1. Introduction
Pancreatic cancer (PC) stands as the tenth most common cancer type globally for both sexes, yet it ranks fourth in mortality rate, reflecting its aggressive nature and limited treatment options [
1]. Approximately 10% of all pancreatic cancer cases are purportedly linked to Familial Pancreatic Cancer (FPC), denoting families exhibiting a notably high incidence of the disease. FPC is recognized as a genetically heterogeneous condition, driven by inherited pathogenic variants within specific genes [
2].
PC manifests within a spectrum of cancer predisposing syndromes, including Hereditary Breast and Ovarian Cancer (HBOC), Peutz-Jeghers, Familial Adenomatous Polyposis (FAP), Lynch, Li-Fraumeni, and Familial Atypical Multiple Mole Melanoma (FAMMM) syndromes. Additionally, clear associations have been established between pancreatic cancer and loss-of-function variants in non-syndromic cancer genes such as
PALB2 and
ATM [
3].
Advancements in genetic testing technologies, particularly Next-Generation sequencing (NGS), have revolutionized the landscape of genetic risk assessment. NGS enables the simultaneous analysis of multiple cancer-associated genes, facilitating the identification of pathogenic alterations [
4]. The National Comprehensive Cancer Network (NCCN) guidelines has underscored the significance of genetic risk assessment by updating its recommendations in 2019 to include pancreatic cancer patients. According to these guidelines, all individuals diagnosed with pancreatic cancer should be offered genetic testing to elucidate potential predisposing genetic alterations [
5].
Panel testing, encompassing a broad array of cancer-risk genes, holds promise in augmenting etiologic insights and refining genetic risk assessment in pancreatic cancer patients, irrespective of their familial history. Furthermore, germline genetic testing in pancreatic cancer serves multifaceted purposes, aiding in the selection of personalized treatment strategies and informing family members about their risk profiles, thus enabling them to pursue appropriate screening and preventive measures [
6].
Our research aimed to investigate the prevalence of germline pathogenic or likely pathogenic variants in cancer predisposing genes among patients diagnosed with pancreatic cancer.
2. Materials and Methods
Between July 2020 and July 2023, individuals diagnosed with pancreatic cancer were directed to Genekor Laboratory for germline genetic testing. A group comprising 184 subjects underwent assessment at the discretion of healthcare providers. Proficient personnel briefed all participants on the rationale behind germline testing and gathered data concerning their personal and familial cancer histories, including both immediate and extended family members affected by the disease. Before undergoing molecular genetic testing, patients consented by signing an informed consent document, granting permission for the anonymous utilization of their data in research endeavors and potential scientific publications.
Genomic DNA was isolated from EDTA whole peripheral blood samples utilizing the MagCore Genomic DNA Whole Blood Kit (RBC Bioscience, New Taipei City, Taipei, Taiwan, ROC) following the manufacturer’s guidelines. The concentration of DNA was quantified using the Qubit fluorometer (Thermo Fisher Scientific, Inc.).
For this study, as previously described, a targeted gene-panel comprising 52 cancer susceptibility genes was employed [
7]. The selection of genes for analysis was based on their association with hereditary cancer as outlined in the NCCN guidelines and corroborated by published research. The custom-designed probe library included coding exons and exon–intron boundaries of each gene within the panel. Library preparation was conducted using the SeqCap EZ Choice Library (Roche NimbleGen) with 150 ng of double-stranded DNA as input material. Sequencing was performed on the DNBSEQ-G400 platform (MGI Tech Co., Ltd., Beishan Industrial Zone, Shenzhen, PR China) in accordance with the manufacturer’s instructions.
Alignment to the reference sequence (hg19), variant calling, and interpretation were executed using optimized algorithms integrated into the commercial SeqPilot suite (JSI medical systems GmbH, Germany). Basecalls with a quality score of 20 or higher were considered for further analysis. Regions of Interest (ROIs), defined as exons ±20-bp intronic sequences for all genes within the panel, underwent automatic screening for homologous regions in the genome. Variants were called with a variant allele frequency (VAF) cutoff of 25% and assessed for pathogenicity according to predefined criteria.
Variant classification followed the recommendations of the American College of Medical Genetics and Genomics, categorizing variants into five groups: Pathogenic, likely pathogenic variants (PVs/LPVs) (> 90% certainty of a variant being disease-causing), variant of uncertain significance (VUS), likely benign (> 90% certainty of a variant being benign), and benign. Variant annotation and interpretation were conducted using an in-house knowledgebase and a proprietary bioinformatics pipeline designed to automate the classification process. All PVs/LPVs were validated using Sanger Sequencing on re-extracted DNA samples.
3. Results
Among the cohort of 184 patients who underwent genetic testing, 91 were women ranging in age from 42 to 82 years, while 93 were men aged between 33 and 84 years. The median age at which the study subjects were diagnosed with pancreatic cancer was 61 years. Out of the 184 cases referred for assessment, 77.7% (143 out of 184) had a personal cancer history. Specifically, 19.6% (36 out of 184) of the individuals had a previous diagnosis of pancreatic cancer. Conversely, 22.3% (41 out of 184) of the subjects had no familial cancer history. Notably, there were instances where patients diagnosed with pancreatic cancer also experienced other types of malignancies, including ovarian cancer, melanoma, glioblastoma, liver cancer, colon cancer, gastric cancer, and breast cancer.
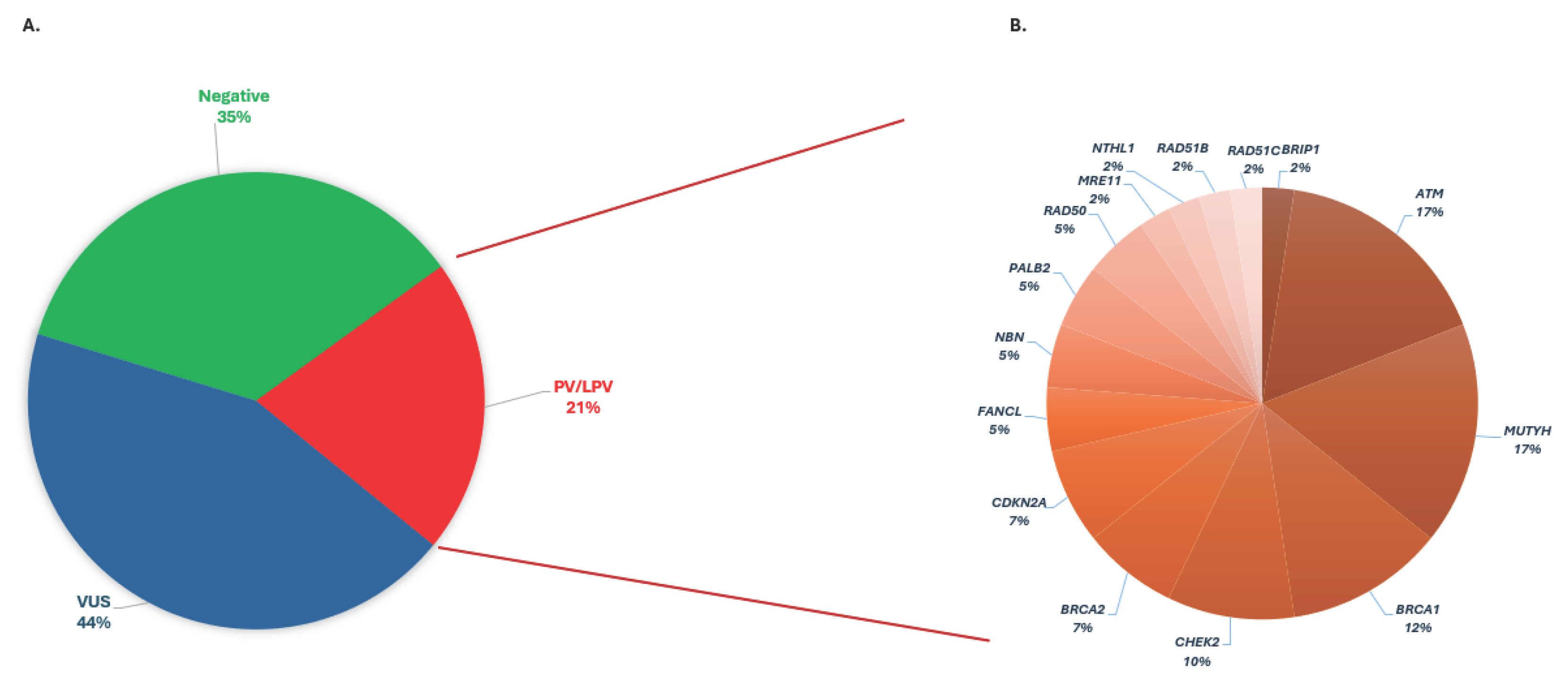
Of the 184 individuals diagnosed with pancreatic cancer, 38 were found to harbor a PV/LPV, constituting approximately 21% of the cases. Remarkably, 35% of the examinees (65 out of 184) did not exhibit any detectable variants within the 52 genes studied. However, in the remaining 44% (81 out of 184) of cases, one or more variants of uncertain significance were identified (
Figure 1A).
PVs/LPVs were identified in 21% of patients diagnosed with pancreatic cancer, spanning several key genes associated with hereditary predisposition to the disease. Among the genes analyzed, including
ATM,
BRCA1,
BRCA2,
BRIP1,
CDKN2A,
CHEK2,
FANCL,
MRE11,
MUTYH,
NBN,
NTHL1,
PALB2,
RAD50,
RAD51B and
RAD51C, significant frequencies of PVs/LPVs were observed. Specifically, PVs/LPVs were detected in
ATM in 15% (6 out of 38) and in
BRCA1 in 12% (5 out of 38) of patients. Additionally,
BRCA2 exhibited PVs/LPVs in 7% (3 out of 38) of patients, while
CHEK2 displayed PVs/LPVs in 10% (4 out of 38) of cases. Moreover,
MUTYH PVs/LPVs were found in 17% (7 out of 38) of patients, in a heterozygous state. Other genes such as
CDKN2A,
FANCL,
NBN,
PALB2, and
RAD50 also harbored PVs/LPVs, at lower frequencies ranging from 5% to 7% within the cohort
(Figure 1B
).
A total of 42 PVs/LPVs were identified across 38 patients. Among these, 16 out of the 42 variants (38%) were detected within high-risk genes associated with pancreatic cancer susceptibility, including BRCA1, BRCA2, CDKN2A, and ATM. Moreover, the majority of the detected variants, specifically 27 out of 42 (64.3%), were found in genes involved in the Homologous Recombination Repair (HRR) pathway (BRCA1, BRCA2, BRIP1, CHEK2, ATM, PALB2, MRE11, NBN, RAD51B, RAD51C, RAD51D, RAD50 and FANCL).
Among the patients identified with PVs/LPVs, 84% had a documented family history of cancer. Remarkably, within this subset, 26.4% of individuals had a family history specifically associated with pancreatic cancer. Among the individuals who tested positive, 13 out of 38 (34.21%) did not report a family history of PC or related cancers. Six out of 38 (15.78%) had a family history of PC, while 19 out of 38 (50%) had a family history involving related cancers. For those identified as having high-risk genes for PC, 4 out of 17 (23.53%) did not have a family history of PC or related cancers. Three out of 17 (17.34%) had a family history of PC, and 10 out of 17 (5.82%) had a family history involving related cancers. In addition, PVs/LPVs were found in 8 individuals heterozygous for the NTHL1 and MUTYH genes, which are inherited recessively. Among these, 4 out of 8 (50%) had no family history of PC or related cancers, 2 out of 8 (25%) had a family history of PC, and 2 out of 8 (25%) had a family history involving related cancers. The CDKN2A gene is linked to both melanoma and pancreatic cancer phenotypes. PVs/LPVs were identified in 3 patients with a family history of melanoma (100%) and in 2 patients with a family history of pancreatic cancer (67%), while those without this gene had a low incidence of melanoma in close relatives (1.65%, 3 out of 181).
In 81 out of 184 cases (44%), at least one variant of uncertain significance (VUS) was identified. Among this group, 38 out of 81 (46.41%) had no reported family history of pancreatic cancer (PC) or related cancers. Thirteen out of 81 (16.04%) had a family history of PC, while 30 out of 81 (37.03%) had a family history involving related cancers. In 65 out of 184 cases (35.32%), no pathogenic variants (PV), likely pathogenic variants (LPV), or VUS were found. Among these cases, 25 (38.46%) had no family history of PC or related cancers, 9 out of 81 (13.84%) had a family history of PC, and 29 out of 81 (44.61%) had a family history involving related cancers.
The presence of a family history of first- and/or second-degree relatives diagnosed with PC did not show a significant correlation with positive results. Across all patient subgroups (negative, VUS, positive, positive-HRR genes), the occurrence of PC in the family ranged from 13.84% to 17.64%. Conversely, High Risk genes such as BRCA1, BRCA2, and ATM were linked to a higher incidence of breast, ovarian, or prostate cancer among close family members compared to non-PV/LPV carriers (58.82% vs. 40.41%), with a significance level of p < 0.05.
4. Discussion
The use of germline genetic testing is increasingly prominent in oncology due to its potential to influence surgical strategies and treatment approaches. NGS technology has been pivotal in uncovering the genetic underpinnings of familial cancers. However, the expanding scope of NGS technology has led to increased testing complexity, including a broader array of genes being analyzed. This expansion brings challenges such as limited understanding of the management implications for newly identified genes and occasional uncertainty surrounding the clinical significance of PVs/LPVs in well-established genes [
8].
Nonetheless, the significance of expanded germline testing in PC patients is underscored by the presence of inherited pathogenic variants associated with various hereditary cancer predisposition syndromes. These syndromes encompass conditions like hereditary Breast and Ovarian cancer syndrome, Lynch syndrome, and Familial Atypical Multiple Mole Melanoma Syndrome, among others. The identification of such variant statuses can profoundly influence clinical management and treatment decisions for patients [
9].
In our current study, 184 pancreatic cancer patients were referred to our laboratory for genetic testing based on physician discretion. The panel utilized for testing comprised 52 genes associated with cancer predisposition and the development of specific cancer types. The findings revealed that 20.65% (38 out of 184) of the patients harbored at least one PV/LPV across 15 genes, some of which are known to be linked to hereditary cancer predisposition syndromes and an elevated risk of pancreatic cancer. Previous studies have documented the presence of PVs/LPVs in 8-20% of pancreatic cancer patients [
10,
11]. The variability in detection rates of PVs/LPVs can be attributed to several factors including the number of genes analyzed in each study, the genetic background of the tested population, and the methodology employed for variant classification. Furthermore, studies have reported a notably higher detection of variants among patients with metastatic disease compared to those with earlier stage disease. This variability underscores the multifaceted nature of genetic predisposition in pancreatic cancer and highlights the need for comprehensive evaluation and classification of variants in clinical practice [
12,
13].
Among the 42 PV/LPV identified, 64.3% (27 out of 42) were in genes associated with homologous recombination repair (HRR), representing 14.7% (27 out of 184) of all patients in the cohort. Our findings align closely with those of a meta-analysis, which reported similar prevalence rates of homologous recombination deficiency in pancreatic cancer patients [
14]. The detection of pathogenic variants within the DNA damage repair pathway holds clinical significance, as tumors with such alterations may exhibit heightened sensitivity to poly-ADP ribose polymerase (PARP) inhibitors, platinum-based chemotherapies, and potentially immunotherapy options [
15].
Among the PVs/LPVs detected, 40.5% (17 out of 42) were in genes strongly linked to pancreatic cancer (
BRCA1, BRCA2, ATM, CDKN2A), constituting 44.7% (17 out of 38) of
PV/LPV carriers or 9.2% (17 out of 184) of all patients. PV/LPV in
CDKN2A accounted for 7.14% (3 out of 42) of the variants detected and were observed in 7.9% (3 out of 38) of carriers. Notably,
CDKN2A PV/LPV were prevalent among patients with a family history of melanomas and pancreatic cancer, with a statistically significant association (p < .00001). The non-syndromic cancer gene
ATM and the cancer predisposition genes
BRCA1 and
BRCA2 are recognized as high penetrance genes for familial pancreatic cancer (FPC). These genes were notably associated with a higher incidence of breast, ovarian, and prostate cancer in close family members compared to PV/LPV carriers, confirming the occurrence of pancreatic cancer within a spectrum of cancer predisposing syndromes, including hereditary breast and ovarian cancer (HBOC) [
16].
PVs/LPVs in genes not conventionally associated with pancreatic cancer susceptibility (
MRE11, CHEK2, MUTYH/MYH, NBN, NTHL1, BRIP1) were also observed in 17 patients, accounting for 45.2% overall or 9.2% of the entire cohort. Among these cases, PVs were detected in heterozygosity in the
NTHL1 and
MUTYH genes in eight cases. These genes are linked to
NTHL1 tumor syndrome and
MUTYH-associated Adenomatous Polyposis (MAP), inherited in a recessive manner, implying that the monoallelic findings in these eight cases present uncertain risks for cancer development. Heterozygous pathogenic variants in the
NTHL1 gene have been associated with low to moderate increased risks for breast cancer, and another study suggested a potential contribution of monoallelic pathogenic
NTHL1 variants to tumorigenesis in certain patient subsets. Similarly, a study indicated a heightened risk for cancer among carriers of deleterious germline variants with only one defective allele in the
MUTYH gene. These incidental findings underscore the necessity for further elucidation regarding genotype–phenotype correlations and the influence of age and gender to better stratify the risk associated with these variants as high-, moderate-, or low-penetrant genes [
17].
Variants of uncertain significance were identified in 44% (81 out of 184) of patients, aligning with findings from recently published studies [
18,
19]. The noteworthy occurrence of VUS underscores the necessity for further in silico and functional investigations to assess the impact of these alterations on protein function, along with segregation analysis within families to elucidate the etiological contribution to cancer development. These data will inform the reassessment of the risk associated with these variants in relation to pancreatic cancer.
The current NCCN guidelines recommend universal germline testing for PC patients [
20]. Previously, according to the 2018 NCCN guidelines, patient selection was based on factors such as personal and family history, age of diagnosis, among others. However, germline variants in cancer susceptibility genes may also be present in patients without an evident history of inherited cancer risk.
In our study, the presence of a first- or second-degree relative diagnosed with pancreatic cancer did not correlate with carrying a PV/LPV, with the incidence of PC in the family ranging from 13.84% to 17.64% across all studied patient subgroups, consistent with recent findings. No statistically significant association was observed between carriers and non-carriers of PGVs concerning other familial cancer types. Ultimately, there were no discernible differences in sex, age, or family history of pancreatic cancer or other familial cancer types between patients based on their variant status, indicating that clinical history alone may be insufficient to identify patients carrying genetic variants.
Interestingly, 12 out of 38 patients (31.6%) with PVs/LPVs would have been overlooked, representing 6.5% of the overall cohort, as they lacked any instances of cancer among close relatives. These individuals presented no family history of pancreatic cancer or other malignancies and thus would not have met the screening recommendations outlined in the 2018 NCCN guidelines, which have since been updated. Among these patients, 3 harbored PGVs in high-risk genes (ATM, BRCA1, BRCA2), leading to distinct clinical management strategies. Furthermore, two of them had clinically actionable findings that would not have been identified based on the testing criteria specified in the 2018 NCCN guidelines.
Upon thorough review of all family history data, it is evident that over 50% of the referred patients have a family history involving close relatives diagnosed with pancreatic cancer or other cancer types associated with familial syndromes (including breast, ovarian, prostate, colon cancer, melanoma, etc.). This suggests that a subset of patients likely undergo genetic testing after meeting specific selection criteria. If guidelines continue to advocate for unselected germline testing, it becomes imperative to establish a long-term strategy to educate all healthcare providers about the new testing criteria. Additionally, it’s crucial to identify other pertinent testing restriction factors, such as patients facing financial constraints or lacking adequate genetic counseling support, among others.
The findings of this study are reinforced by a sizable cohort comprising 184 pancreatic cancer patients of diverse sexes and ages, with the utilization of an expanded 52-gene panel encompassing cancer predisposition genes and risk genes for non-syndromic cancers. Nonetheless, this research is susceptible to several limitations. The examined group primarily comprises individuals of Greek ancestry, alongside some patients of Turkish and Romanian heritage, leading to a diverse population concerning identified variants. Additionally, critical clinicopathologic features like grade, histological subtype, metastases occurrence, and follow-up information were unavailable. Moreover, germline testing was administered at the discretion of attending physicians, each possibly employing distinct referral criteria. Finally, operating as a private institution, the socioeconomic standing of the patients could impede access to germline testing.
5. Conclusions
Multigene germline testing represents a pivotal approach that should be extended to all patients diagnosed with PC, irrespective of stage, family history, or age at onset. The insights gleaned from such testing can significantly impact disease prognosis, inform treatment selection, and guide familial cancer counseling initiatives. Our study underscores and amplifies the outcomes observed in prior research endeavors, shedding light on the underutilization of universal germline testing. By integrating multigene germline testing into routine clinical practice, healthcare providers gain valuable insights into the genetic underpinnings of pancreatic cancer, which can inform personalized treatment strategies and enhance patient care outcomes. Moreover, the identification of hereditary cancer predisposition syndromes through germline testing enables the implementation of proactive screening measures for at-risk family members, thereby facilitating early detection and intervention. Despite the clear benefits and implications of universal germline testing, its widespread adoption remains limited. Addressing barriers to implementation, such as access to testing, provider education, and patient awareness, is crucial to maximize the utility of germline testing in the management of pancreatic cancer and improve outcomes for affected individuals and their families. Through concerted efforts to integrate universal germline testing into standard clinical practice, we can enhance our understanding of the genetic landscape of pancreatic cancer and optimize patient care pathways.
Author Contributions
Conceptualization, A.K. and M.O.; methodology, K.P., A.K., K.A., C.N. and N.T.; software, G.N.T.; validation, K.P., A.K., K.A., C.N. and N.T.; formal analysis, K.P., A.K., K.A., C.N. and N.T.; investigation, K.P., A.K., K.A., C.N. and N.T.; data curation, A.E.C., D.Z., M.T., C.C., E.S., D.J., I.A., A.K. writing—original draft preparation, K.P., A.K., K.A.; writing—review and editing, K.P., A.K., K.A., C.N., N.T., E.P., G.N., O.K., M.O.C., M.O.;supervision, E.P.; project administration, G.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethical Committee of Antalya Memorial Hospital (2024).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J Clin 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Liew, S.Z.H.; Ng, K.W.; Ishak, N.D.B.; Lee, S.Y.; Zhang, Z.; Chiang, J.; Ngeow, J.Y.Y. Geographical, ethnic, and genetic differences in pancreatic cancer predisposition. Chin Clin Oncol 2023, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Pflüger, M.J.; Brosens, LAA, Hruban, RH. Precursor lesions in familial and hereditary pancreatic cancer. Fam Cancer 2024. [Google Scholar]
- Hainaut, P.; Plymoth, A. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr Opin Oncol 2013, 25, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.B.; Pilarski, R.; Yurgelun, M.B.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Garber, J.E.; Goggins, M.; Hutton, M.L.; Khan, S.; Klein, C.; Kohlmann, W.; Kurian, A.W.; Laronga, C.; Litton, J.K.; Mak, J.S.; Menendez, C.S.; Merajver, S.D.; Norquist, B.S.; Offit, K.; Pal, T.; Pederson, H.J.; Reiser, G.; Shannon, K.M.; Visvanathan, K.; Weitzel, J.N.; Wick, M.J.; Wisinski, K.B.; Dwyer, M.A.; Darlow, S.D. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J Natl Compr Canc Netw 2020, 18, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.; Deluca, J.; Abenavoli, L.; Boccuto, L. Pancreatic Cancer and the Family Connection: The Role of Advanced Practitioners in Screening and Educating Genetically At-Risk Individuals. J Adv Pract Oncol 2023, 14, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Aginnitopoulos, K.; Pepe, G.; Tsaousis, G.N.; Potska, K.; Bouzarelou, D.; Katseli, A.; Ntogka, C.; Meintani, A.; Tsoulos, N.; Giassas, S.; et al. Copy Number Variations (CNVs) account for 10.8% of pathogenic variants in patients referred for hereditary cancer testing. Cancer Genom Proteom 2023, 20, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Pak, H.Y.; Heo, J.Y.; Lim, H.; Choi, Y.L.; Shim, H.S.; Kim, E.K. Trends and Clinical Characteristics of Next-Generation Sequencing-Based Genetic Panel Tests: An Analysis of Korean Nationwide Claims Data. Cancer Res Treat 2024, 56, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Anaclerio, F.; Pilenzi, L.; Dell’Elice, A.; Ferrante, R.; Grossi, S.; Ferlito, L.M.; Marinelli, C.; Gildetti, S.; Calabrese, G.; Stuppia, L.; Antonucci, I. Clinical usefulness of NGS multi-gene panel testing in hereditary cancer analysis. Front Genet 2023, 14:1060504.
- Uson, P.L.S. Jr; Samadder, N.J.; Riegert-Johnson, D.; Boardman, L.; Borad, M.J.; Ahn, D.; Sonbol, M.B.; Faigel, D.O.; Fukami, N.; Pannala, R.; Kunze, K.; Golafshar, M.; Klint, M.; Esplin, E.D.; Nussbaum, R.L.; Stewart, A.K.; Bekaii-Saab, T. Clinical Impact of Pathogenic Germline Variants in Pancreatic Cancer: Results From a Multicenter, Prospective, Universal Genetic Testing Study. Clin Transl Gastroenterol 2021, 12, e00414. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Polley, E.C.; Gnanaolivu, R.; Shimelis, H.; Lee, K.Y.; Lilyquist, J.; Na, J.; Moore, R.; Antwi, S.O.; Bamlet, W.R.; Chaffee, K.G.; DiCarlo, J.; Wu, Z.; Samara, R. Kasi, P.M.; McWilliams, R.R.; Petersen, G.M.; Couch, F.J. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA 2018, 319, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, N.J.; Furniss, C.S.; Yurgelun, M.B.; Ukaegbu, C.; Constantinou, P.E.; Fortes, I.; Caruso, A.; Schwartz, A.N.; Stopfer, J.E.; Underhill-Blazey, M.; Kenner, B.; Nelson, S.H.; Okumura, S.; Zhou, A.Y.; Coffin, T.B.; Uno, H.; Horiguchi, M.; Ocean, A.J.; McAllister, F.; Lowy, A.M.; Klein, A.P.; Madlensky, L; Petersen, G. M.; Garber, J.E.; Lippman, S.M.; Goggins, M.G.; Maitra, A.; Syngal, S. A Randomized Trial of Two Remote Health Care Delivery Models on the Uptake of Genetic Testing and Impact on Patient-Reported Psychological Outcomes in Families With Pancreatic Cancer: The Genetic Education, Risk Assessment, and Testing (GENERATE) Study. Gastroenterology 2024, S0016, 5085(24)00129-X. [Google Scholar]
- Lowery, M.A.; Wong, W.; Jordan, E.J.; Lee, J.W.; Kemel, Y.; Vijai, J.; Mandelker, D.; Zehir, A.; Capanu, M.; Salo-Mullen, E.; Arnold, A.G.; Yu, K.H.; Varghese, A.M.; Kelsen, D.P.; Brenner, R.; Kaufmann, E.; Ravichandran, V.; Mukherjee, S.; Berger, M.F.; Hyman, D.M.; Klimstra, D.S.; Abou-Alfa, G.K.; Tjan, C.; Covington, C.; Maynard, H.; Allen, P.J.; Askan, G.; Leach, S.D.; Iacobuzio-Donahue, C.A.; Robson, M.E.; Offit, K.; Stadler, Z.K.; O’Reilly, E.M. Prospective Evaluation of Germline Alterations in Patients With Exocrine Pancreatic Neoplasms. J Natl Cancer Inst 2018, 110, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, A.; Forte, G.; Fasano, C.; Lepore Signorile, M.; Sanese, P.; De Marco, K.; Di Nicola, E.; Latrofa, M.; Grossi, V.; Disciglio, V.; Simone, C. Understanding the Genetic Landscape of Pancreatic Ductal Adenocarcinoma to Support Personalized Medicine: A Systematic Review. Cancers (Basel) 2023, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- LaRose, M.; Manji, G.A.; Bates, S.E. Beyond BRCA: Diagnosis and management of homologous recombination repair deficient pancreatic cancer. Semin Oncol 2023, S0093, 7754(23)00081-7. [Google Scholar] [CrossRef] [PubMed]
- Barili V, Ambrosini E, Bortesi B, Minari R, De Sensi E, Cannizzaro IR, Taiani A, Michiara M, Sikokis A, Boggiani D, Tommasi C, Serra O, Bonatti F, Adorni A, Luberto A, Caggiati P, Martorana D, Uliana V, Percesepe A, Musolino A, Pellegrino B. Genetic Basis of Breast and Ovarian Cancer: Approaches and Lessons Learnt from Three Decades of Inherited Predisposition Testing. Genes (Basel) 2024, 15, 219. [Google Scholar] [CrossRef] [PubMed]
- Hryhorowicz, S.; Kaczmarek-Ryś, M.; Lis-Tanaś, E.; Porowski, J.; Szuman, M.; Grot, N.; Kryszczyńska, A.; Paszkowski, J.; Banasiewicz, T.; Pławski, A. Strong Hereditary Predispositions to Colorectal Cancer. Genes (Basel) 2022, 13, 2326. [Google Scholar] [CrossRef] [PubMed]
- Terashima, T.; Morizane, C.; Ushiama, M.; Shiba, S.; Takahashi, H.; Ikeda, M.; Mizuno, N.; Tsuji, K.; Yasui, K.; Azemoto, N.; Satake, H.; Nomura, S.; Yachida, S.; Sugano, K.; Furuse, J. Germline variants in cancer-predisposing genes in pancreatic cancer patients with a family history of cancer. Jpn J Clin Oncol 2022, 52, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Ceyhan-Birsoy, O.; Jayakumaran, G.; Kemel, Y.; Misyura, M.; Aypar, U.; Jairam, S.; Yang, C.; Li, Y.; Mehta, N.; Maio, A.; Arnold, A.; Salo-Mullen, E.; Sheehan, M.; Syed, A.; Walsh, M.; Carlo, M.; Robson, M.; Offit, K.; Ladanyi, M.; Reis-Filho, J.S.; Stadler, Z.K.; Zhang, L.; Latham, A.; Zehir, A.; Mandelker, D. Diagnostic yield and clinical relevance of expanded genetic testing for cancer patients. Genome Med 2022, 14, 92. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.B.; Pal, T.; Maxwell, K.N.; Churpek, J.; Kohlmann, W.; AlHilli, Z.; Arun, B.; Buys, S.S.; Cheng, H.; Domchek, S.M.; Friedman, S.; Giri, V.; Goggins, M.; Hagemann, A.; Hendrix, A.; Hutton, M.L.; Karlan, B.Y.; Kassem, N.; Khan, S.; Khoury, K.; Kurian, A.W.; Laronga, C.; Mak, J.S.; Mansour, J.; McDonnell, K.; Menendez, C.S.; Merajver, S.D.; Norquist, B.S.; Offit, K.; Rash, D.; Reiser, G.; Senter-Jamieson, L.; Shannon, K.M.; Visvanathan, K.; Welborn, J.; Wick, M.J.; Wood, M.; Yurgelun, M.B.; Dwyer, M.A.; Darlow, S.D. NCCN Guidelines® Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2024. J Natl Compr Canc Netw 2023, 21, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).