Submitted:
26 March 2024
Posted:
26 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Statistical Optimization Strategies Applied for the Extraction of Bioactive Molecules from Seafood Byproducts
2.1. Classical versus Multivariate Optimization Techniques Applied for the Extraction of Bioactive Molecules from Seafood Byproducts
2.2. Screening Extraction Parameters Used for the Extraction of Bioactive Compounds from Seafood Byproducts
2.3. Screening Used for Selecting Potential Extraction Solvents and Hydrolyzing Enzymes
2.4. Multivariate Regression Model Selection and Optimization of Screened Extraction Parameters of Bioactive Compounds
2.4.1. Response Surface Optimization (RSM) as a Tool to Optimize the Extraction Parameters of Bioactive Compounds
Choice of the RSM Experimental Design
2.4.2. Coding the Factor Levels
2.4.3. Central Composite Design
2.4.4. Box-Behnken Design
2.4.5. Full Factorial Design
2.4.6. Doehlert Design
2.4.7. Presentation of the Model and Determination of Optimal Conditions
2.4.8. Robustness, Validation and Verification of Predicted Models/Optimized Extraction Conditions
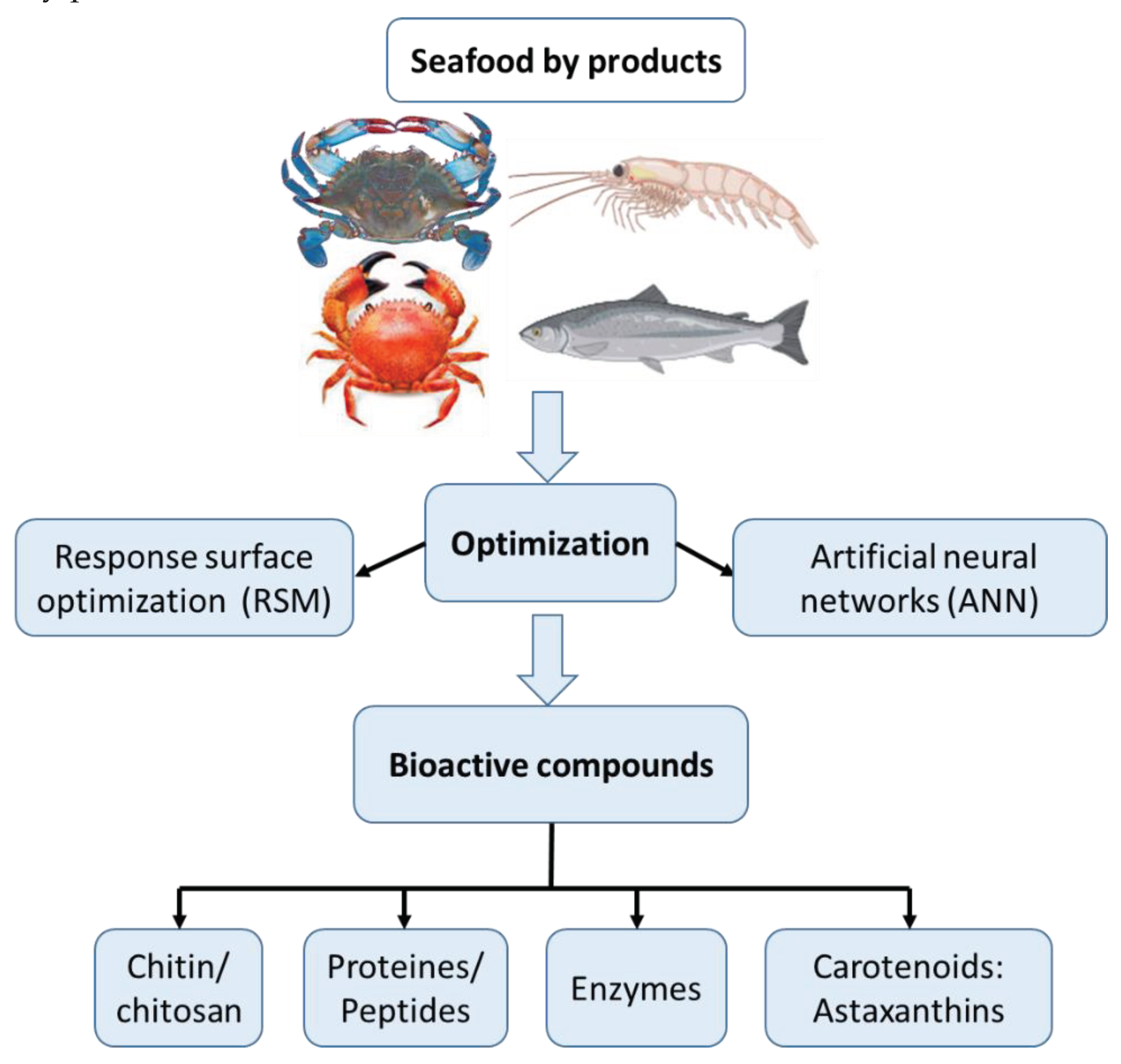
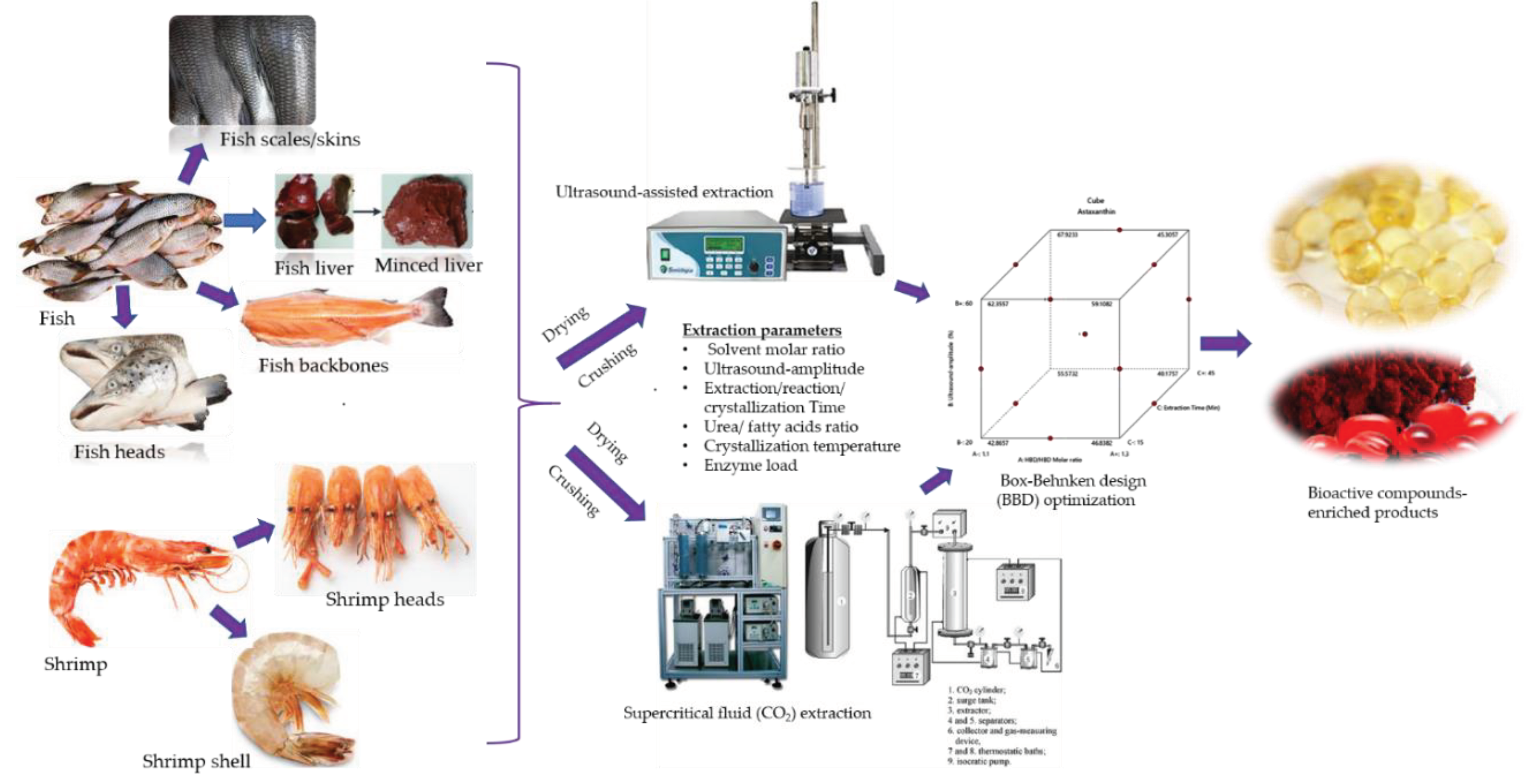
3. Extraction Process Parameters Considered for Bioactive Molecules from Seafood Byproducts
3.1. Chitin and Chitosan
3.2. Proteins and Peptides
3.3. Enzymes
3.4. Carotenoids: Astaxanthins
4. Economic and Quality Considered Statistical Optimization Methods
4.1. Optimization Strategies Considered Processing Costs, Quality and Efficiency
4.2. Best Optimization Strategies that Favour the Production of Potential Bioactive Molecules
4.3. Statistical Optimizations on Emerging Green Extraction Technologies
4.3.1. Green Solvent Extraction Parameters Optimization
4.3.2. Optimizing Physical Processing (Cell Wall Breakdown) Extraction Parameters
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, A.; Kumar, D.; George, N.; Sharma, P.; Gupta, N. A process for complete biodegradation of shrimp waste by a novel marine isolate Paenibacillus sp. AD with simultaneous production of chitinase and chitin oligosaccharides. Int. J. Biol. Macromol. 2018, 109, 263–272. [Google Scholar] [CrossRef]
- Coelho, T.L.S.; Silva, D.S.N.; Junior, J.M.d.S.; Dantas, C.; Nogueira, A.R.d.A.; Júnior, C.A.L.; Vieira, E.C. Multivariate optimization and comparison between conventional extraction (CE) and ultrasonic-assisted extraction (UAE) of carotenoid extraction from cashew apple. Ultrason. Sonochemistry 2022, 84, 105980. [Google Scholar] [CrossRef] [PubMed]
- Pinela, J.; de la Fuente, B.; Rodrigues, M.; Pires, T.C.S.P.; Mandim, F.; Almeida, A.; Dias, M.I.; Caleja, C.; Barros, L. Upcycling Fish By-Products into Bioactive Fish Oil: The Suitability of Microwave-Assisted Extraction. Biomolecules 2023, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Ozogul, F.; Cagalj, M.; Šimat, V.; Ozogul, Y.; Tkaczewska, J.; Hassoun, A.; Kaddour, A.A.; Kuley, E.; Rathod, N.B.; Phadke, G.G. Recent developments in valorisation of bioactive ingredients in discard/seafood processing by-products. Trends Food Sci. Technol. 2021, 116, 559–582. [Google Scholar] [CrossRef]
- Roy, V.C.; Islam, R.; Sadia, S.; Yeasmin, M.; Park, J.-S.; Lee, H.-J.; Chun, B.-S. Trash to Treasure: An Up-to-Date Understanding of the Valorization of Seafood By-Products, Targeting the Major Bioactive Compounds. Mar. Drugs 2023, 21, 485. [Google Scholar] [CrossRef] [PubMed]
- Nirmal, N.P.; Santivarangkna, C.; Rajput, M.S.; Benjakul, S.; Maqsood, S. Valorization of fish byproducts: Sources to end-product applications of bioactive protein hydrolysate. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1803–1842. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Wei, S.; Liu, Z.; Fan, X.; Sun, Q.; Xia, Q.; Liu, S.; Hao, J.; Deng, C. Non-thermal processing technologies for the recovery of bioactive compounds from marine by-products. LWT 2021, 147, 111549. [Google Scholar] [CrossRef]
- Lefebvre, T.; Destandau, E.; Lesellier, E. Selective extraction of bioactive compounds from plants using recent extraction techniques: A review. J. Chromatogr. A 2021, 1635, 461770. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, J.; Kannuchamy, N.; Kannaiyan, S.; Chakraborti, R.; Gudipati, V. Protein Hydrolysates from Shrimp (Metapenaeus dobsoni) Head Waste: Optimization of Extraction Conditions by Response Surface Methodology. J. Aquat. Food Prod. Technol. 2015, 24, 429–442. [Google Scholar] [CrossRef]
- Suryawanshi, N.; Eswari, J.S. Chitin from seafood waste: particle swarm optimization and neural network study for the improved chitinase production. J. Chem. Technol. Biotechnol. 2022, 97, 509–519. [Google Scholar] [CrossRef]
- Sharayei, P.; Azarpazhooh, E.; Zomorodi, S.; Einafshar, S.; Ramaswamy, H.S. Optimization of ultrasonic-assisted extraction of astaxanthin from green tiger (Penaeus semisulcatus) shrimp shell. Ultrason. Sonochemistry 2021, 76, 105666. [Google Scholar] [CrossRef]
- Martins, R.; Barbosa, A.; Advinha, B.; Sales, H.; Pontes, R.; Nunes, J. Green Extraction Techniques of Bioactive Compounds: A State-of-the-Art Review. Processes 2023, 11, 2255. [Google Scholar] [CrossRef]
- Picot-Allain, C.; Mahomoodally, M.F.; Ak, G.; Zengin, G. Conventional versus green extraction techniques — a comparative perspective. Curr. Opin. Food Sci. 2021, 40, 144–156. [Google Scholar] [CrossRef]
- Ojha, K.S.; Aznar, R.; O'Donnell, C.; Tiwari, B.K. Ultrasound technology for the extraction of biologically active molecules from plant, animal and marine sources. TrAC Trends Anal. Chem. 2020, 122, 115663. [Google Scholar] [CrossRef]
- Ghalamara, S.; Silva, S.; Brazinha, C.; Pintado, M. Valorization of Fish by-Products: Purification of Bioactive Peptides from Codfish Blood and Sardine Cooking Wastewaters by Membrane Processing. Membranes 2020, 10, 44. [Google Scholar] [CrossRef]
- Chemat, F.; Vian, M.A.; Cravotto, G. Green Extraction of Natural Products: Concept and Principles. Int. J. Mol. Sci. 2012, 13, 8615–8627. [Google Scholar] [CrossRef]
- Benvenutti, L.; Zielinski, A.A.F.; Ferreira, S.R.S. Which is the best food emerging solvent: IL, DES or NADES? Trends Food Sci. Technol. 2019, 90, 133–146. [Google Scholar] [CrossRef]
- Saini, A.; Panesar, P.S. Beneficiation of food processing by-products through extraction of bioactive compounds using neoteric solvents. LWT 2020, 134, 110263. [Google Scholar] [CrossRef]
- El-Shamy, S.; Farag, M.A. Novel trends in extraction and optimization methods of bioactives recovery from pomegranate fruit biowastes: Valorization purposes for industrial applications. Food Chem. 2021, 365, 130465. [Google Scholar] [CrossRef]
- Dayakar, B.; Xavier, M.; Ngasotter, S.; Dhanabalan, V.; Porayil, L.; Balange, A.K.; Nayak, B.B. Extraction, optimization, and functional quality evaluation of carotenoproteins from shrimp processing side streams through enzymatic process. Environ. Sci. Pollut. Res. 2023, 1–14. [Google Scholar] [CrossRef]
- Ahmadkelayeh, S.; Cheema, S.K.; Hawboldt, K. Supercritical CO2 extraction of lipids and astaxanthin from Atlantic shrimp by-products with static co-solvents: Process optimization and mathematical modeling studies. J. CO2 Util. 2022, 58. [Google Scholar] [CrossRef]
- Vanaja, K.; Shobha Rani, R.H. Design of Experiments: Concept and Applications of Plackett Burman Design. Clin. Res. Regul. Aff. 2007, 24, 1–23. [Google Scholar] [CrossRef]
- Yolmeh, M.; Jafari, S.M. Applications of Response Surface Methodology in the Food Industry Processes. Food Bioprocess Technol. 2017, 10, 413–433. [Google Scholar] [CrossRef]
- Honrado, A.; Rubio, S.; Beltrán, J.A.; Calanche, J. Fish By-Product Valorization as Source of Bioactive Compounds for Food Enrichment: Characterization, Suitability and Shelf Life. Foods 2022, 11, 3656. [Google Scholar] [CrossRef]
- He, C.; Cao, J.; Bao, Y.; Sun, Z.; Liu, Z.; Li, C. Characterization of lipid profiling in three parts (muscle, head and viscera) of tilapia (Oreochromis niloticus) using lipidomics with UPLC-ESI-Q-TOF-MS. Food Chem. 2021, 347, 129057. [Google Scholar] [CrossRef] [PubMed]
- Latha, S.; Sivaranjani, G.; Dhanasekaran, D. Response surface methodology: A non-conventional statistical tool to maximize the throughput of Streptomyces species biomass and their bioactive metabolites. Crit. Rev. Microbiol. 2017, 43, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Trocine, L. and L.C. Malone. Finding important independent variables through screening designs: a comparison of methods. in 2000 Winter simulation conference proceedings (Cat. No. 00CH37165). 2000. IEEE.
- Khalil, M.; Darusman, L.K.; Syafitri, U.D. Application of fractional factorial design to optimize extraction method of artemisinin from Artemisia annua. ScienceAsia 2011, 37, 219–224. [Google Scholar] [CrossRef]
- Ye, Z.; Wang, W.; Yuan, Q.; Ye, H.; Sun, Y.; Zhang, H.; Zeng, X. Box–Behnken design for extraction optimization, characterization and in vitro antioxidant activity of Cicer arietinum L. hull polysaccharides. Carbohydr. Polym. 2016, 147, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Nidheesh, T.; Suresh, P.V. Optimization of conditions for isolation of high quality chitin from shrimp processing raw byproducts using response surface methodology and its characterization. J. Food Sci. Technol. 2015, 52, 3812–3823. [Google Scholar] [CrossRef] [PubMed]
- Gamal, R.F.; El-Tayeb, T.S.; Raffat, E.I.; Ibrahim, H.M.; Bashandy, A. Optimization of chitin yield from shrimp shell waste by Bacillus subtilis and impact of gamma irradiation on production of low molecular weight chitosan. Int. J. Biol. Macromol. 2016, 91, 598–608. [Google Scholar] [CrossRef]
- Ismail, S.A. Microbial valorization of shrimp byproducts via the production of thermostable chitosanase and antioxidant chitooligosaccharides. Biocatal. Agric. Biotechnol. 2019, 20. [Google Scholar] [CrossRef]
- Chen, X.; Ding, J.; Ji, D.; He, S.; Ma, H. Optimization of ultrasonic-assisted extraction conditions for bioactive components from coffee leaves using the Taguchi design and response surface methodology. J. Food Sci. 2020, 85, 1742–1751. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; K, S.; Joseph, J.; George, D. Optimization of extraction parameters of bioactive components from Moringa oleifera leaves using Taguchi method. Biomass- Convers. Biorefinery 2023, 13, 11973–11982. [Google Scholar] [CrossRef]
- Jabeur, F.; Mechri, S.; Kriaa, M.; Gharbi, I.; Bejaoui, N.; Sadok, S.; Jaouadi, B. Statistical Experimental Design Optimization of Microbial Proteases Production under Co-Culture Conditions for Chitin Recovery from Speckled Shrimp Metapenaeus monoceros By-Product. BioMed Res. Int. 2020, 2020, 3707804. [Google Scholar] [CrossRef] [PubMed]
- Dave, J.; Ali, A.M.M.; Kudre, T.; Nukhthamna, P.; Kumar, N.; Kieliszek, M.; Bavisetty, S.C.B. Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach. Open Life Sci. 2023, 18, 20220789. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; You, J.; Kang, J.; Nie, F.; Ji, H.; Liu, S. Recovery of astaxanthin from shrimp (Penaeus vannamei) waste by ultrasonic-assisted extraction using ionic liquid-in-water microemulsions. Food Chem. 2020, 325, 126850. [Google Scholar] [CrossRef] [PubMed]
- Iñarra, B.; Bald, C.; Gutierrez, M.; Martin, D.S.; Zufía, J.; Ibarruri, J. Production of Bioactive Peptides from Hake By-Catches: Optimization and Scale-Up of Enzymatic Hydrolysis Process. Mar. Drugs 2023, 21, 552. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.L.; Lemos, V.A.; de Carvalho, V.S.; da Silva, E.G.; Queiroz, A.F.; Felix, C.S.; da Silva, D.L.; Dourado, G.B.; Oliveira, R.V. Multivariate optimization techniques in analytical chemistry - an overview. Microchem. J. 2018, 140, 176–182. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Menduíña, A.; Nogueira, M.; Durán, A.I.; Sanz, N.; Valcarcel, J. Optimal Production of Protein Hydrolysates from Monkfish By-Products: Chemical Features and Associated Biological Activities. Molecules 2020, 25, 4068. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Blanco, M.; Massa, A.E.; Amado, I.R.; Pérez-Martín, R.I. Production of Fish Protein Hydrolysates from Scyliorhinus canicula Discards with Antihypertensive and Antioxidant Activities by Enzymatic Hydrolysis and Mathematical Optimization Using Response Surface Methodology. Mar. Drugs 2017, 15, 306. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, J.A.; Sotelo, C.G.; Sanz, N.; Pérez-Martín, R.I.; Rodríguez-Amado, I.; Valcarcel, J. Valorization of Aquaculture By-Products of Salmonids to Produce Enzymatic Hydrolysates: Process Optimization, Chemical Characterization and Evaluation of Bioactives. Mar. Drugs 2019, 17, 676. [Google Scholar] [CrossRef]
- Pandian, P.S.; Sindhanaiselvan, S.; Subathira, A.; Saravanan, S. A correlative algorithmic optimization study for an integrated soft computing technique in aqueous two-phase protein extraction from Litopenaeus vannamei waste. Biomass- Convers. Biorefinery 2023, 13, 16819–16833. [Google Scholar] [CrossRef]
- Kennedy, J. and R. Eberhart. Particle swarm optimization. in Proceedings of ICNN'95-international conference on neural networks. 1995. ieee.
- Selber, K.; Nellen, F.; Steffen, B.; Thömmes, J.; Kula, M.-R. Investigation of mathematical methods for efficient optimisation of aqueous two-phase extraction. J. Chromatogr. B: Biomed. Sci. Appl. 2000, 743, 21–30. [Google Scholar] [CrossRef]
- Ferreira, S.L.; Junior, M.M.S.; Felix, C.S.; da Silva, D.L.; Santos, A.S.; Neto, J.H.S.; de Souza, C.T.; Junior, R.A.C.; Souza, A.S. Multivariate optimization techniques in food analysis – A review. Food Chem. 2019, 273, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Tsiaka, T.; Zoumpoulakis, P.; Sinanoglou, V.J.; Makris, C.; Heropoulos, G.A.; Calokerinos, A.C. Response surface methodology toward the optimization of high-energy carotenoid extraction from Aristeus antennatus shrimp. Anal. Chim. Acta 2015, 877, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.; Vázquez, J.A.; Pérez-Martín, R.I.; Sotelo, C.G. Collagen Extraction Optimization from the Skin of the Small-Spotted Catshark (S. canicula) by Response Surface Methodology. Marine Drugs 2019, 17, 40. [Google Scholar] [CrossRef]
- Paulo, F.; Tavares, L.; Santos, L. Response Surface Modeling and Optimization of the Extraction of Phenolic Antioxidants from Olive Mill Pomace. Molecules 2022, 27, 8620. [Google Scholar] [CrossRef]
- Maran, J.P.; Manikandan, S.; Thirugnanasambandham, K.; Nivetha, C.V.; Dinesh, R. Box–Behnken design based statistical modeling for ultrasound-assisted extraction of corn silk polysaccharide. Carbohydr. Polym. 2013, 92, 604–611. [Google Scholar] [CrossRef]
- Khataee, A.; Fathinia, M.; Aber, S.; Zarei, M. Optimization of photocatalytic treatment of dye solution on supported TiO2 nanoparticles by central composite design: Intermediates identification. J. Hazard. Mater. 2010, 181, 886–897. [Google Scholar] [CrossRef]
- Abdessalem, A.K.; Oturan, N.; Bellakhal, N.; Dachraoui, M.; Oturan, M.A. Experimental design methodology applied to electro-Fenton treatment for degradation of herbicide chlortoluron. Appl. Catal. B: Environ. 2008, 78, 334–341. [Google Scholar] [CrossRef]
- Yetilmezsoy, K.; Demirel, S.; Vanderbei, R.J. Response surface modeling of Pb (II) removal from aqueous solution by Pistacia vera L.: Box–Behnken experimental design. J. Hazard. Mater. 2009, 171, 551–562. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, B.; Pinela, J.; Mandim, F.; Heleno, S.A.; Ferreira, I.C.; Barba, F.J.; Berrada, H.; Caleja, C.; Barros, L. Nutritional and bioactive oils from salmon (Salmo salar) side streams obtained by Soxhlet and optimized microwave-assisted extraction. Food Chem. 2022, 386, 132778. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.V.; Dave, D.; Liu, Y.; Routray, W.; Murphy, W. Statistical Optimization of Biodiesel Production from Salmon Oil via Enzymatic Transesterification: Investigation of the Effects of Various Operational Parameters. Processes 2021, 9, 700. [Google Scholar] [CrossRef]
- Haq, M.; Pendleton, P.; Chun, B.-S. Utilization of Atlantic Salmon By-product Oil for Omega-3 Fatty Acids Rich 2-Monoacylglycerol Production: Optimization of Enzymatic Reaction Parameters. Waste Biomass- Valorization 2020, 11, 153–163. [Google Scholar] [CrossRef]
- Nouri, M.; Khodaiyan, F.; Razavi, S.H.; Mousavi, M. Improvement of chitosan production from Persian Gulf shrimp waste by response surface methodology. Food Hydrocoll. 2016, 59, 50–58. [Google Scholar] [CrossRef]
- Roy, V.C.; Ho, T.C.; Lee, H.-J.; Park, J.-S.; Nam, S.Y.; Lee, H.; Getachew, A.T.; Chun, B.-S. Extraction of astaxanthin using ultrasound-assisted natural deep eutectic solvents from shrimp wastes and its application in bioactive films. J. Clean. Prod. 2021, 284, 125417. [Google Scholar] [CrossRef]
- Chasquibol, N.; Gonzales, B.F.; Alarcón, R.; Sotelo, A.; Márquez-López, J.C.; Rodríguez-Martin, N.M.; Millán-Linares, M.d.C.; Millán, F.; Pedroche, J. Optimisation and Characterisation of the Protein Hydrolysate of Scallops (Argopecten purpuratus) Visceral By-Products. Foods 2023, 12, 2003. [Google Scholar] [CrossRef]
- Haq, M.; Getachew, A.T.; Saravana, P.S.; Cho, Y.-J.; Park, S.-K.; Kim, M.-J.; Chun, B.-S. Effects of process parameters on EPA and DHA concentrate production from Atlantic salmon by-product oil: Optimization and characterization. Korean J. Chem. Eng. 2017, 34, 2255–2264. [Google Scholar] [CrossRef]
- Koutsoukos, S.; Tsiaka, T.; Tzani, A.; Zoumpoulakis, P.; Detsi, A. Choline chloride and tartaric acid, a Natural Deep Eutectic Solvent for the efficient extraction of phenolic and carotenoid compounds. J. Clean. Prod. 2019, 241. [Google Scholar] [CrossRef]
- Alves, F.E.D.S.B.; Carpiné, D.; Teixeira, G.L.; Goedert, A.C.; de Paula Scheer, A.; Ribani, R.H. Valorization of an Abundant Slaughterhouse By-product as a Source of Highly Technofunctional and Antioxidant Protein Hydrolysates. Waste Biomass Valori. 2021, 12, 263–279. [Google Scholar] [CrossRef]
- Baş, D.; Boyacı, .H. Modeling and optimization I: Usability of response surface methodology. J. Food Eng. 2007, 78, 836–845. [Google Scholar] [CrossRef]
- Paul, T., S.K. Halder, A. Das, K. Ghosh, A. Mandal, P. Payra, . . . K.C. Mondal, Production of chitin and bioactive materials from Black tiger shrimp (Penaeus monodon) shell waste by the treatment of bacterial protease cocktail. 3 Biotech, 2015. 5(4): p. 483-493.
- Mousavi, L.; Tamiji, Z.; Khoshayand, M.R. Applications and opportunities of experimental design for the dispersive liquid–liquid microextraction method – A review. Talanta 2018, 190, 335–356. [Google Scholar] [CrossRef] [PubMed]
- Ghorbannezhad, P.; Bay, A.; Yolmeh, M.; Yadollahi, R.; Moghadam, J.Y. Optimization of coagulation–flocculation process for medium density fiberboard (MDF) wastewater through response surface methodology. Desalination Water Treat. 2016, 57, 26916–26931. [Google Scholar] [CrossRef]
- Sindhu, S. and P. Sherief. Extraction, characterization, antioxidant and anti-inflammatory properties of carotenoids from the shell waste of arabian red shrimp Aristeus alcocki, ramadan 1938. in The open Conference proceedings journal. 2011.
- Bernardo, B.d.S.; Kopplin, B.W.; Daroit, D.J. Bioconversion of Fish Scales and Feather Wastes by Bacillus sp. CL18 to Obtain Protease and Bioactive Hydrolysates. Waste Biomass- Valorization 2023, 14, 1045–1056. [Google Scholar] [CrossRef]
- Martí-Quijal, F.J.; Tornos, A.; Príncep, A.; Luz, C.; Meca, G.; Tedeschi, P.; Ruiz, M.-J.; Barba, F.J. Impact of Fermentation on the Recovery of Antioxidant Bioactive Compounds from Sea Bass Byproducts. Antioxidants 2020, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Roy, V.C.; Ho, T.C.; Park, J.-S.; Jeong, Y.-R.; Lee, S.-C.; Kim, S.-Y.; Chun, B.-S. Amino Acid Profiles and Biopotentiality of Hydrolysates Obtained from Comb Penshell (Atrina pectinata) Viscera Using Subcritical Water Hydrolysis. Mar. Drugs 2021, 19, 137. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.; Rakshit, S.; Mondal, K.C.; Halder, S.K. Microbial decomposition of crustacean shell for production of bioactive metabolites and study of its fertilizing potential. Environ. Sci. Pollut. Res. 2021, 28, 58915–58928. [Google Scholar] [CrossRef]
- Mechri, S.; Sellem, I.; Bouacem, K.; Jabeur, F.; Laribi-Habchi, H.; Mellouli, L.; Hacène, H.; Bouanane-Darenfed, A.; Jaouadi, B. A biological clean processing approach for the valorization of speckled shrimp Metapenaeus monoceros by-product as a source of bioactive compounds. Environ. Sci. Pollut. Res. 2020, 27, 15842–15855. [Google Scholar] [CrossRef]
- Hamdi, M.; Hammami, A.; Hajji, S.; Jridi, M.; Nasri, M.; Nasri, R. Chitin extraction from blue crab ( Portunus segnis ) and shrimp ( Penaeus kerathurus ) shells using digestive alkaline proteases from P. segnis viscera. Int. J. Biol. Macromol. 2017, 101, 455–463. [Google Scholar] [CrossRef]
- Messina, C.M., S. Manuguerra, R. Arena, G. Renda, G. Ficano, M. Randazzo, . . . A. Santulli, In Vitro Bioactivity of Astaxanthin and Peptides from Hydrolisates of Shrimp (Parapenaeus longirostris) By-Products: From the Extraction Process to Biological Effect Evaluation, as Pilot Actions for the Strategy “From Waste to Profit”. Marine Drugs, 2021. 19(4): p. 216.
- Messina, C.M.; Manuguerra, S.; Renda, G.; Santulli, A. Biotechnological Applications for the Sustainable Use of Marine By-products: In Vitro Antioxidant and Pro-apoptotic Effects of Astaxanthin Extracted with Supercritical CO2 from Parapeneus longirostris. Mar. Biotechnol. 2019, 21, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Sedaghat, F.; Yousefzadi, M.; Toiserkani, H.; Najafipour, S. Chitin from Penaeus merguiensis via microbial fermentation processing and antioxidant activity. Int. J. Biol. Macromol. 2016, 82, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fang, J.; Deng, Y.; Zhao, Y. Optimized production of Serratia marcescens B742 mutants for preparing chitin from shrimp shells powders. Int. J. Biol. Macromol. 2014, 69, 319–328. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, J.; Collado, M.C.; Barba, F.J. Accelerated Solvent Extraction and Pulsed Electric Fields for Valorization of Rainbow Trout (Oncorhynchus mykiss) and Sole (Dover sole) By-Products: Protein Content, Molecular Weight Distribution and Antioxidant Potential of the Extracts. Marine Drugs 2021, 19, 207. [Google Scholar] [CrossRef]
- Hamdi, M.; Nasri, R.; Dridi, N.; Li, S.; Nasri, M. Development of novel high-selective extraction approach of carotenoproteins from blue crab (Portunus segnis) shells, contribution to the qualitative analysis of bioactive compounds by HR-ESI-MS. Food Chem. 2020, 302, 125334. [Google Scholar] [CrossRef]
- Lee, S.-C.; Nkurunziza, D.; Kim, S.-Y.; Surendhiran, D.; Singh, A.A.; Chun, B.-S. Supercritical carbon dioxide extraction of squalene rich cod liver oil: Optimization, characterization and functional properties. J. Supercrit. Fluids 2022, 188, 105693. [Google Scholar] [CrossRef]
- Islam, N.; Hoque, M.; Taharat, S.F. Recent advances in extraction of chitin and chitosan. World J. Microbiol. Biotechnol. 2023, 39, 1–17. [Google Scholar] [CrossRef]
- Ibram, A.; Ionescu, A.-M.; Cadar, E. Comparison of Extraction Methods of Chitin and Chitosan from Different Sources. Eur. J. Nat. Sci. Med. 2019, 2, 23–36. [Google Scholar] [CrossRef]
- Kumari, S.; Rath, P.K. Extraction and Characterization of Chitin and Chitosan from (Labeo rohit) Fish Scales. Procedia Mater. Sci. 2014, 6, 482–489. [Google Scholar] [CrossRef]
- Srinivasan, H.; Kanayairam, V.; Ravichandran, R. Chitin and chitosan preparation from shrimp shells Penaeus monodon and its human ovarian cancer cell line, PA-1. Int. J. Biol. Macromol. 2018, 107, 662–667. [Google Scholar] [CrossRef]
- Arancibia, M.Y.; Alemán, A.; Calvo, M.M.; López-Caballero, M.E.; Montero, P.; Gómez-Guillén, M.C. Antimicrobial and antioxidant chitosan solutions enriched with active shrimp (Litopenaeus vannamei) waste materials. Food Hydrocoll. 2014, 35, 710–717. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.; Liu, S.; Li, Y. Establishment of successive co-fermentation by Bacillus subtilis and Acetobacter pasteurianus for extracting chitin from shrimp shells. Carbohydr. Polym. 2021, 258, 117720. [Google Scholar] [CrossRef] [PubMed]
- Bahasan, S.H.O.; Satheesh, S.; Ba-Akdah, M.A. Extraction of Chitin from the Shell Wastes of Two Shrimp Species Fenneropenaeus semisulcatus and Fenneropenaeus indicus using Microorganisms. J. Aquat. Food Prod. Technol. 2017, 26, 390–405. [Google Scholar] [CrossRef]
- Liu, Y.; Xing, R.; Yang, H.; Liu, S.; Qin, Y.; Li, K.; Yu, H.; Li, P. Chitin extraction from shrimp (Litopenaeus vannamei) shells by successive two-step fermentation with Lactobacillus rhamnoides and Bacillus amyloliquefaciens. Int. J. Biol. Macromol. 2020, 148, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Arbia, W., L. Arbia, L. Adour, A. Amrane, H. Lounici, and N. Mameri, Kinetic study of bio-demineralization and bio-deproteinization of shrimp biowaste for chitin recovery. Algerian Journal of Environmental Science and Technology, 2017. 3(1).
- Arbia, W., L. Arbia, L. Adour, A. Amrane, A. Benhadji, and H. Lounici, Characterization by spectrometric methods of chitin produced from white shrimp shells of Parapenaeus longirostris byLactobacillus helveticus cultivated on glucose or date waste. Algerian Journal of Environmental Science and Technology, 2019. 5(2).
- Aranday-García, R.; Saimoto, H.; Shirai, K.; Ifuku, S. Chitin biological extraction from shrimp wastes and its fibrillation for elastic nanofiber sheets preparation. Carbohydr. Polym. 2019, 213, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Siddik, M.A.; Howieson, J.; Fotedar, R.; Partridge, G.J. Enzymatic fish protein hydrolysates in finfish aquaculture: a review. Rev. Aquac. 2021, 13, 406–430. [Google Scholar] [CrossRef]
- Aspevik, T., Å. Oterhals, S.B. Rønning, T. Altintzoglou, S.G. Wubshet, A. Gildberg,... D. Lindberg, Valorization of proteins from co-and by-products from the fish and meat industry. Chemistry and chemical technologies in waste valorization, 2018: p. 123-150.
- Gao, R.; Yu, Q.; Shen, Y.; Chu, Q.; Chen, G.; Fen, S.; Yang, M.; Yuan, L.; McClements, D.J.; Sun, Q. Production, bioactive properties, and potential applications of fish protein hydrolysates: Developments and challenges. Trends Food Sci. Technol. 2021, 110, 687–699. [Google Scholar] [CrossRef]
- Mathew, G.M., C. C. Huang, R. Sindhu, P. Binod, and A. Pandey, Enzymes in seafood processing, in Value-Addition in Food Products and Processing Through Enzyme Technology. 2022, Elsevier. p. 189-204.
- Halim, N.R.A.; Yusof, H.M.; Sarbon, N.M. Functional and bioactive properties of fish protein hydolysates and peptides: A comprehensive review. Trends Food Sci. Technol. 2016, 51, 24–33. [Google Scholar] [CrossRef]
- Dinakarkumar, Y.; Krishnamoorthy, S.; Margavelu, G.; Ramakrishnan, G.; Chandran, M. Production and characterization of fish protein hydrolysate: Effective utilization of trawl by-catch. Food Chem. Adv. 2022, 1. [Google Scholar] [CrossRef]
- Muzaddadi, A.U., S. Devatkal, and H.S. Oberoi, Chapter 9 - Seafood Enzymes and Their Application in Food Processing, in Agro-Industrial Wastes as Feedstock for Enzyme Production, G.S. Dhillon and S. Kaur, Editors. 2016, Academic Press: San Diego. p. 201-232.
- R. , S.; J., J.; A., T.S. Purification, characterization, molecular modeling and docking study of fish waste protease. Int. J. Biol. Macromol. 2018, 118, 569–583. [Google Scholar] [CrossRef]
- Murthy, L.N.; Phadke, G.G.; Unnikrishnan, P.; Annamalai, J.; Joshy, C.G.; Zynudheen, A.A.; Ravishankar, C.N. Valorization of Fish Viscera for Crude Proteases Production and Its Use in Bioactive Protein Hydrolysate Preparation. Waste Biomass- Valorization 2017, 9, 1735–1746. [Google Scholar] [CrossRef]
- Subramanian, K.; Sadaiappan, B.; Aruni, W.; Kumarappan, A.; Thirunavukarasu, R.; Srinivasan, G.P.; Bharathi, S.; Nainangu, P.; Renuga, P.S.; Elamaran, A.; et al. Bioconversion of chitin and concomitant production of chitinase and N-acetylglucosamine by novel Achromobacter xylosoxidans isolated from shrimp waste disposal area. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Affes, S.; Aranaz, I.; Hamdi, M.; Acosta, N.; Ghorbel-Bellaaj, O.; Heras, .; Nasri, M.; Maalej, H. ; Nasri, M.; Maalej, H. Preparation of a crude chitosanase from blue crab viscera as well as its application in the production of biologically active chito-oligosaccharides from shrimp shells chitosan. Int. J. Biol. Macromol. 2019, 139, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Tran, T.D.; Nguyen, A.D.; Wang, S.-L. Bioprocessing of Squid Pens Waste into Chitosanase by Paenibacillus sp. TKU047 and Its Application in Low-Molecular Weight Chitosan Oligosaccharides Production. Polymers 2020, 12, 1163. [Google Scholar] [CrossRef]
- Silva, A.K.N.d.; Rodrigues, B.D.; L.H.M.d. SILVA, and A.M.d.C. RODRIGUES. Drying and extraction of astaxanthin from pink shrimp waste (Farfantepenaeus subtilis): the applicability of spouted beds. Food Sci. Technol. 2018, 38, 454–461. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Q.; Zhang, D.; Wei, S.; Sun, Q.; Xia, Q.; Shi, W.; Ji, H.; Liu, S. Comparison of the Proximate Composition and Nutritional Profile of Byproducts and Edible Parts of Five Species of Shrimp. Foods 2021, 10, 2603. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Lu, W.; Lv, M.; Wang, Y.; Ding, R.; Wang, L. Extraction and purification of astaxanthin from shrimp shells and the effects of different treatments on its content. Rev. Bras. de Farm. 2019, 29, 24–29. [Google Scholar] [CrossRef]
- Li, J.; Sun, W.; Ramaswamy, H.S.; Yu, Y.; Zhu, S.; Wang, J.; Li, H. High Pressure Extraction of Astaxanthin from Shrimp Waste (Penaeus Vannamei Boone): Effect on Yield and Antioxidant Activity. J. Food Process. Eng. 2017, 40. [Google Scholar] [CrossRef]
- Shazana, A.R.; Masturah, M.; Badlishah, S.B.; Rashidi, O.; Russly, A. Optimisation of supercritical fluid extraction of astaxanthin from Penaeus monodon waste using ethanol-modified carbon dioxide. J. Eng. Sci. Technol 2016, 11, 722–736. [Google Scholar]
- Sánchez-Camargo, A.P.; Meireles, M. .A.; Ferreira, A.L.; Saito, E.; Cabral, F.A. Extraction of ω-3 fatty acids and astaxanthin from Brazilian redspotted shrimp waste using supercritical CO2+ethanol mixtures. J. Supercrit. Fluids 2012, 61, 71–77. [Google Scholar] [CrossRef]
- Cabanillas-Bojórquez, L.A.; Gutiérrez-Grijalva, E.P.; González-Aguilar, G.A.; López-Martinez, L.X.; Castillo-López, R.I.; Bastidas-Bastidas, P.d.J.; Heredia, J.B. Valorization of Fermented Shrimp Waste with Supercritical CO2 Conditions: Extraction of Astaxanthin and Effect of Simulated Gastrointestinal Digestion on Its Antioxidant Capacity. Molecules 2021, 26, 4465. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, M.; Fawzy, S.; Xue, Y.; Wu, M.; Huang, X.; Yi, G.; Lin, Q. Effects of Dietary Phaffia rhodozyma Astaxanthin on Growth Performance, Carotenoid Analysis, Biochemical and Immune-Physiological Parameters, Intestinal Microbiota, and Disease Resistance in Penaeus monodon. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Gulzar, S.; Benjakul, S. Impact of pulsed electric field pretreatment on yield and quality of lipid extracted from cephalothorax of Pacific white shrimp (Litopenaeus vannamei) by ultrasound-assisted process. Int. J. Food Sci. Technol. 2020, 55, 619–630. [Google Scholar] [CrossRef]
- Poojary, M.M.; Barba, F.J.; Aliakbarian, B.; Donsì, F.; Pataro, G.; Dias, D.A.; Juliano, P. Innovative Alternative Technologies to Extract Carotenoids from Microalgae and Seaweeds. Mar. Drugs 2016, 14, 214. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.d.S.V.; Bragagnolo, N. Development and validation of a novel microwave assisted extraction method for fish lipids. Eur. J. Lipid Sci. Technol. 2017, 119, 1600108. [Google Scholar] [CrossRef]
- Barba, F.J.; Grimi, N.; Vorobiev, E. New Approaches for the Use of Non-conventional Cell Disruption Technologies to Extract Potential Food Additives and Nutraceuticals from Microalgae. Food Eng. Rev. 2015, 7, 45–62. [Google Scholar] [CrossRef]
- Schuur, B.; Brouwer, T.; Smink, D.; Sprakel, L.M. Green solvents for sustainable separation processes. Curr. Opin. Green Sustain. Chem. 2019, 18, 57–65. [Google Scholar] [CrossRef]
- Carpentieri, S.; Soltanipour, F.; Ferrari, G.; Pataro, G.; Donsì, F. Emerging Green Techniques for the Extraction of Antioxidants from Agri-Food By-Products as Promising Ingredients for the Food Industry. Antioxidants 2021, 10, 1417. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.M.; Kumar, P.; Ismail-Fitry, M.R.; Jusoh, S.; Ab Aziz, M.F.; Sazili, A.Q. Green Extraction of Bioactive Compounds from Plant Biomass and Their Application in Meat as Natural Antioxidant. Antioxidants 2021, 10, 1465. [Google Scholar] [CrossRef] [PubMed]
- Saravana, P.S.; Ho, T.C.; Chae, S.-J.; Cho, Y.-J.; Park, J.-S.; Lee, H.-J.; Chun, B.-S. Deep eutectic solvent-based extraction and fabrication of chitin films from crustacean waste. Carbohydr. Polym. 2018, 195, 622–630. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Meduíña, A.; Durán, A.I.; Nogueira, M.; Fernández-Compás, A.; Pérez-Martín, R.I.; Rodríguez-Amado, I. Production of Valuable Compounds and Bioactive Metabolites from By-Products of Fish Discards Using Chemical Processing, Enzymatic Hydrolysis, and Bacterial Fermentation. Mar. Drugs 2019, 17, 139. [Google Scholar] [CrossRef] [PubMed]
- El-Bialy, H.A.A.; Abd El-Khalek, H.H. A comparative study on astaxanthin recovery from shrimp wastes using lactic fermentation and green solvents:an applied model on minced Tilapia. J. Radiat. Res. Appl. Sci. 2020, 13, 594–605. [Google Scholar] [CrossRef]
- Vernès, L.; Abert-Vian, M.; El Maâtaoui, M.; Tao, Y.; Bornard, I.; Chemat, F. Application of ultrasound for green extraction of proteins from spirulina. Mechanism, optimization, modeling, and industrial prospects. Ultrason. Sonochemistry 2019, 54, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Gu, S.; Liu, S.; Zhang, J.; Ding, Y.; Liu, J. Extraction of oil from high-moisture tuna liver by subcritical dimethyl ether: feasibility and optimization by the response surface method. RSC Adv. 2018, 8, 2723–2732. [Google Scholar] [CrossRef]
- Dutta, S.; Priyadarshini, S.R.; Moses, J.A.; Anandharamakrishnan, C. Supercritical Fluid and Ultrasound-assisted Green Extraction Technologies for Catechin Recovery. ChemBioEng Rev. 2021, 8, 654–664. [Google Scholar] [CrossRef]
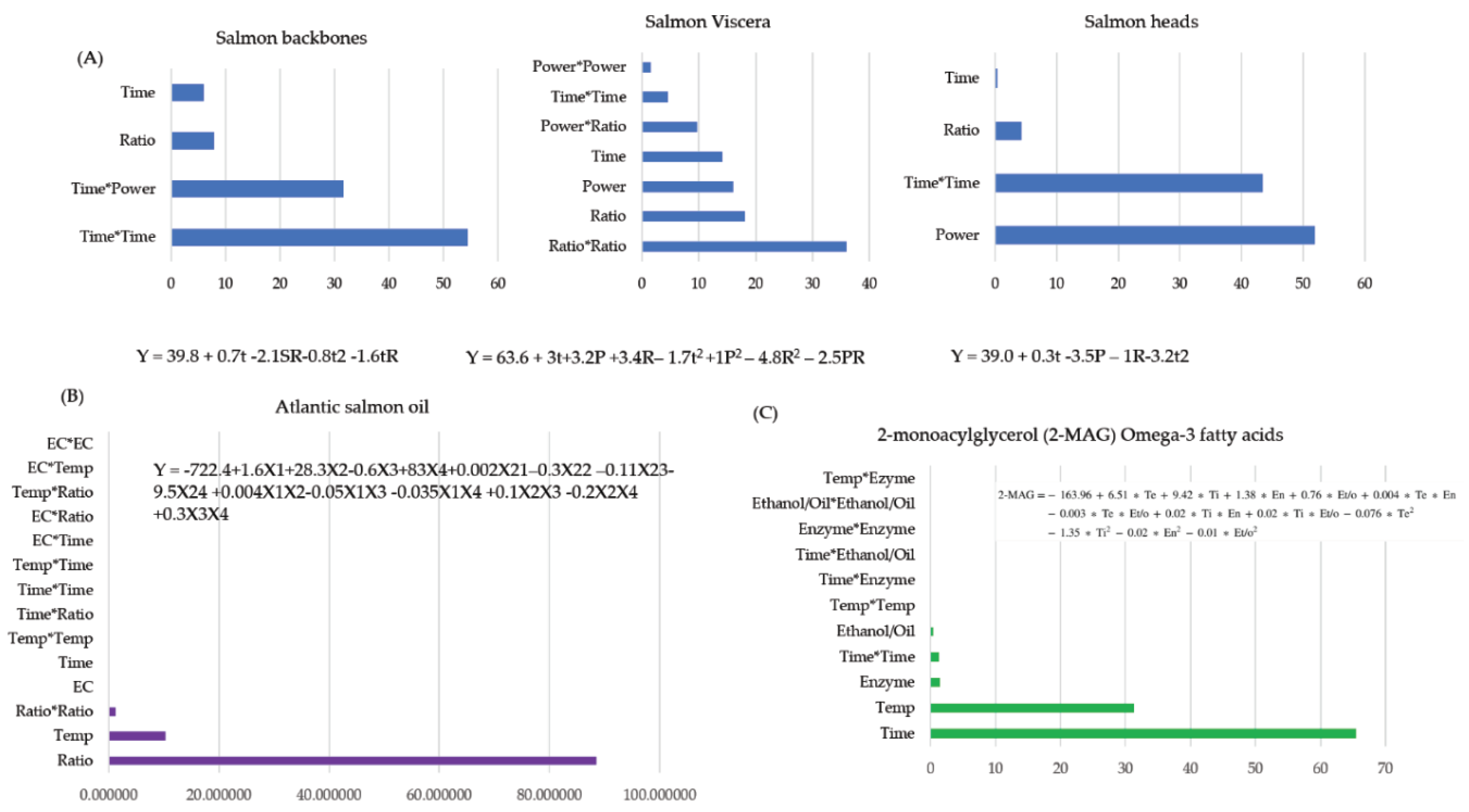
| DoE | Developed Equation | Number of factors | p | Percentage contribution of variables (%) | Reference | ||
|---|---|---|---|---|---|---|---|
| TPCi | TPCii | TPCij | |||||
| CCD | Y = 39.2 + 9.3X1 + 3.1X2 + 4.1X3 – 3.4X23 | 3 | 5 | 86.5 | 13.5 | [49] | |
| Y = 82.46-2.43X1 +5.23X2 +7.02X3 + 0.64X4 +0.31X1X2 +0.35X1X3 +0.33X1X4 -0.16X2X3 +0.1X2X4 – 0.5X3X4–10.6X21 +0.4X22 –0.051X23 +16.62X24 | 4 | 15 | 59.5 | 40.1 | 0.4 | [44] | |
| Y = -120.3 + 416X1 + 2.8X2 + 9.2X3 -3.6X21 – 0.01X22 -0.2X23 – 0.14X1X2 -0.8X1X3 – -0.04X2X3 | 3 | 10 | 80.8 | 16.2 | 3.0 | [30] | |
| Y = 14.4 + 0.8X1 +0.04X2 -1.5X3–0.45X23 – 0.3X1X3 +0.4X2X3 Y = 19.5 + 0.52X1 +1.2X2 -1X3–1.25X21 -1 X22–0.3X2X3 |
3 3 |
7 7 |
88.7 51.2 |
6.8 47.6 |
4.5 1.2 |
[3] | |
| BBD | Y = 39.2 + 21.2X1 - 3.7X2 -0.066X3 +0.154X1X2 + 0.045 X1X3 + 0.003 X2X3 – 0.64 X21 – 0.1 X22 + 0.004X23 | 3 | 10 | 86.2 | 13.5 | 0.3 | [61] |
| Y = -33.1+0.81X1 + 0.6X2 + 85.3X3 – 0.008X21 – 0.003X22 -91.2X23 – 0.003X1X2 – 0.3X1X3 | 3 | 9 | 51.1 | 44.3 | 4.6 | [60] | |
| Y = -9.9 + 11.5X1 +1.7X2 +1.7X3 -0.09X1X2 -0.32X1X3 -0.006X2X3 – 0.7X21 – 0.01X22 - 0.1X23 | 3 | 10 | 64.2 | 19.7 | 16.1 | [59] | |
| Y = 4.9+0.9X1 +0.5X2 -0.4X3– 0.3X21 – 1X22 - 0.4X23 Y = 7.1+0.9X1 +0.5X2 -0.4X3– 0.5X21 – 1.2X22 - 0.5X23 |
3 3 |
7 7 |
68.2 52.2 |
30.9 43.0 |
0.9 4.8 |
[58] | |
| Y = -18.1+3.2X1 -580.2X2 +0.02X3 –0.05X21 +49269.7X22 +0.27X23 -16.6X1X2 +0.13X1X3 -47.5X2X3 | 3 | 10 | 72.9 | 26.2 | 0.9 | [32] | |
| Y = 63.7-63.7X1 -5.8X2 -3X3 + 16.6X4 +5.8X1X2 +6.14X1X3 -2.9X1X4 -0.24X2X4 -0.3X3X4–1.4X21– 4.7X22 – 4.34X23 + 1.8X24 | 4 | 14 | 86.2 | 6.6 | 7.2 | [31] | |
| Y = 10.7+1.3X1 +0.1X2 +2.2X3 +0.4X21 -0.6X22 +1X23 -0.8X1X2 +0.25X1 X22 +0.8X21X2 +0.7X1X3 – 0.3X21X3 +0.4X2X3 Y=15.9+0.6X1+0.5X2+0.7X3-0.9X21-0.4X22+0.4X23+0.23X1X2-1.55X1X22+0.5X21X2+1X1X3+1X21X3 -0.6X2X3 |
3 3 |
13 13 |
18.1 30.6 |
68.0 14.0 |
13.9 55.4 |
[48] | |
| Full Factorial Design | 1. Y = 528.9 –29.04X1+0.87X21 -164.8X3 +23.2 X23 2. Y = 28.8 –0.0013X21 – 0.1X2 -12.7X3 + 1.8X23 3. Y = 121.1 –78.4X1+49.3X21 -44.2X3 +31.9X23 |
3 | 5 5 5 |
98.1 98.0 70.1 |
0.003 0.006 21.0 |
1.9 2.0 8.8 |
[62] |
| Y = -722.4+1.6X1+28.3X2-0.6X3+83X4+0.002X21–0.3X22 –0.11X23-9.5X24 +0.004X1X2-0.05X1X3 -0.035X1X4 +0.1X2X3 -0.2X2X4 +0.3X3X4 | 4 | 15 | 10.3 | 1.2 | 88.5 | [56] | |
| Statistical parameter | Value |
|---|---|
| Std.dev. | 1.19 |
| C.V. % | 2.09 |
| R² | 0.9870 |
| Adjusted R² | 0.9702 |
| Predicted R² | 0.8990 |
| Adeq Precision | 24.6656 |
| PRESS | 77.19 |
| AAD (%) | 1.07 |
| Seafood byproduct type | Design method of experiments (DoE) | Employed software | Extraction method | Targeted Bioactive molecule | Considered Extraction parameter/s | Reference |
|---|---|---|---|---|---|---|
| Shrimp chitinaceous waste | Central composite design | Design Expert 8.0.7.1 | Enzymatic Digestion | Chitinase and chitin oligosaccharides | Incubation time, different media, pH, temperature, carbon source, nitrogen source and metal ions | [1] |
| Red Shrimp (Aristeus alcocki) shell waste | Analysis of variance technique | SPSS 15 | Non-deproteinization of enzymatic digestion |
Carotenoids | Different organic solvents Three different vegetable oils |
[68] |
| Fish scales and feather wastes | Analysis of variance technique | Bacillus sp. CL18 as a bioconverter | Protease, Bioactive hydrolysates | Twelve substrates and co-substrates | [69] | |
| Sea bass skinhead, tail, thorns, and backbone) | Analysis of variance technique | InfoStatfi and StatAdvisorfi version 2018 | Bacterial fermentation | Phenolic acids | Fermentation time (in hours) | [70] |
| Comb penshell (Atrina pectinata) | One-way analysis of variance | SPSS version 23 | Subcritical Water Hydrolysis | Amino acids and marine bioactive peptides | Extraction temperatures | [71] |
| Crustacean shell waste | One-way analysis of variance | Sigma Plot 14.0 | Submerged fermentation | Chitinase, protease | fermentation time, pH, and temperature | [72] |
| Speckled shrimp Metapenaeus monoceros shells | One-way analysis of variance | SPSS Version 11.0.1.2001 | Flask based hydrolysis | Protease | Concentrations of shrimp, sugar | [35] |
| Speckled shrimp Metapenaeus monoceros shells | One-way analysis of variance | SPSS ver.17.0 | Deproteinization of enzymatic digestion | Deproteinized bioactive hydrolysate | enzyme/substrate ratios | [73] |
| shrimp (P. kerathurus) shells and blue crabs (P. segnis) Viscera | One-way analysis of variance | SPSS ver.17.0 | Deproteinization of enzymatic digestion | Chitin | pH and temperature | [74] |
| Shrimp (Parapenaeus longirostris) heads, thorax, appendix cephalothorax and abdominal parts | One-way analysis of variance | SPSS version 20.0 | Supercritical CO2 Extraction | Astaxanthin and Peptides Carotenoid astaxanthin |
Extraction rate | [75, 76] |
| Shrimp (Penaeus merguiensis) shells | One-way analysis of variance | SPSS version 19.0 | Fermentation | Chitin, chitosan | Differences in bacterial strains | [77] |
| Shrimp shells powders | One-way analysis of variance | SPSS version 19.0 | Submerged fermentation | Chitin | Time, Dilution, 2% diethyl sulfate, UV-irradiation, microwave heating treatments | [78] |
| Head, skins and viscera of Rainbow Trout (Oncorhynchus mykiss) and Sole (Dover sole) | One-way analysis of variance | SPSS | Accelerated solvent extraction and pulsed electric fields | Protein content | Temperature, time, pH and pressure | [79] |
| Blue crab (Portunus segnis) shells | One-way analysis of variance | SPSS ver. 17.0 | Enzymatic pretreatment combined with solvent maceration | Carotenoproteins | Time intervals and concentration Portunus segnis proteases |
[80] |
| Seafood byproduct type | Statistical Methodology |
Design method of experiments (DoE) | Employed software | Extraction method | Targeted Bioactive molecule | Considered Extraction parameters | Reference |
|---|---|---|---|---|---|---|---|
| Cod fish liver | RSM | Conventional hexane and supercritical carbon dioxide | Cod liver oil | temperature, pressure, and CO2 flow rate | [81] | ||
| Shrimp shell waste | Particle swarm optimization algorithm and artificial neural network | Central composite design | MATLAB R2016a | Fermentation | Chitinase | Colloidal chitin, glucose, Tween 80 (common surfactant micelles), yeast extract | [10] |
| Shrimp (Penaeus sp.) cephalothoraxes and carapaces | RSM | Fractional factorial design (FFD) CCD |
Statsoft 1997 | Thermochemical treatments | Chitin | Concentration of HCl solution, solid liquid ratio of HCl solution, number of treatments, Concentration of NaOH solution, reaction time, reaction temperature, solid liquid ratio of NaOH solution | [30] |
| Shrimp Litopenaeus vannamei waste | RSM Genetic algorithm and Particle Swarm |
Central composite design | Design-Expert software (version 10.0.1.0 | Aqueous two-phase system | Protein recovery | Polyethylene glycol concentration, trisodium citrate concentration, pH and temperature | [44] |
| Speckled shrimp Metapenaeus monoceros shells | RSM | Taguchi’s L27, Box–Behnken Design |
SPSS Version 11.0.1.2001 | Flask based hydrolysis | Chitin | Temperature, Inoculum size of strain, Culture volume | [35] |
| Shrimp heads | RSM | 3-level fractional factorial | Statistica software Version 10 | Ultrasound and Microwave Assisted Extraction | Phenolic and Carotenoids | Extraction time, solvent-to-propolis and Choline Chloride: Tartaric Acid-to-H2O ratio | [62] |
| Atlantic salmon frame bone | RSM | Box-Behnken Design (BBD) | Design-Expert v. 7 Trail | Supercritical carbon dioxide (SC-CO2) | Oil | Urea/ fatty acids ratio, crystallization temperature and crystallization time | [61] |
| Small-Spotted Catshark (S. canicula) skin | RSM | CCRD | Microsoft Excel spreadsheet | Alkaline pre-treatment, Acid-soluble collagen extraction |
Collagen | Chemical treatment (NaOH) concentration, temperature and time, concentration of acetic acid | [49] |
| Scallops (Argopecten purpuratus) byproducts | RSM | Box–Behnken Design | Minitab 19 | Enzymatic Hydrolysis | Protein Hydrolysate | Temperature, time, and enzyme concentration (enzyme/substrate level) | [60] |
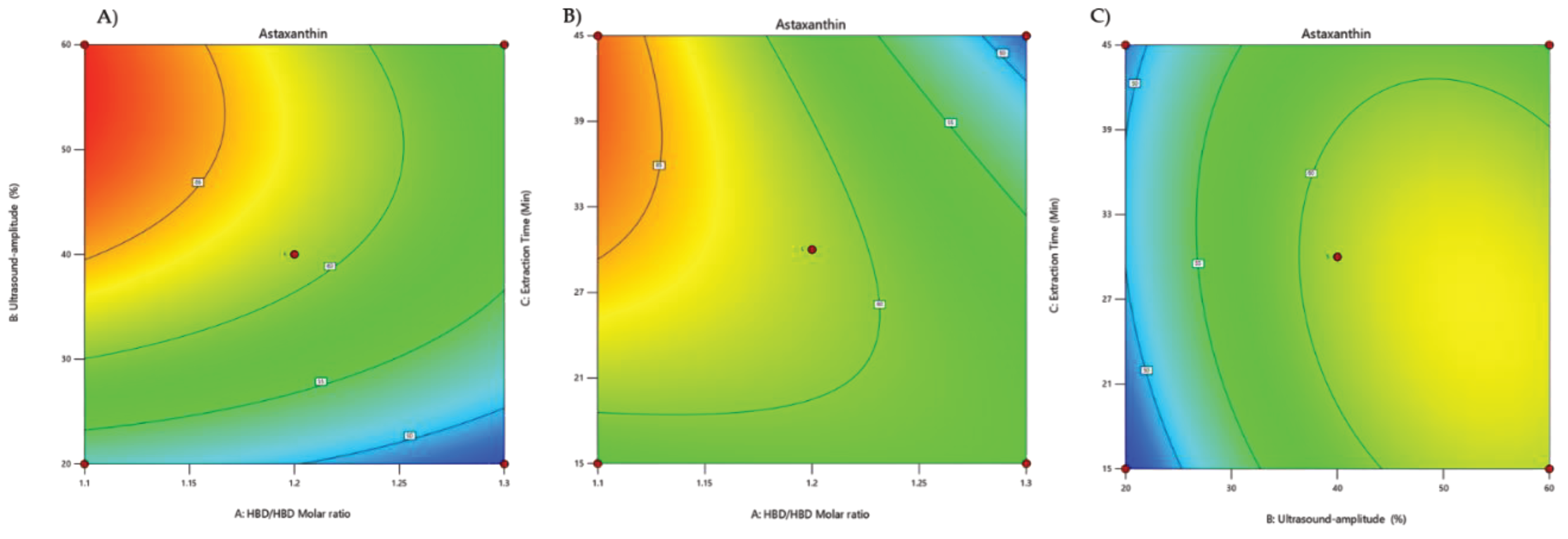
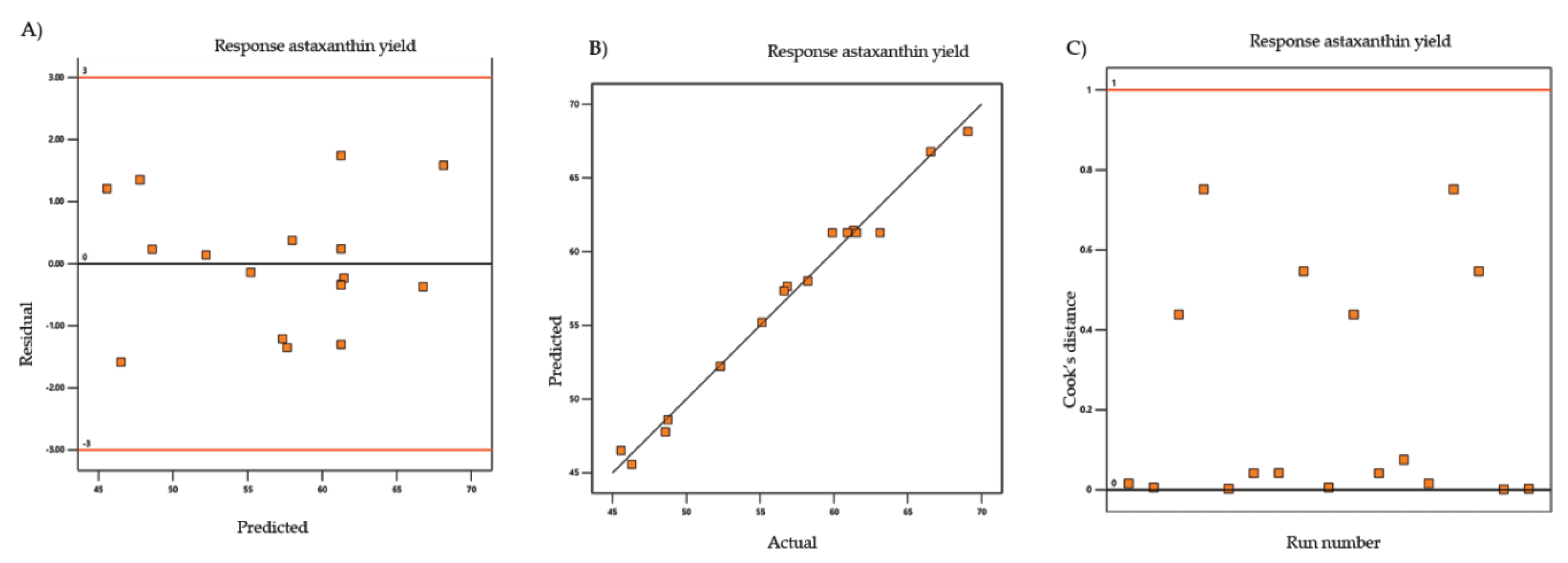
| Shrimp (Penaeus monodon) shells | RSM | Box-Behnken design | Design-Expert software (version 7.0.0) | Ultrasound-assisted natural deep eutectic solvents | Astaxanthin | Natural deep eutectic solvents molar ratio, Ultrasound-amplitude, Extraction Time | [59] |
| Indian white shrimp waste | RSM | BoxBehnken Design | Design Expert 7.1.6 and Minitab 16 statistical software | Chemical and Microwave method | Chitosan | Temperature, concentration of alkaline, time of reaction, power of microwave, Irradiation time | [58] |
| Marine shrimp processing raw byproducts | Plackett-Burman and BBD | Fermentation | Chitosanase | fermentation period, temperature, period of microwave pretreatment, K2HPO4 (%), MgSO4 (%), KCl (%), FeSO4·7H2O | [32] | ||
| Salmon (Salmo salar) backbones, heads, and viscera | RSM | Central composite rotatable design | Design-Expert Version 11 | Soxhlet and microwave-assisted extraction | Bioactive oils | Time, microwave power, and solid/liquid ratio | [55] |
| Monkfish (Lophius piscatorius) heads and viscera | Non-linear least-squares (quasi-Newton) method | Data-fitting and parametric estimations | Solver of Excel spreadsheet | Proteolytic digestion | Protein hydrolysates | pH, temperature, and protease concentration | [41] |
| Shrimp (Parapenaeus longirostris) shells waste | RSM | Box-Behnken Design | STATISTICA | Fermentation | Chitin and chitosan | Sucrose concentration, Shrimp shells waste concentration, inoculum size, incubation period | [31] |
| Undersized hakes (fish by-catch) | RSM | Box–Behnken Design | Statgraphics Centurion XVI | Enzymatic Hydrolysis | Protein hydrolysates | Enzyme/substrate (protein) ratio, %solids, time | [38] |
| Black tiger shrimp (Penaeus monodon) shells | RSM | Box–Behnken Design | Sigmaplot-11 Excel |
Enzymatic Hydrolysis | chitin | pH, temperature, agitation speed, enzyme substrate ratio, incubation time | [65] |
| Scyliorhinus canicula Discards | Non-linear least-squares (quasi-Newton) method | Rotatable second order design | SolverAid, Microsoft Excel spreadsheet | Enzymatic Hydrolysis | Protein Hydrolysates | Temperature and pH | [42] |
| Red shrimps, A. antennatus head | RSM | Box-Behnken Design | Statistica Version 10 | ultrasound assisted, microwave assisted extraction | Carotenoids | extraction time, ultrasound, microwave power and solvent/material ratio | [48] |
| Atlantic salmon (Salmo salar) heads, frames, viscera | RSM | Factorial design | Minitab 17.1 | Enzymatic Transesterification | Oil, biodiesel | Enzyme concentration, oil/alcohol molar ratio, time, and temperature | [56] |
| Fish by-product: heads, fins | RSM | CCRD | Design-Expert, Version 11 | Microwave-Assisted Extraction | Bioactive Fish Oil | Time, microwave power, and solid/liquid ratio | [3] |
| Salmonids (rainbow trout and salmon) heads, trimmings, frames | Nonlinear least-squares (quasi-Newton) method |
Second order rotatable design | Solver, Microsoft Excel spreadsheet | Enzymatic Hydrolysis | Protein hydrolysates | Enzyme concentration, pH, ratio (Solid:Liquid, time of hydrolysis, agitation speed | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





