.; Law, M.; Mariuzza, R.A.; Toth, E.A.; Pierce, B.G.; et al. Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies. Nat. Commun. 2023, 14, 3980. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, C.; Yusim, K.; Boykin, L.; Richardson, R. The Los Alamos hepatitis C sequence database. Bioinformatics 2005, 21, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Keck, Z.Y.; Wang, R.; Lau, P.; Garagusi, K.; Elkholy, K.; Toth, E.A.; Urbanowicz, R.A.; Guest, J.D.; Agnihotri, P.; et al. Structure-Based Design of Hepatitis C Virus E2 Glycoprotein Improves Serum Binding and Cross-Neutralization. Journal of virology 2020, 94, epublish. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- WHO, Hepatitis C Key Facts (WHO, 2022).
- Rosen, H.R. Chronic Hepatitis C Infection. New England Journal of Medicine 2011, 364, 2429–2438. [Google Scholar] [CrossRef]
- Ly, K.N.; Hughes, E.M.; Jiles, R.B.; Holmberg, S.D. Rising Mortality Associated With Hepatitis C Virus in the United States, 2003-2013. Clin Infect Dis 2016, 62, 1287–1288. [Google Scholar] [CrossRef] [PubMed]
- Spach, D.H. HCV Epidemiology in the United States. 2024.
- Chung, R.T.; Baumert, T.F. Curing chronic hepatitis C--the arc of a medical triumph. New England Journal of Medicine 2014, 370, 1576–1578. [Google Scholar] [CrossRef]
- Cox, A.L.; Thomas, D.L. Hepatitis C virus vaccines among people who inject drugs. Clin. Infect. Dis. 2013, 57 Suppl. 2, S46–S50. [Google Scholar] [CrossRef]
- Callaway, E. Hepatitis C drugs not reaching poor. Nature 2014, 508, 295–296. [Google Scholar] [CrossRef]
- Cox, A.L. MEDICINE. Global control of hepatitis C virus. Science 2015, 349, 790–791. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Giang, E.; Nieusma, T.; Kadam, R.U.; Cogburn, K.E.; Hua, Y.; Dai, X.; Stanfield, R.L.; Burton, D.R.; Ward, A.B.; et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 2013, 342, 1090–1094. [Google Scholar] [CrossRef]
- Khan, A.G.; Whidby, J.; Miller, M.T.; Scarborough, H.; Zatorski, A.V.; Cygan, A.; Price, A.A.; Yost, S.A.; Bohannon, C.D.; Jacob, J.; et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 2014, 509, 381–384. [Google Scholar] [CrossRef]
- Flyak, A.I.; Ruiz, S.; Colbert, M.D.; Luong, T.; Crowe, J.E., Jr.; Bailey, J.R.; Bjorkman, P.J. HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design. Cell Host Microbe 2018, 24, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Tzarum, N.; Giang, E.; Kong, L.; He, L.; Prentoe, J.; Augestad, E.; Hua, Y.; Castillo, S.; Lauer, G.M.; Bukh, J.; et al. Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies. Sci. Adv. 2019, 5, eaav1882. [Google Scholar] [CrossRef]
- Kong, L.; Lee, D.E.; Kadam, R.U.; Liu, T.; Giang, E.; Nieusma, T.; Garces, F.; Tzarum, N.; Woods, V.L., Jr.; Ward, A.B.; et al. Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen. Proc. Natl. Acad. Sci. U S A 2016, 113, 12768–12773. [Google Scholar] [CrossRef]
- Tzarum, N.; Wilson, I.A.; Law, M. The Neutralizing Face of Hepatitis C Virus E2 Envelope Glycoprotein. Frontiers in immunology 2018, 9, 1315. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pierce, B.G.; Wang, Q.; Keck, Z.Y.; Fuerst, T.R.; Foung, S.K.; Mariuzza, R.A. Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody. J. Biol. Chem. 2015, 290, 10117–10125. [Google Scholar] [CrossRef]
- Yechezkel, I.; Law, M.; Tzarum, N. From Structural Studies to HCV Vaccine Design. Viruses 2021, 13. [Google Scholar] [CrossRef]
- Aleman, F.; Tzarum, N.; Kong, L.; Nagy, K.; Zhu, J.; Wilson, I.A.; Law, M. Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors. Proc. Natl. Acad. Sci. U S A 2018, 115, 7569–7574. [Google Scholar] [CrossRef]
- Torrents de la Pena, A.; Sliepen, K.; Eshun-Wilson, L.; Newby, M.L.; Allen, J.D.; Zon, I.; Koekkoek, S.; Chumbe, A.; Crispin, M.; Schinkel, J.; et al. Structure of the hepatitis C virus E1E2 glycoprotein complex. Science 2022, 378, 263–269. [Google Scholar] [CrossRef]
- Fuerst, T.R.; Pierce, B.G.; Keck, Z.Y.; Foung, S.K.H. Designing a B Cell-Based Vaccine against a Highly Variable Hepatitis C Virus. Frontiers in microbiology 2017, 8, 2692. [Google Scholar] [CrossRef]
- Bailey, J.R.; Barnes, E.; Cox, A.L. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology 2019, 156, 418–430. [Google Scholar] [CrossRef]
- Duncan, J.D.; Urbanowicz, R.A.; Tarr, A.W.; Ball, J.K. Hepatitis C Virus Vaccine: Challenges and Prospects. Vaccines 2020, 8. [Google Scholar] [CrossRef]
- Law, M. Antibody Responses in Hepatitis C Infection. Cold Spring Harb. Perspect. Med. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.M. Designing an HCV vaccine: a unique convergence of prevention and therapy? Curr. Opin. Virol. 2017, 23, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Lavie, M.; Hanoulle, X.; Dubuisson, J. Glycan Shielding and Modulation of Hepatitis C Virus Neutralizing Antibodies. Frontiers in immunology 2018, 9, 910. [Google Scholar] [CrossRef]
- Cashman, S.B.; Marsden, B.D.; Dustin, L.B. The Humoral Immune Response to HCV: Understanding is Key to Vaccine Development. Frontiers in immunology 2014, 5, 550. [Google Scholar] [CrossRef] [PubMed]
- Prentoe, J.; Bukh, J. Hypervariable Region 1 in Envelope Protein 2 of Hepatitis C Virus: A Linchpin in Neutralizing Antibody Evasion and Viral Entry. Frontiers in immunology 2018, 9, 2146. [Google Scholar] [CrossRef]
- Bhattarai, N.; McLinden, J.H.; Xiang, J.; Kaufman, T.M.; Stapleton, J.T. Conserved Motifs within Hepatitis C Virus Envelope (E2) RNA and Protein Independently Inhibit T Cell Activation. PLoS Pathog. 2015, 11, e1005183. [Google Scholar] [CrossRef]
- Vijayamahantesh, V.; Patra, T.; Meyer, K.; Alameh, M.G.; Reagan, E.K.; Weissman, D.; Ray, R. Modified E2 Glycoprotein of Hepatitis C Virus Enhances Proinflammatory Cytokines and Protective Immune Response. J. Virol. 2022, 96, e0052322. [Google Scholar] [CrossRef]
- Dunlop, J.; Owsianka, A.; Cowton, V.; Patel, A. Current and future prophylactic vaccines for hepatitis C virus. Vaccine: Development and Therapy 2015, 2015, 31–44. [Google Scholar] [CrossRef]
- Pierce, B.G.; Keck, Z.Y.; Foung, S.K. Viral evasion and challenges of hepatitis C virus vaccine development. Curr. Opin. Virol. 2016, 20, 55–63. [Google Scholar] [CrossRef]
- Andre, P.; Komurian-Pradel, F.; Deforges, S.; Perret, M.; Berland, J.L.; Sodoyer, M.; Pol, S.; Brechot, C.; Paranhos-Baccala, G.; Lotteau, V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 2002, 76, 6919–6928. [Google Scholar] [CrossRef] [PubMed]
- Fauvelle, C.; Colpitts, C.C.; Keck, Z.Y.; Pierce, B.G.; Foung, S.K.; Baumert, T.F. Hepatitis C virus vaccine candidates inducing protective neutralizing antibodies. Expert. Rev. Vaccines 2016, 15, 1535–1544. [Google Scholar] [CrossRef]
- Walker, C.M.; Grakoui, A. Hepatitis C virus: why do we need a vaccine to prevent a curable persistent infection? Current opinion in immunology 2015, 35, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Carlsen, T.H.; Pedersen, J.; Prentoe, J.C.; Giang, E.; Keck, Z.Y.; Mikkelsen, L.S.; Law, M.; Foung, S.K.; Bukh, J. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology 2014, 60, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- de Jong, Y.P.; Dorner, M.; Mommersteeg, M.C.; Xiao, J.W.; Balazs, A.B.; Robbins, J.B.; Winer, B.Y.; Gerges, S.; Vega, K.; Labitt, R.N.; et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci. Transl. Med. 2014, 6, 254ra129. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, V.J.; Massaccesi, G.; Flyak, A.I.; Mankowski, M.C.; Colbert, M.D.; Osburn, W.O.; Ray, S.C.; Cox, A.L.; Crowe, J.E., Jr.; Bailey, J.R. Plasma deconvolution identifies broadly neutralizing antibodies associated with hepatitis C virus clearance. J. Clin. Invest. 2019, 129, 4786–4796. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Maruyama, T.; Lewis, J.; Giang, E.; Tarr, A.W.; Stamataki, Z.; Gastaminza, P.; Chisari, F.V.; Jones, I.M.; Fox, R.I.; et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 2008, 14, 25–27. [Google Scholar] [CrossRef]
- Mankowski, M.C.; Kinchen, V.J.; Wasilewski, L.N.; Flyak, A.I.; Ray, S.C.; Crowe, J.E., Jr.; Bailey, J.R. Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms. Proc. Natl. Acad. Sci. U S A 2018, 115, E82–E91. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.R.; Laskey, S.; Wasilewski, L.N.; Munshaw, S.; Fanning, L.J.; Kenny-Walsh, E.; Ray, S.C. Constraints on viral evolution during chronic hepatitis C virus infection arising from a common-source exposure. J. Virol. 2012, 86, 12582–12590. [Google Scholar] [CrossRef]
- Logvinoff, C.; Major, M.E.; Oldach, D.; Heyward, S.; Talal, A.; Balfe, P.; Feinstone, S.M.; Alter, H.; Rice, C.M.; McKeating, J.A. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. U S A 2004, 101, 10149–10154. [Google Scholar] [CrossRef] [PubMed]
- Osburn, W.O.; Snider, A.E.; Wells, B.L.; Latanich, R.; Bailey, J.R.; Thomas, D.L.; Cox, A.L.; Ray, S.C. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology 2014, 59, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.M.; Zeisel, M.B.; Blaser, E.; Schurmann, P.; Bartosch, B.; Cosset, F.L.; Patel, A.H.; Meisel, H.; Baumert, J.; Viazov, S.; et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. U S A 2007, 104, 6025–6030. [Google Scholar] [CrossRef] [PubMed]
- Bowen, D.G.; Walker, C.M. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 2005, 436, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.H.; Cox, A.; Hoover, D.R.; Wang, X.H.; Mao, Q.; Ray, S.; Strathdee, S.A.; Vlahov, D.; Thomas, D.L. Protection against persistence of hepatitis C. Lancet 2002, 359, 1478–1483. [Google Scholar] [CrossRef]
- Osburn, W.O.; Fisher, B.E.; Dowd, K.A.; Urban, G.; Liu, L.; Ray, S.C.; Thomas, D.L.; Cox, A.L. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology 2010, 138, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Page, K.; Hahn, J.A.; Evans, J.; Shiboski, S.; Lum, P.; Delwart, E.; Tobler, L.; Andrews, W.; Avanesyan, L.; Cooper, S.; et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J. Infect. Dis. 2009, 200, 1216–1226. [Google Scholar] [CrossRef]
- Grebely, J.; Prins, M.; Hellard, M.; Cox, A.L.; Osburn, W.O.; Lauer, G.; Page, K.; Lloyd, A.R.; Dore, G.J.; International Collaboration of Incident, H.I.V.; et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect. Dis. 2012, 12, 408–414. [Google Scholar] [CrossRef]
- Keck, Z.Y.; Wang, Y.; Lau, P.; Lund, G.; Rangarajan, S.; Fauvelle, C.; Liao, G.C.; Holtsberg, F.W.; Warfield, K.L.; Aman, M.J.; et al. Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice. Hepatology 2016, 64, 1922–1933. [Google Scholar] [CrossRef] [PubMed]
- Morin, T.J.; Broering, T.J.; Leav, B.A.; Blair, B.M.; Rowley, K.J.; Boucher, E.N.; Wang, Y.; Cheslock, P.S.; Knauber, M.; Olsen, D.B.; et al. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog. 2012, 8, e1002895. [Google Scholar] [CrossRef]
- Bukh, J.; Engle, R.E.; Faulk, K.; Wang, R.Y.; Farci, P.; Alter, H.J.; Purcell, R.H. Immunoglobulin with High-Titer In Vitro Cross-Neutralizing Hepatitis C Virus Antibodies Passively Protects Chimpanzees from Homologous, but Not Heterologous, Challenge. J. Virol. 2015, 89, 9128–9132. [Google Scholar] [CrossRef] [PubMed]
- Dorner, M.; Horwitz, J.A.; Robbins, J.B.; Barry, W.T.; Feng, Q.; Mu, K.; Jones, C.T.; Schoggins, J.W.; Catanese, M.T.; Burton, D.R.; et al. A genetically humanized mouse model for hepatitis C virus infection. Nature 2011, 474, 208–211. [Google Scholar] [CrossRef]
- Farci, P.; Shimoda, A.; Wong, D.; Cabezon, T.; De Gioannis, D.; Strazzera, A.; Shimizu, Y.; Shapiro, M.; Alter, H.J.; Purcell, R.H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. U S A 1996, 93, 15394–15399. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Maruyama, T.; Lewis, J.; Giang, E.; Tarr, A.W.; Stamataki, Z.; Gastaminza, P.; Chisari, F.V.; Jones, I.M.; Fox, R.I.; et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 2008, 14, 25–27. [Google Scholar] [CrossRef]
- Law, J.L.; Chen, C.; Wong, J.; Hockman, D.; Santer, D.M.; Frey, S.E.; Belshe, R.B.; Wakita, T.; Bukh, J.; Jones, C.T.; et al. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One 2013, 8, e59776. [Google Scholar] [CrossRef]
- Stamataki, Z.; Coates, S.; Abrignani, S.; Houghton, M.; McKeating, J.A. Immunization of human volunteers with hepatitis C virus envelope glycoproteins elicits antibodies that cross-neutralize heterologous virus strains. The Journal of infectious diseases 2011, 204, 811–813. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Keck, Z.Y.; Lau, P.; Fauvelle, C.; Gowthaman, R.; Baumert, T.F.; Fuerst, T.R.; Mariuzza, R.A.; Foung, S.K.H. Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design. Proc. Natl. Acad. Sci. U S A 2016, 113, E6946–E6954. [Google Scholar] [CrossRef]
- Gopal, R.; Jackson, K.; Tzarum, N.; Kong, L.; Ettenger, A.; Guest, J.; Pfaff, J.M.; Barnes, T.; Honda, A.; Giang, E.; et al. Probing the antigenicity of hepatitis C virus envelope glycoprotein complex by high-throughput mutagenesis. PLoS Pathog. 2017, 13, e1006735. [Google Scholar] [CrossRef]
- Pfaff-Kilgore, J.M.; Davidson, E.; Kadash-Edmondson, K.; Hernandez, M.; Rosenberg, E.; Chambers, R.; Castelli, M.; Clementi, N.; Mancini, N.; Bailey, J.R.; et al. Sites of vulnerability in HCV E1E2 identified by comprehensive functional screening. Cell Rep. 2022, 39, 110859. [Google Scholar] [CrossRef] [PubMed]
- Petruzziello, A.; Marigliano, S.; Loquercio, G.; Cozzolino, A.; Cacciapuoti, C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J. Gastroenterol. 2016, 22, 7824–7840. [Google Scholar] [CrossRef] [PubMed]
- Augestad, E.H.; Olesen, C.H.; Grønberg, C.; Soerensen, A.; Velázquez-Moctezuma, R.; Fanalista, M.; Bukh, J.; Wang, K.; Gourdon, P.; Prentoe, J. Structure of the hepatitis C virus E1/E2 envelope proteins in a homodimeric complex. bioRxiv 2023. [Google Scholar] [CrossRef]
- von Hahn, T.; Yoon, J.C.; Alter, H.; Rice, C.M.; Rehermann, B.; Balfe, P.; McKeating, J.A. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 2007, 132, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Broering, T.J.; Garrity, K.A.; Boatright, N.K.; Sloan, S.E.; Sandor, F.; Thomas, W.D., Jr.; Szabo, G.; Finberg, R.W.; Ambrosino, D.M.; Babcock, G.J. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J. Virol. 2009, 83, 12473–12482. [Google Scholar] [CrossRef]
- Chung, R.T.; Gordon, F.D.; Curry, M.P.; Schiano, T.D.; Emre, S.; Corey, K.; Markmann, J.F.; Hertl, M.; Pomposelli, J.J.; Pomfret, E.A.; et al. Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant. Surgeons 2013, 13, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Babcock, G.J.; Iyer, S.; Smith, H.L.; Wang, Y.; Rowley, K.; Ambrosino, D.M.; Zamore, P.D.; Pierce, B.G.; Molrine, D.C.; Weng, Z. High-throughput sequencing analysis of post-liver transplantation HCV E2 glycoprotein evolution in the presence and absence of neutralizing monoclonal antibody. PLoS One 2014, 9, e100325. [Google Scholar] [CrossRef]
- Pantua, H.; Diao, J.; Ultsch, M.; Hazen, M.; Mathieu, M.; McCutcheon, K.; Takeda, K.; Date, S.; Cheung, T.K.; Phung, Q.; et al. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J. Mol. Biol. 2013, 425, 1899–1914. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.R.; Wasilewski, L.N.; Snider, A.E.; El-Diwany, R.; Osburn, W.O.; Keck, Z.; Foung, S.K.; Ray, S.C. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J. Clin. Invest. 2015, 125, 437–447. [Google Scholar] [CrossRef]
- El-Diwany, R.; Cohen, V.J.; Mankowski, M.C.; Wasilewski, L.N.; Brady, J.K.; Snider, A.E.; Osburn, W.O.; Murrell, B.; Ray, S.C.; Bailey, J.R. Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog. 2017, 13, e1006235. [Google Scholar] [CrossRef]
- Augestad, E.H.; Castelli, M.; Clementi, N.; Stroh, L.J.; Krey, T.; Burioni, R.; Mancini, N.; Bukh, J.; Prentoe, J. Global and local envelope protein dynamics of hepatitis C virus determine broad antibody sensitivity. Sci. Adv. 2020, 6, eabb5938. [Google Scholar] [CrossRef]
- Salas, J.H.; Urbanowicz, R.A.; Guest, J.D.; Frumento, N.; Figueroa, A.; Clark, K.E.; Keck, Z.; Cowton, V.M.; Cole, S.J.; Patel, A.H.; et al. An Antigenically Diverse, Representative Panel of Envelope Glycoproteins for Hepatitis C Virus Vaccine Development. Gastroenterology 2022, 162, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Bankwitz, D.; Bahai, A.; Labuhn, M.; Doepke, M.; Ginkel, C.; Khera, T.; Todt, D.; Stroh, L.J.; Dold, L.; Klein, F.; et al. Hepatitis C reference viruses highlight potent antibody responses and diverse viral functional interactions with neutralising antibodies. Gut 2020. [Google Scholar] [CrossRef]
- Chumbe, A.; Grobben, M.; Capella-Pujol, J.; Koekkoek, S.M.; Zon, I.; Slamanig, S.; Merat, S.J.; Beaumont, T.; Sliepen, K.; Schinkel, J.; et al. A panel of hepatitis C virus glycoproteins for the characterization of antibody responses using antibodies with diverse recognition and neutralization patterns. Virus Res. 2024, 341, 199308. [Google Scholar] [CrossRef]
- deCamp, A.; Hraber, P.; Bailer, R.T.; Seaman, M.S.; Ochsenbauer, C.; Kappes, J.; Gottardo, R.; Edlefsen, P.; Self, S.; Tang, H.; et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2014, 88, 2489–2507. [Google Scholar] [CrossRef] [PubMed]
- Sevvana, M.; Keck, Z.; Foung, S.K.; Kuhn, R.J. Structural perspectives on HCV humoral immune evasion mechanisms. Curr. Opin. Virol. 2021, 49, 92–101. [Google Scholar] [CrossRef]
- Keck, M.L.; Wrensch, F.; Pierce, B.G.; Baumert, T.F.; Foung, S.K.H. Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine. Frontiers in immunology 2018, 9, 1194. [Google Scholar] [CrossRef]
- Kong, L.; Giang, E.; Nieusma, T.; Robbins, J.B.; Deller, M.C.; Stanfield, R.L.; Wilson, I.A.; Law, M. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 2012, 86, 13085–13088. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Giang, E.; Robbins, J.B.; Stanfield, R.L.; Burton, D.R.; Wilson, I.A.; Law, M. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl. Acad. Sci. U S A 2012, 109, 9499–9504. [Google Scholar] [CrossRef]
- Meunier, J.C.; Russell, R.S.; Goossens, V.; Priem, S.; Walter, H.; Depla, E.; Union, A.; Faulk, K.N.; Bukh, J.; Emerson, S.U.; et al. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J. Virol. 2008, 82, 966–973. [Google Scholar] [CrossRef]
- Wahid, A.; Helle, F.; Descamps, V.; Duverlie, G.; Penin, F.; Dubuisson, J. Disulfide bonds in hepatitis C virus glycoprotein E1 control the assembly and entry functions of E2 glycoprotein. J. Virol. 2013, 87, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Giang, E.; Dorner, M.; Prentoe, J.C.; Dreux, M.; Evans, M.J.; Bukh, J.; Rice, C.M.; Ploss, A.; Burton, D.R.; Law, M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. U S A 2012, 109, 6205–6210. [Google Scholar] [CrossRef] [PubMed]
- Flyak, A.I.; Ruiz, S.E.; Salas, J.; Rho, S.; Bailey, J.R.; Bjorkman, P.J. An ultralong CDRH2 in HCV neutralizing antibody demonstrates structural plasticity of antibodies against E2 glycoprotein. eLife 2020, 9. [Google Scholar] [CrossRef]
- Kumar, A.; Rohe, T.C.; Elrod, E.J.; Khan, A.G.; Dearborn, A.D.; Kissinger, R.; Grakoui, A.; Marcotrigiano, J. Regions of hepatitis C virus E2 required for membrane association. Nat. Commun. 2023, 14, 433. [Google Scholar] [CrossRef] [PubMed]
- Tzarum, N.; Giang, E.; Kadam, R.U.; Chen, F.; Nagy, K.; Augestad, E.H.; Velazquez-Moctezuma, R.; Keck, Z.Y.; Hua, Y.; Stanfield, R.L.; et al. An alternate conformation of HCV E2 neutralizing face as an additional vaccine target. Sci. Adv. 2020, 6, eabb5642. [Google Scholar] [CrossRef] [PubMed]
- Tzarum, N.; Giang, E.; Kong, L.; He, L.; Prentoe, J.; Augestad, E.; Hua, Y.; Castillo, S.; Lauer, G.M.; Bukh, J.; et al. Genetic and structural insights into broad neutralization of hepatitis C virus by human V(H)1-69 antibodies. Sci. Adv. 2019, 5, eaav1882. [Google Scholar] [CrossRef]
- Kumar, A.; Hossain, R.A.; Yost, S.A.; Bu, W.; Wang, Y.; Dearborn, A.D.; Grakoui, A.; Cohen, J.I.; Marcotrigiano, J. Structural insights into hepatitis C virus receptor binding and entry. Nature 2021, 598, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Stejskal, L.; Kalemera, M.D.; Lewis, C.B.; Palor, M.; Walker, L.; Daviter, T.; Lees, W.D.; Moss, D.S.; Kremyda-Vlachou, M.; Kozlakidis, Z.; et al. An entropic safety catch controls hepatitis C virus entry and antibody resistance. eLife 2022, 11. [Google Scholar] [CrossRef]
- Stejskal, L.; Lees, W.D.; Moss, D.S.; Palor, M.; Bingham, R.J.; Shepherd, A.J.; Grove, J. Flexibility and intrinsic disorder are conserved features of hepatitis C virus E2 glycoprotein. PLoS Comput. Biol. 2020, 16, e1007710. [Google Scholar] [CrossRef]
- Helle, F.; Vieyres, G.; Elkrief, L.; Popescu, C.I.; Wychowski, C.; Descamps, V.; Castelain, S.; Roingeard, P.; Duverlie, G.; Dubuisson, J. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 2010, 84, 11905–11915. [Google Scholar] [CrossRef]
- Drummer, H.E.; Boo, I.; Poumbourios, P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 2007, 88 Pt 4, 1144–1148. [Google Scholar] [CrossRef]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of hepatitis C virus to CD81. Science 1998, 282, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Scarselli, E.; Ansuini, H.; Cerino, R.; Roccasecca, R.M.; Acali, S.; Filocamo, G.; Traboni, C.; Nicosia, A.; Cortese, R.; Vitelli, A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002, 21, 5017–5025. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R.; Bottomley, M.J.; D’Oro, U.; Finco, O.; De Gregorio, E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 2016, 213, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Byrne, P.O.; McLellan, J.S. Principles and practical applications of structure-based vaccine design. Curr. Opin. Immunol. 2022, 77, 102209. [Google Scholar] [CrossRef] [PubMed]
- Guest, J.D.; Pierce, B.G. Structure-Based and Rational Design of a Hepatitis C Virus Vaccine. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.S.; Chen, M.; Joyce, M.G.; Sastry, M.; Stewart-Jones, G.B.; Yang, Y.; Zhang, B.; Chen, L.; Srivatsan, S.; Zheng, A.; et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 2013, 342, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.J.; Kanekiyo, M.; Kong, W.P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Widge, A.T.; Hofstetter, A.R.; Houser, K.V.; Awan, S.F.; Chen, G.L.; Burgos Florez, M.C.; Berkowitz, N.M.; Mendoza, F.; Hendel, C.S.; Holman, L.A.; et al. An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med. 2023, 15, eade4790. [Google Scholar] [CrossRef]
- Ruckwardt, T.J. The road to approved vaccines for respiratory syncytial virus. NPJ Vaccines 2023, 8, 138. [Google Scholar] [CrossRef]
- Sandomenico, A.; Leonardi, A.; Berisio, R.; Sanguigno, L.; Foca, G.; Foca, A.; Ruggiero, A.; Doti, N.; Muscariello, L.; Barone, D.; et al. Generation and Characterization of Monoclonal Antibodies against a Cyclic Variant of Hepatitis C Virus E2 Epitope 412-422. J. Virol. 2016, 90, 3745–3759. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Cheng, Y.; Kong, L.; Azadnia, P.; Giang, E.; Kim, J.; Wood, M.R.; Wilson, I.A.; Law, M.; Zhu, J. Approaching rational epitope vaccine design for hepatitis C virus with meta-server and multivalent scaffolding. Sci. Rep. 2015, 5, 12501. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Boucher, E.N.; Piepenbrink, K.H.; Ejemel, M.; Rapp, C.A.; Thomas, W.D.; Sundberg, E.J.; Weng, Z.; Wang, Y. Structure-Based Design of Hepatitis C Virus Vaccines That Elicit Neutralizing Antibody Responses to a Conserved Epitope. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Tzarum, N.; Lin, X.; Shapero, B.; Sou, C.; Mann, C.J.; Stano, A.; Zhang, L.; Nagy, K.; Giang, E.; et al. Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines. Sci. Adv. 2020, 6, eaaz6225. [Google Scholar] [CrossRef]
- Vietheer, P.T.; Boo, I.; Gu, J.; McCaffrey, K.; Edwards, S.; Owczarek, C.; Hardy, M.P.; Fabri, L.; Center, R.J.; Poumbourios, P.; et al. The core domain of hepatitis C virus glycoprotein E2 generates potent cross-neutralizing antibodies in guinea pigs. Hepatology 2017, 65, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Keck, Z.Y.; Wang, R.; Lau, P.; Garagusi, K.; Elkholy, K.; Toth, E.A.; Urbanowicz, R.A.; Guest, J.D.; Agnihotri, P. Structure-Based Design of Hepatitis C Virus E2 Glycoprotein Improves Serum Binding and Cross-Neutralization. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Frumento, N.; Figueroa, A.; Wang, T.; Zahid, M.N.; Wang, S.; Massaccesi, G.; Stavrakis, G.; Crowe, J.E.; Flyak, A.I.; Ji, H. Repeated exposure to heterologous hepatitis C viruses associates with enhanced neutralizing antibody breadth and potency. J. Clin. Invest. 2022, 132. [Google Scholar] [CrossRef]
- Tarr, A.W.; Backx, M.; Hamed, M.R.; Urbanowicz, R.A.; McClure, C.P.; Brown, R.J.P.; Ball, J.K. Immunization with a synthetic consensus hepatitis C virus E2 glycoprotein ectodomain elicits virus-neutralizing antibodies. Antiviral research 2018, 160, 25–37. [Google Scholar] [CrossRef]
- Liao, H.X.; Tsao, C.Y.; Alam, S.M.; Muldoon, M.; Vandergrift, N.; Ma, B.J.; Lu, X.; Sutherland, L.L.; Scearce, R.M.; Bowman, C.; et al. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J. Virol. 2013, 87, 4185–4201. [Google Scholar] [CrossRef]
- Sliepen, K.; Han, B.W.; Bontjer, I.; Mooij, P.; Garces, F.; Behrens, A.J.; Rantalainen, K.; Kumar, S.; Sarkar, A.; Brouwer, P.J.M.; et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 2019, 10, 2355. [Google Scholar] [CrossRef]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.J.; Howarth, M.; West, A.P.; et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines Against Respiratory Viruses. Front. Immunol. 2019, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Wu, Y.; Kim, Y.B.; Oh, Y.K. Advances in vaccine delivery systems against viral infectious diseases. Drug Deliv. Transl. Res. 2021, 11, 1401–1419. [Google Scholar] [CrossRef] [PubMed]
- Kushnir, N.; Streatfield, S.J.; Yusibov, V. Virus-like particles as a highly efficient vaccine platform: diversity of targets and production systems and advances in clinical development. Vaccine 2012, 31, 58–83. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kroll, A.V.; Holay, M.; Fang, R.H.; Zhang, L. Biomimetic Nanotechnology toward Personalized Vaccines. Adv. Mater. 2020, 32, e1901255. [Google Scholar] [CrossRef] [PubMed]
- De Temmerman, M.L.; Rejman, J.; Demeester, J.; Irvine, D.J.; Gander, B.; De Smedt, S.C. Particulate vaccines: on the quest for optimal delivery and immune response. Drug Discov. Today 2011, 16, 569–582. [Google Scholar] [CrossRef]
- Aung, A.; Cui, A.; Maiorino, L.; Amini, A.P.; Gregory, J.R.; Bukenya, M.; Zhang, Y.; Lee, H.; Cottrell, C.A.; Morgan, D.M.; et al. Low protease activity in B cell follicles promotes retention of intact antigens after immunization. Science 2023, 379, eabn8934. [Google Scholar] [CrossRef] [PubMed]
- Agger, E.M.; Rosenkrands, I.; Hansen, J.; Brahimi, K.; Vandahl, B.S.; Aagaard, C.; Werninghaus, K.; Kirschning, C.; Lang, R.; Christensen, D.; et al. Cationic liposomes formulated with synthetic mycobacterial cordfactor (CAF01): a versatile adjuvant for vaccines with different immunological requirements. PLoS One 2008, 3, e3116. [Google Scholar] [CrossRef]
- Coeshott, C.M.; Smithson, S.L.; Verderber, E.; Samaniego, A.; Blonder, J.M.; Rosenthal, G.J.; Westerink, M.A. Pluronic F127-based systemic vaccine delivery systems. Vaccine 2004, 22, 2396–2405. [Google Scholar] [CrossRef]
- Cox, M.M.; Patriarca, P.A.; Treanor, J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respir. Viruses 2008, 2, 211–219. [Google Scholar] [CrossRef]
- Didierlaurent, A.M.; Laupeze, B.; Di Pasquale, A.; Hergli, N.; Collignon, C.; Garcon, N. Adjuvant system AS01: helping to overcome the challenges of modern vaccines. Expert. review of vaccines 2017, 16, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Holtz, K.M.; Robinson, P.S.; Matthews, E.E.; Hashimoto, Y.; McPherson, C.E.; Khramtsov, N.; Reifler, M.J.; Meghrous, J.; Rhodes, D.G.; Cox, M.M.; et al. Modifications of cysteine residues in the transmembrane and cytoplasmic domains of a recombinant hemagglutinin protein prevent cross-linked multimer formation and potency loss. BMC Biotechnol. 2014, 14, 111. [Google Scholar] [CrossRef] [PubMed]
- Schwendener, R.A. Liposomes as vaccine delivery systems: a review of the recent advances. Ther. Adv. Vaccines 2014, 2, 159–182. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.H.; Patel, N.; Haupt, R.; Zhou, H.; Weston, S.; Hammond, H.; Logue, J.; Portnoff, A.D.; Norton, J.; Guebre-Xabier, M.; et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Bale, J.B.; Gonen, S.; Liu, Y.; Sheffler, W.; Ellis, D.; Thomas, C.; Cascio, D.; Yeates, T.O.; Gonen, T.; King, N.P.; et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 2016, 353, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Bruun, T.U.J.; Andersson, A.C.; Draper, S.J.; Howarth, M. Engineering a Rugged Nanoscaffold To Enhance Plug-and-Display Vaccination. ACS Nano 2018, 12, 8855–8866. [Google Scholar] [CrossRef] [PubMed]
- Domingo, G.J.; Orru, S.; Perham, R.N. Multiple display of peptides and proteins on a macromolecular scaffold derived from a multienzyme complex. J. Mol. Biol. 2001, 305, 259–267. [Google Scholar] [CrossRef]
- Hsia, Y.; Bale, J.B.; Gonen, S.; Shi, D.; Sheffler, W.; Fong, K.K.; Nattermann, U.; Xu, C.; Huang, P.S.; Ravichandran, R.; et al. Design of a hypersTable 60-subunit protein dodecahedron. [corrected]. Nature 2016, 535, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sagaseta, J.; Malito, E.; Rappuoli, R.; Bottomley, M.J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 2016, 14, 58–68. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Z.; Zhou, X.; Guo, Z.; Zhang, J.; Zhu, P.; Yao, S.; Zhu, M. Ferritin nanoparticle-based SpyTag/SpyCatcher-enabled click vaccine for tumor immunotherapy. Nanomedicine 2019, 16, 69–78. [Google Scholar] [CrossRef]
- Wei, Y.; Kumar, P.; Wahome, N.; Mantis, N.J.; Middaugh, C.R. Biomedical Applications of Lumazine Synthase. J. Pharm. Sci. 2018, 107, 2283–2296. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; Radic, L.; Capella-Pujol, J.; Watanabe, Y.; Zon, I.; Chumbe, A.; Lee, W.H.; de Gast, M.; Koopsen, J.; Koekkoek, S.; et al. Induction of cross-neutralizing antibodies by a permuted hepatitis C virus glycoprotein nanoparticle vaccine candidate. Nat. Commun. 2022, 13, 7271. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, X.; Lou, P.; Hu, Z.; Qu, P.; Li, D.; Li, Q.; Xu, Y.; Niu, J.; He, Y.; et al. A Nanoparticle-Based Hepatitis C Virus Vaccine With Enhanced Potency. J. Infect. Dis. 2020, 221, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Frumento, N.; Figueroa, A.; Wang, T.; Zahid, M.N.; Wang, S.; Massaccesi, G.; Stavrakis, G.; Crowe, J.E., Jr.; Flyak, A.I.; Ji, H.; et al. Repeated exposure to heterologous hepatitis C viruses associates with enhanced neutralizing antibody breadth and potency. J. Clin. Invest. 2022, 132. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; van Doremalen, N.; Greaney, A.J.; Andersen, H.; Sharma, A.; Starr, T.N.; Keeffe, J.R.; Fan, C.; Schulz, J.E.; Gnanapragasam, P.N.P.; et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 2022, 377, eabq0839. [Google Scholar] [CrossRef]
- Cohen, A.A.; Yang, Z.; Gnanapragasam, P.N.P.; Ou, S.; Dam, K.A.; Wang, H.; Bjorkman, P.J. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS One 2021, 16, e0247963. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.B.; Kim, H.; Jeong, J.H.; Jang, U.S.; Jang, Y.; Roh, S.; Jeon, H.; Kim, E.J.; Han, S.Y.; Maeng, J.Y.; et al. Mosaic RBD nanoparticles induce intergenus cross-reactive antibodies and protect against SARS-CoV-2 challenge. Proc. Natl. Acad. Sci. U S A 2023, 120, e2208425120. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, S.; Zhang, Y.; Yuan, L.; Zheng, Q.; Wei, M.; Shi, Y.; Wang, Z.; Ma, J.; Wang, K.; et al. Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine. Cell Host Microbe 2022, 30, 1732–1744. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, S.; Liu, J.; Chen, R.; Zhang, Y.; Lin, Y.; Xi, Z.; Deng, J.; Pu, Z.; Liang, C.; et al. A Mosaic Nanoparticle Vaccine Elicits Potent Mucosal Immune Response with Significant Cross-Protection Activity against Multiple SARS-CoV-2 Sublineages. Adv Sci (Weinh) 2023, 10, e2301034. [Google Scholar] [CrossRef]
- Baden, L.R.; Walsh, S.R.; Seaman, M.S.; Cohen, Y.Z.; Johnson, J.A.; Licona, J.H.; Filter, R.D.; Kleinjan, J.A.; Gothing, J.A.; Jennings, J.; et al. First-in-Human Randomized, Controlled Trial of Mosaic HIV-1 Immunogens Delivered via a Modified Vaccinia Ankara Vector. J. Infect. Dis. 2018, 218, 633–644. [Google Scholar] [CrossRef]
- Barouch, D.H.; Tomaka, F.L.; Wegmann, F.; Stieh, D.J.; Alter, G.; Robb, M.L.; Michael, N.L.; Peter, L.; Nkolola, J.P.; Borducchi, E.N.; et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19). Lancet 2018, 392, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Maciel, M., Jr.; Amara, R.R.; Bar, K.J.; Crotty, S.; Deeks, S.G.; Duplessis, C.; Gaiha, G.; McElrath, M.J.; McMichael, A.; Palin, A.; et al. Exploring synergies between B- and T-cell vaccine approaches to optimize immune responses against HIV-workshop report. NPJ Vaccines 2024, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Murji, A.A.; Qin, J.S.; Hermanus, T.; Morris, L.; Georgiev, I.S. Elicitation of Neutralizing Antibody Responses to HIV-1 Immunization with Nanoparticle Vaccine Platforms. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Allander, T.; Drakenberg, K.; Beyene, A.; Rosa, D.; Abrignani, S.; Houghton, M.; Widell, A.; Grillner, L.; Persson, M.A. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J. Gen. Virol. 2000, 81 Pt 10, 2451–2459. [Google Scholar] [CrossRef]
- Bugli, F.; Mancini, N.; Kang, C.Y.; Di Campli, C.; Grieco, A.; Manzin, A.; Gabrielli, A.; Gasbarrini, A.; Fadda, G.; Varaldo, P.E.; et al. Mapping B-cell epitopes of hepatitis C virus E2 glycoprotein using human monoclonal antibodies from phage display libraries. J. Virol. 2001, 75, 9986–9990. [Google Scholar] [CrossRef] [PubMed]
- Drummer, H.E. Challenges to the development of vaccines to hepatitis C virus that elicit neutralizing antibodies. Frontiers in microbiology 2014, 5, 329. [Google Scholar] [CrossRef]
- Habersetzer, F.; Fournillier, A.; Dubuisson, J.; Rosa, D.; Abrignani, S.; Wychowski, C.; Nakano, I.; Trepo, C.; Desgranges, C.; Inchauspe, G. Characterization of human monoclonal antibodies specific to the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology 1998, 249, 32–41. [Google Scholar] [CrossRef]
- Hadlock, K.G.; Lanford, R.E.; Perkins, S.; Rowe, J.; Yang, Q.; Levy, S.; Pileri, P.; Abrignani, S.; Foung, S.K. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 2000, 74, 10407–10416. [Google Scholar] [CrossRef]
- Keck, Z.; Wang, W.; Wang, Y.; Lau, P.; Carlsen, T.H.; Prentoe, J.; Xia, J.; Patel, A.H.; Bukh, J.; Foung, S.K. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J. Virol. 2013, 87, 37–51. [Google Scholar] [CrossRef]
- Keck, Z.Y.; Li, T.K.; Xia, J.; Bartosch, B.; Cosset, F.L.; Dubuisson, J.; Foung, S.K. Analysis of a highly flexible conformational immunogenic domain a in hepatitis C virus E2. J. Virol. 2005, 79, 13199–13208. [Google Scholar] [CrossRef] [PubMed]
- Keck, Z.Y.; Li, T.K.; Xia, J.; Gal-Tanamy, M.; Olson, O.; Li, S.H.; Patel, A.H.; Ball, J.K.; Lemon, S.M.; Foung, S.K. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J. Virol. 2008, 82, 6061–6066. [Google Scholar] [CrossRef] [PubMed]
- Keck, Z.Y.; Xia, J.; Wang, Y.; Wang, W.; Krey, T.; Prentoe, J.; Carlsen, T.; Li, A.Y.; Patel, A.H.; Lemon, S.M.; et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012, 8, e1002653. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Sidney, J.; Southwood, S.; Ruppert, J.; Oseroff, C.; Maewal, A.; Snoke, K.; Serra, H.M.; Kubo, R.T.; Sette, A.; et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1994, 1, 751–761. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chaudhary, A.; Lin, X.; Sou, C.; Alkutkar, T.; Kumar, S.; Ngo, T.; Kosviner, E.; Ozorowski, G.; Stanfield, R.L.; et al. Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines. Nat. Commun. 2021, 12, 2633. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lin, X.; Wang, Y.; Abraham, C.; Sou, C.; Ngo, T.; Zhang, Y.; Wilson, I.A.; Zhu, J. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Paynter, J.; Antanasijevic, A.; Allen, J.D.; Eldad, M.; Lee, Y.Z.; Copps, J.; Newby, M.L.; He, L.; Chavez, D.; et al. Single-component multilayered self-assembling protein nanoparticles presenting glycan-trimmed uncleaved prefusion optimized envelope trimmers as HIV-1 vaccine candidates. Nat. Commun. 2023, 14, 1985. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




