1. Introduction
Citrus Bacterial Canker (CBC), caused by
Xanthomonas citri pv.
citri (
Xcc), stands as a significant bacterial disease with profound economic implications in tropical and subtropical citrus-growing regions [
1,
2]. The severity of symptoms varies across regions, potentially resulting in production losses and exportation restrictions. The pathogen was recently described in Burkina Faso by [
3]. Subsequent research by [
4] uncovered prevalent occurrences of bacterial canker in nurseries and production areas, with incidences ranging from 50 % to 90 % and corresponding fruit yield losses per hectare due to CBC is ranging from 18.45 % to 90.8 %, depending on the species.
Due to its economic significance, regulatory implications, and global distribution, the
Xcc strain has been the focus of numerous epidemiological and population structure studies [
5,
6]. Over the past decade, international research efforts have contributed to elucidating the global genetic diversity of
Xcc [
7,
8]. The foundation of
Xcc strain characterization lies in the type 3 secretion system (T3SS) and the associated proteins, particularly type 3 effectors (T3E), secreted during interactions with plant cells. Each Xanthomonas strain can produce a minimum of 15 different T3Es [
9], with a total of 26 T3Es identified in
Xcc through bioinformatics analysis [
10]. Notably, among Xanthomonas T3Es, a specific class known as TAL (Transcription Activator-Like) effectors act as transcription factors transferred to the nucleus, where they regulate host gene expression in favor of the pathogen [
11,
12]. The number of TALEs varies among strains of the Xanthomonas genus.
While extensive information exists on the global genetic structure of
Xcc populations, research specific to Burkina Faso is limited. Notably, only [
13] study has explored genetic connections between strains from Burkina Faso and Mali.
This study aims to investigate the presence of T3Es and TALEs in Xcc strains isolated in 2012, 2020, and 2021 from diverse citrus production sites in Burkina Faso. Additionally, the study seeks to analyze the correlation between T3Es content in Xcc strains, their geographical origins, and the years of their isolation.
2. Materials and Methods
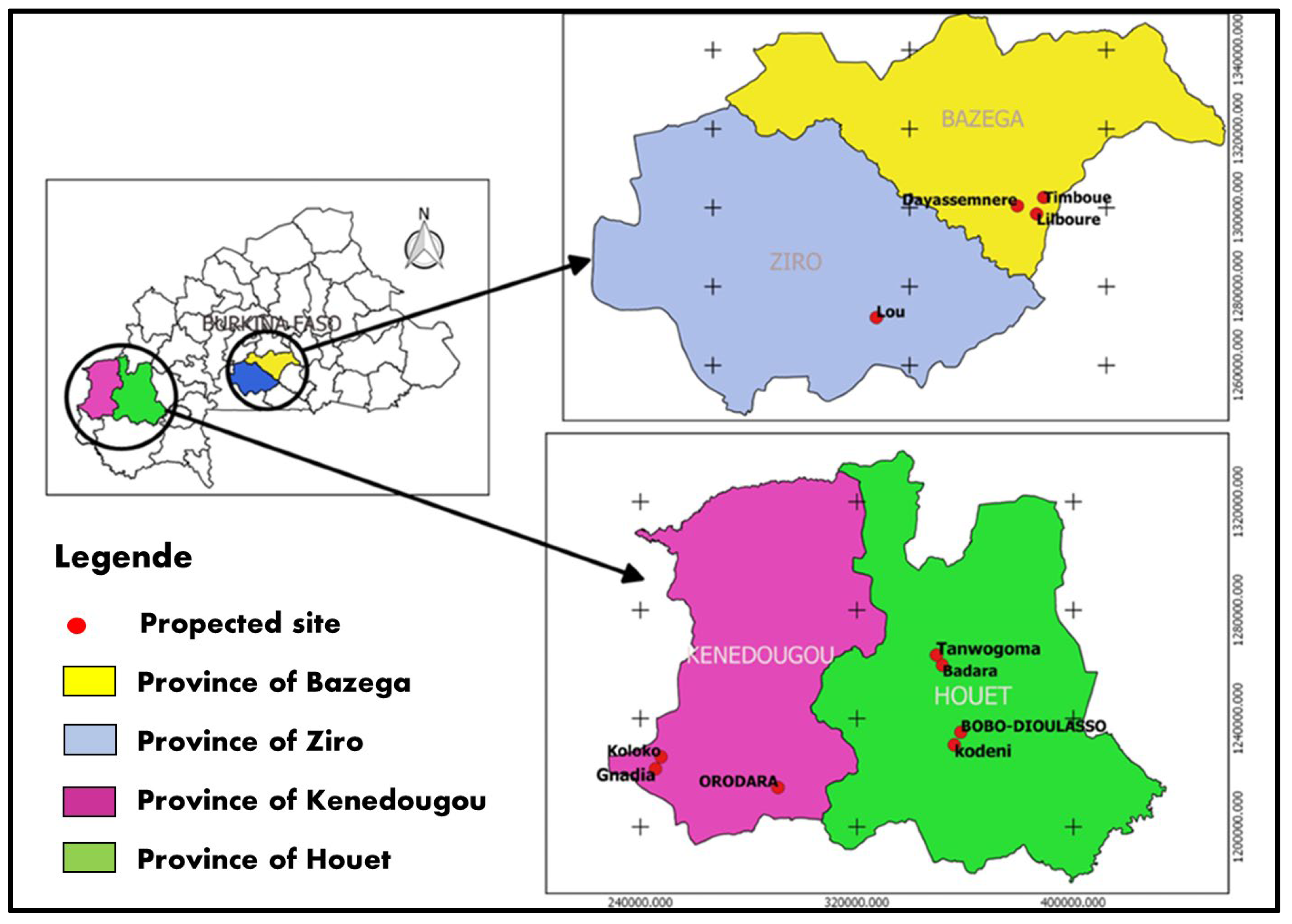
2.1. Collection Site
The study was carried out in four provinces belonging to three administrative regions of Burkina Faso, which are the main fruit production areas. These were the provinces of Bazega, Houet, Kénédougou and Ziro. In all, eleven (11) citrus production sites were surveyed. These were the Kodéni, Bobo Dioulasso, Badara and Tanwogoma production sites in Houet ; the Orodara, Koloko and Gnadia sites in Kénédougou ; the Lou site in Ziro and the Lilbouré, Timboué and Dayasmnere sites in Bazega (
Figure 1). All these sites belong to two agro-ecological zones in Burkina Faso. These are the northern Sudanian zone, with annual rainfall of between 700 and 900 mm, and the southern Sudanian zone, with annual rainfall of between 900 and 1200 mm.
2.2. Material
The study focused on
Xcc strains isolated from samples collected from the main citrus production sites in Burkina Faso. The samples were collected from all the citrus species hosting
Xcc produced at the sites. Surveys and isolations were carried out by the Phytopathology/Bacteriology Laboratory team at Institute for the Environment and Agricultural Research (INERA) Farakoba during 2012. Thirty-one (31)
Xcc strains isolated from their collection were used in our study. In addition, this collection was enriched with
Xcc strains isolated during our study.
Table (1) presents the characteristics of the 2012
Xcc strains used.
2.3. Methods
2.3.1. Isolation
The isolation process involved collecting fragments from symptomatic organ tissues, encompassing both affected and healthy portions. These fragments underwent successive disinfection steps in ethanol (70 %), bleach (0.1 %) and were subsequently rinsed in sterile distilled water. Following this, the fragments were ground and suspended in 1 ml of sterile distilled water. The suspension was allowed to stand for a minimum of 30 minutes at room temperature, with intermittent stirring to facilitate bacterial diffusion. A 50 µl aliquot of the suspension was then evenly spread into Yeast Peptone Glucose Agar (LPGA) nutrient medium, supplemented with antibiotics such as kasugamycin (20 mg/l), cephalexin (40 mg/l), and propiconazole (fungicide at 40 mg/l).
Incubation was carried out for 72 to 96 hours, during which bacterial colonies exhibiting morphological characteristics (ivory-white) akin to the reference strain were selected and purified for pathogenicity tests.
2.3.2. Pathogenicity Testing
To assess the pathogenicity of
Xcc strains, citrus plants cultivated under semi-controlled conditions were employed. Leaves were inoculated through infiltration with a bacterial suspension containing 10
8 bacteria/ml, corresponding to an optical density (OD) of 0.2 at 600 nm. The OD measurement was determined by placing 1 ml of the bacterial suspension into a spectrophotometer cuvette, calibrated at 0.2 with an uncertainty of 10 %, as per the following formula :
Vr = Volume of bacterial suspension remaining after sampling 1 ml to measure OD.
A citrus plant was inoculated with sterile distilled water to serve as a control. Inoculated plants were incubated under semi-natural conditions : 28-30 °C under an average relative humidity of 75 %. Symptoms were observed visually from 7 days after inoculation.
2.3.3. Identification of Xcc Strains
For the identification of Xcc strains, specific primers were used in PCR amplification. The primer sequences were REP_FW ATG-TCC-GAC-ATG-AAA-GTT-AAT-T and REP_RV CGC-TTC-TCC-TGC-ATT. The PCR reaction mixture, with a volume of 25 µl per reaction, comprised 1X PCR buffer colored with 5X at 3 mM MgCl2 (25 mM), 1 µM REP_FW primer (10 µM), 1 µM REP_RV primer (10 µM), 0.2 mM of each deoxynucleotide triphosphate (dNTP) (2.5 mM), 0.25 U (5 U/µl) of Taq DNA polymerase and 2.5 µl of DNA sample at 20 ng/µl. The PCR program involved an initial denaturation phase at 95 ºC for 2 min, followed by 35 cycles at 95 ºC for 60 sec, 58 ºC for 70 sec, and 72 ºC for 75 sec, concluding with a final elongation phase at 72 ºC for 10 min. The resulting amplification products, expected to be 222 bp in size, were analyzed on a 2 % agarose gel in 0.5X Tris- Borate-EDTA (TBE) buffer after staining with Ethidium Bromide (BET).
2.3.4. Analysis of Type 3 Effectors
From all the isolated
Xcc strains, 11 highly polymorphic T3E genes, as per [
14], were characterized (Table 2). DésoxyriboNucleic acid (DNA) extracted using the Promega Kit underwent PCR amplifications with a 20 ul reaction mixture consisting of 1X Go Taq Buffer (Promega), 200 mM dNTP, 0.5 µM of each primer, 0.4 U Go Taq Polymerase (final concentrations), and 1 µl of DNA (20 ng/µl).
The amplification conditions using T3E primers involved an initial denaturation phase of 2 min at 94 °C, followed by 30 reaction cycles comprising denaturation at 94 °C for 1 min, hybridization at 60 °C for 1 min, elongation at 72 °C for 2 min, and a final extension of 10 min at 72 °C [
14]. The resulting amplicons were separated by 1 % agarose gel electrophoresis containing 2 % BET in 0.5X TAE buffer (Tris Acetate 50 mM, 1 mM) at 100 Volt for 40 min.
Table 2.
List of T3Es used in this study.
Table 2.
List of T3Es used in this study.
| Oligo name |
Sequence |
Th °C |
Size
(bp) |
| xopB-F |
ATG-AAG-GCA-GAG-CTC-ACA-CGA |
49,2 |
1835 |
| xopB-R |
TCA-GGC-GCG-GGT-TGG-TGC-GAA-GTA |
57,4 |
| xopD-F |
ATG-GAA-TAT-ATA-CCA-AGA-ATG-AAG-C |
46 |
1638 |
| xopD-R |
CTA-GAA-CTT-TTT-CCA-CCA-CTT-GC |
48,4 |
| xopE2-F |
ATG-GGG-CTA-TGC-AGT-TCA-AAG-C |
49,7 |
393 |
| xopE2-R |
TCA-CCA-ACT-CAA-GGG-TGG-GC |
64 |
| xopO-F |
ATG-ATC-AAC-ACT-TCC-GTC-AAG |
45,3 |
636 |
| xopO-R |
TCA-CCT-GTT-TAT-CCG-ACG-AC |
60 |
| xopN-F |
ATG-AAG-TCA-TCC-GCA-TCC-GTC |
49,2 |
2090 |
| xopN-R |
GTG-CAG-TTC-CGT-CCC-CAA-GTT-CC |
55,5 |
| xopQ-F |
ATG-CAG-CCC-ACC-GCA-ATC |
58 |
1224 |
| xopQ-R |
TCA-GCG-CCC-GCG-TTG-CC |
60 |
| xopX-F |
ATG-GAG-ATC-AAG-AAA-CAG-CAA-ACC |
48,9 |
1865 |
| xopX-R |
TCA-GGA-CGA-AGG-CAC-AGT-GC |
64 |
| avrBs2-F |
GGA-CTA-GTC-CTG-CCG-GTG-TTG-ATG-CAC-GA |
60,7 |
2118 |
| avrBs2-R |
CCG-CTC-GAG-CGG-TGA-TCG-GTC-AAC-AGG-CTT-TC |
64,4 |
| avrBs1-F |
ATG-TCC-GAC-ATG-AAA-GTT-AAT-T |
42,3 |
1338 |
| avrBs1-R |
TTA-CGC-TTC-TCC-TGC-ATT-TG |
58 |
| avrXccA1-F |
ATG-CCC-AAC-GCA-TCG-CCT-GCA |
53,1 |
813 |
| avrXccA1-R |
CGC-AGG-TCT-GCA-CCG-AAC-TCA-G |
55,3 |
| avrXccA2-F |
GTG-CTG-GAG-AGT-GCC-G |
54 |
1442 |
| avrXccA2-R |
TCA-TCG-GCA-GGG-TTG-TGG |
58 |
2.3.5. Amplification of TAL Effectors
The primers REP_FW (GAGTTGAGAGGTCCACCGTTAC) and REP_RV (GGGAAGACGGCGATTGGTTC) described by [
8] were used to amplify the TAL effectors of
Xcc strains. They are positioned in the conserved regions of the TALEs close to the repeats. PCR assays were performed using 50 μl of reaction mixture containing 1 μl of DNA at 20 ng/µl, 10 μl of 5X Buffer, 1 µl of dNTPs (10 mM), 0.5µl of betaine (100 mM), 0.32 µl of 10X GO Taq buffer (Promega) and 1.5 µl of each primer (10 mM). The PCR programme included DNA denaturation at 95 °C for 2 min, 25 cycles comprising DNA denaturation at 95 °C for 40 sec, hybridisation at 58 °C for 40 sec and extension at 72 °C for 8 sec. PCR products were migrated by electrophoresis in a 1 % agarose gel in TAE 1X (TRIS-acetate 40 mM, EDTA 1 mM) for 2 h at 70 Volt and stained with BET (2 µg. mL-1).
2.3.6. Analysis of Strain Structuring
To analyse the structure of the strains and determine the links between them, classification and correlation parameters were studied. These were the Hierarchical Ascending Classification (HAC) and the correlation matrix (PCA). The AHC was represented by a dendrogram. The parameters concerned were the years of collection of the strains, their geographical origins and their T3E repertoires.
2.4. Data Analysis
To interpret the results a reaction was considered positive if a single clear band of the expected size was detected on the agarose gel; if no band was detected, the reaction was considered negative. The distribution was then presented according to the criterion of presence or absence of the characterised effectors. Minitab software 2018 and XLSTAT 2016 software packages were used for the statistical analysis of the structuring data and the representation of the graphs.
Results
3.1. Strains Isolated
Based on the symptoms shown in
Figure 2A, laboratory isolations showed strains with pale yellow colonies, circular in shape and mucous in appearance (
Figure 2B).
The pathogenicity test on the strains enabled us to observe symptoms characteristic of Xcc on C. reticulata × C. paradisi. This confirms that all the strains are pathogenic.
Following surveys carried out during the 2020 and 2021 periods in citrus-growing areas in Burkina Faso, 17 strains were isolated, including 7 strains isolated in 2020 and 10 strains isolated in 2021. Twelve (12) strains were isolated to C. reticulata × C. paradisi, tree (3) strains to Citrus maxima and two (2) to Citrus limon. Most of the strains isolated come from the Houet locality. The small number of strains used is linked to the extreme difficulty encountered during isolation and also to the storage conditions during which many strains were lost.
Table 3.
Characteristics of strains used.
Table 3.
Characteristics of strains used.
| Strain code |
Host plant |
Provence |
Site |
Year of collection |
| 80 |
C. maxima |
Bazega |
Timboué |
2020 |
| 81 |
C. limon |
Ziro |
Lou |
2020 |
| 82 |
C. reticulata × C. paradisi |
Houet |
Kodéni |
2020 |
| 83 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2020 |
| 84 |
C. reticulata × C. paradisi |
Houet |
Bobo Dioulasso |
2020 |
| 85 |
C. reticulata × C. paradisi |
Houet |
Kodéni |
2020 |
| 86 |
C. reticulata × C. paradisi |
Houet |
Bobo Dioulasso |
2020 |
| 92 |
C. maxima |
Bazega |
Dayasmeré |
2021 |
| 94 |
C. reticulata × C. paradisi |
Houet |
Kodéni |
2021 |
| 95 |
C. reticulata × C. paradisi |
Houet |
Kodéni |
2021 |
| 98 |
C. reticulata × C. paradisi |
Ziro |
Lou |
2021 |
| 99 |
C. maxima |
Bazega |
Lilbouré |
2021 |
| 100 |
C. reticulata × C. paradisi |
Bazega |
Lilbouré |
2021 |
| 102 |
C. reticulata × C. paradisi |
Houet |
Bobo Dioulasso |
2021 |
| 104 |
C. limon |
Houet |
Kodéni |
2021 |
| 106 |
C. reticulata × C. paradisi |
Houet |
Bobo Dioulasso |
2021 |
| 107 |
C. reticulata × C. paradisi |
Ziro |
Lou |
2021 |
3.2. Distribution of Non-TAL Type 3 Effectors in Xcc Strains
The distribution pattern of the 11 T3Es among the 48 Xcc strains is outlined in table 4. Broadly, strains isolated in 2020 and 2021 exhibit a more diverse range of T3E repertoires compared to those from 2012. Specifically, the genes XopX, XopQ, XopD, XopO, and AvrBS1 were absent in strains collected in 2012 but were present in those isolated in 2020 and 2021. Moreover, all 11 screened effectors were identified in three strains, including one from 2020 (Xcc85) and two from 2021 (Xcc102 and Xcc104). The distribution of T3Es within strains isolated in 2012 demonstrated a nearly uniform pattern.
The percentage representation of T3Es indicates a widespread presence of three effectors, namely XopE2, XopN, and AvrBs2, across the strains. Conversely, the least prevalent effectors are AvrBS1 (16.4 %), XopO (16.7 %), and XopX (18.7 %).
Table 4.
Distribution of the 11 T3Es within Xcc strains.
Table 4.
Distribution of the 11 T3Es within Xcc strains.
| Strains Code |
Type 3 effectors |
| XopB |
XopE2 |
XopN |
XopX |
XopQ |
XopD |
XopO |
AvrxccA1 |
AvrxccA2 |
AvrBS1 |
AvrBs2 |
| Strains isolated in 2020 |
| Xcc80 |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
- |
+ |
| Xcc81 |
- |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
- |
+ |
| Xcc82 |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
- |
+ |
| Xcc83 |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
- |
+ |
| Xcc84 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
| Xcc85 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| Xcc86 |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
| Strains isolated in 2021 |
| Xcc92 |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
- |
+ |
| Xcc94 |
+ |
+ |
+ |
- |
|
- |
- |
- |
- |
- |
+ |
| Xcc95 |
+ |
+ |
+ |
- |
+ |
- |
- |
- |
- |
- |
+ |
| Xcc98 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
| Xcc99 |
- |
+ |
+ |
- |
+ |
- |
- |
- |
+ |
- |
+ |
| Xcc100 |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| Xcc102 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| Xcc104 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| Xcc106 |
+ |
+ |
+ |
- |
+ |
+ |
- |
+ |
+ |
+ |
+ |
| Xcc107 |
- |
+ |
+ |
- |
- |
+ |
- |
+ |
+ |
+ |
+ |
| Strains isolated in 2022 |
| Xcc864 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc865 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc866 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc867 |
+ |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
+ |
| Xcc868 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc869 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc870 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc871 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc872 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc873 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc874 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc875 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc876 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc877 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc878 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc879 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc880 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc881 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc882 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc883 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc884 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc885 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc886 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc887 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc888 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc889 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc890 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc891 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc895 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc896 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Xcc897 |
+ |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
| Attendance percentage (%) |
| + |
91,6 |
100 |
100 |
18,7 |
27,1 |
27,1 |
16,7 |
91,6 |
84,4 |
16,7 |
100 |
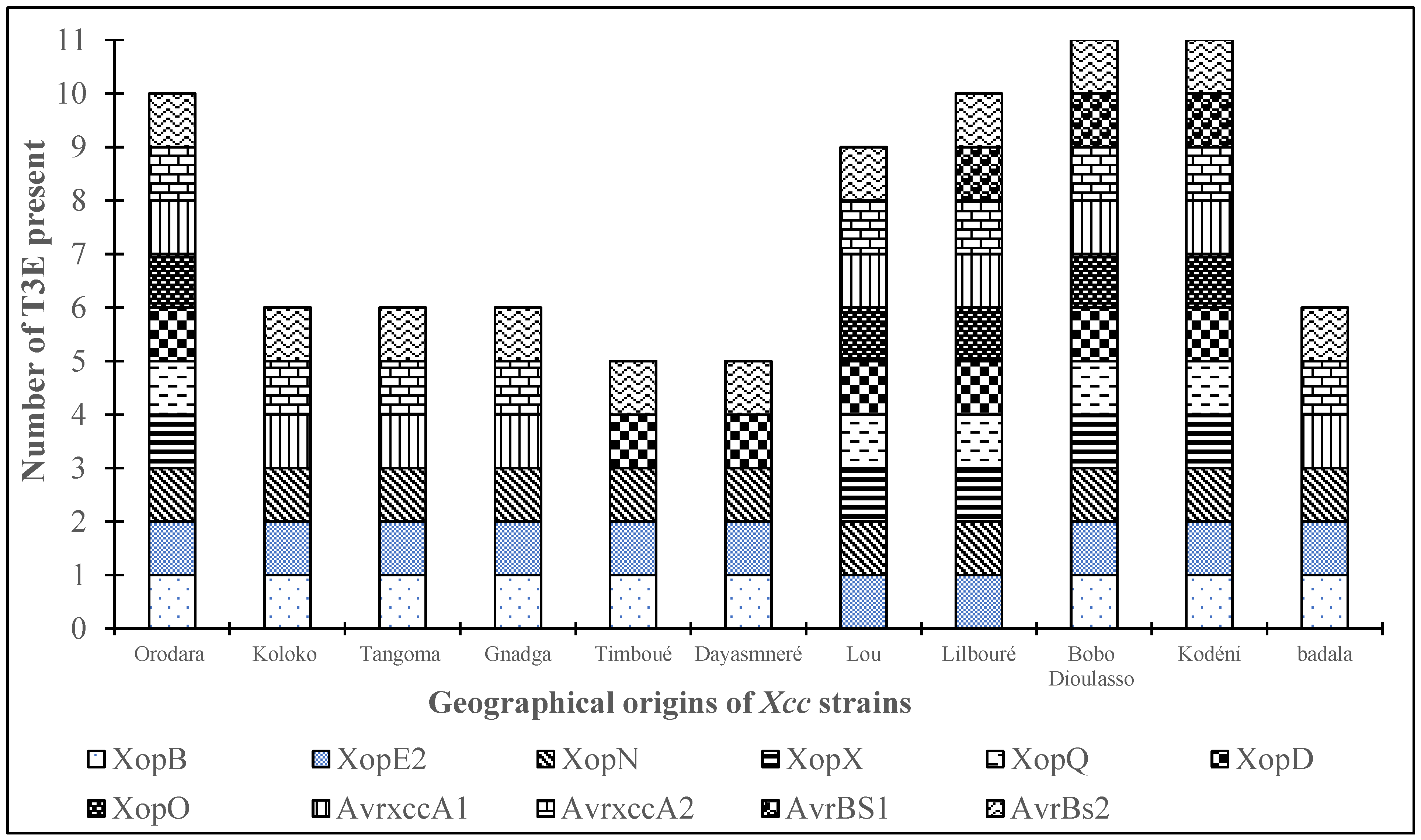
3.3. Geographical Distribution of T3Es
The distribution of T3Es based on the geographical origins of the strains is illustrated in
Figure 4. The graph reveals that all 11 screened T3Es were identified in the Bobo-Dioulasso and Kodéni sites, showcasing the highest diversity. Following closely, the Orodara and Lilbouré sites recorded the presence of 10 T3Es, while the Timboué and Dayasmnaré sites exhibited a lower diversity with five (05) T3Es. The Koloko, Tangoma and Gnadga sites each registered six (06) T3Es.
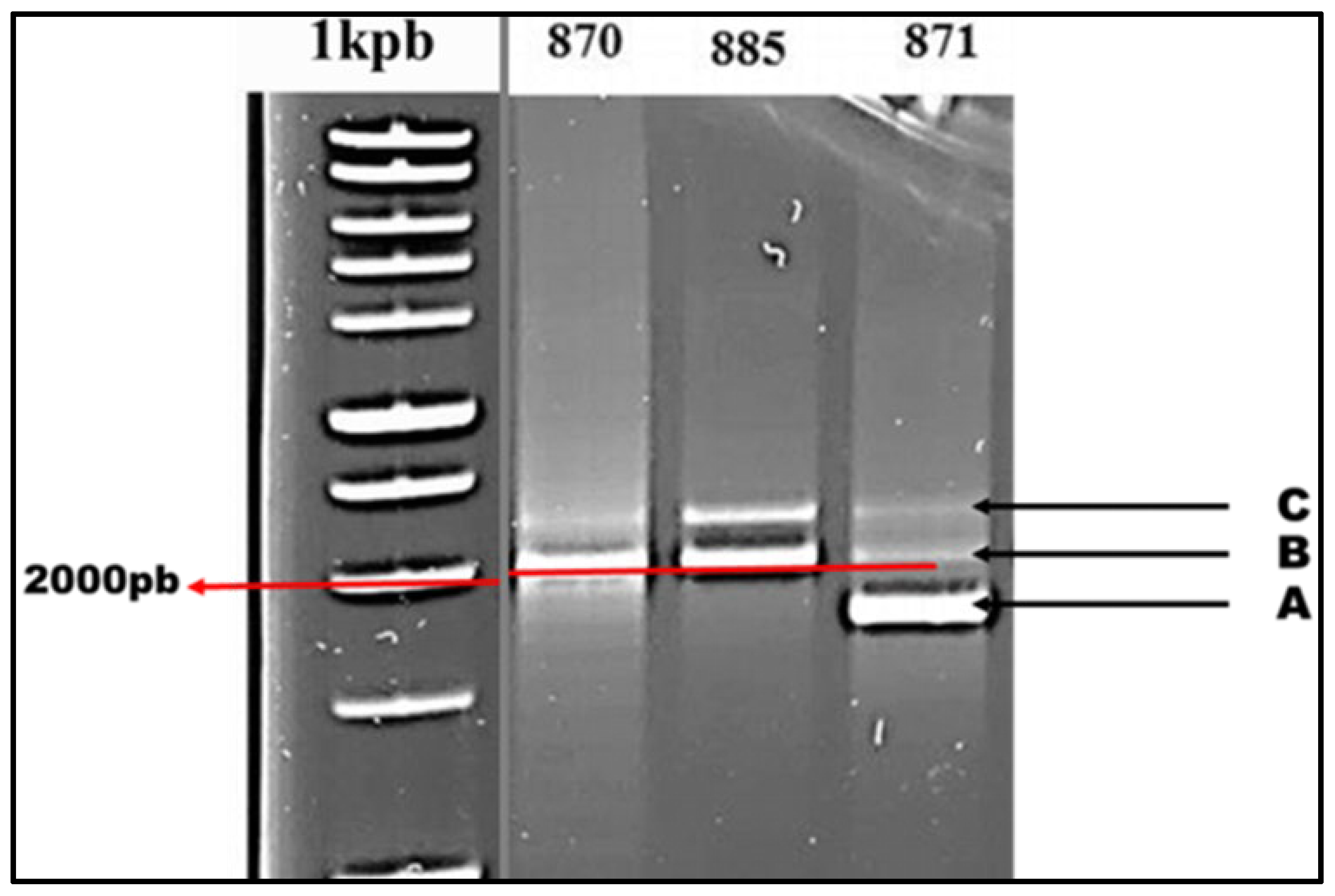
3.4. TALEs Diversity in Xcc Strains
A study of the distribution of TALEs within the collection of
Xcc strains reveals the presence of two distinct profiles, labeled as Profile 1 and Profile 2, with varying numbers of TALEs ranging from two to three (
Figure 5). Strains within Profile 1 exhibit two TALEs (B and C), with TALE B amplified at 2000 bp and TALE C slightly above 2000 bp. Conversely, Profile 2 strains have an additional TALE (A) compared to Profile 1 strains, and its amplification occurs below 2000 bp
3.5. Distribution of TALEs by Strain
The descriptive analysis of TALE distribution, as presented in table 4, highlights that TALEs B and C are universally distributed (100 %) and are common to all isolated Xcc strains. TALE A, on the other hand, was present in 68.7 % of the isolates.
Profile characterization reveals that out of the 48 tested Xcc strains, 37 strains exhibit Profile 1, while the remaining 11 strains display Profile 2. Among the 11 Profile 2, Xcc80 and Xcc84 strains, collected in 2020, Xcc92 and Xcc102 in 2021, and the remaining seven (Xcc871, Xcc879, Xcc881, Xcc882, Xcc887, Xcc896, Xcc897) in 2012.
Table 5.
Distribution of TALEs by Xcc strain.
Table 5.
Distribution of TALEs by Xcc strain.
| Years |
Strains |
TALE |
Profil |
| A |
B |
C |
| 2020 |
Xcc80 |
+ |
+ |
+ |
2 |
| Xcc81 |
- |
+ |
+ |
1 |
| Xcc82 |
- |
+ |
+ |
1 |
| Xcc83 |
- |
+ |
+ |
1 |
| Xcc84 |
+ |
+ |
+ |
2 |
| Xcc85 |
- |
+ |
+ |
1 |
| Xcc86 |
- |
+ |
+ |
1 |
| 2021 |
Xcc92 |
+ |
+ |
+ |
2 |
| Xcc94 |
- |
+ |
+ |
2 |
| Xcc95 |
- |
+ |
+ |
1 |
| Xcc98 |
- |
+ |
+ |
1 |
| Xcc99 |
- |
+ |
+ |
1 |
| Xcc100 |
- |
+ |
+ |
1 |
| Xcc102 |
+ |
+ |
+ |
1 |
| Xcc104 |
- |
+ |
+ |
1 |
| Xcc106 |
- |
+ |
+ |
1 |
| Xcc107 |
- |
+ |
+ |
1 |
| 2012 |
Xcc864 |
- |
+ |
+ |
1 |
| Xcc865 |
- |
+ |
+ |
1 |
| Xcc866 |
- |
+ |
+ |
1 |
| Xcc867 |
- |
+ |
+ |
1 |
| Xcc868 |
- |
+ |
+ |
1 |
| Xcc869 |
- |
+ |
+ |
1 |
| Xcc870 |
- |
+ |
+ |
1 |
| Xcc871 |
+ |
+ |
+ |
2 |
| Xcc872 |
- |
+ |
+ |
1 |
| Xcc873 |
- |
+ |
+ |
1 |
| Xcc874 |
- |
+ |
+ |
1 |
| Xcc875 |
- |
+ |
+ |
1 |
| Xcc876 |
- |
+ |
+ |
1 |
| Xcc877 |
- |
+ |
+ |
1 |
| Xcc878 |
- |
+ |
+ |
1 |
| Xcc879 |
+ |
+ |
+ |
2 |
| Xcc880 |
- |
+ |
+ |
1 |
| Xcc881 |
+ |
+ |
+ |
2 |
| Xcc882 |
+ |
+ |
+ |
2 |
| Xcc883 |
- |
+ |
+ |
1 |
| Xcc884 |
- |
+ |
+ |
1 |
| Xcc885 |
- |
+ |
+ |
1 |
| Xcc886 |
- |
+ |
+ |
1 |
| Xcc887 |
+ |
+ |
+ |
2 |
| Xcc888 |
- |
+ |
+ |
1 |
| Xcc889 |
- |
+ |
+ |
1 |
| Xcc890 |
- |
+ |
+ |
1 |
| Xcc891 |
- |
+ |
+ |
1 |
| Xcc895 |
- |
+ |
+ |
1 |
| Xcc896 |
+ |
+ |
+ |
2 |
| Xcc897 |
+ |
+ |
+ |
2 |
| Attendance percentage (%) |
|
| Present |
68,7 |
100 |
100 |
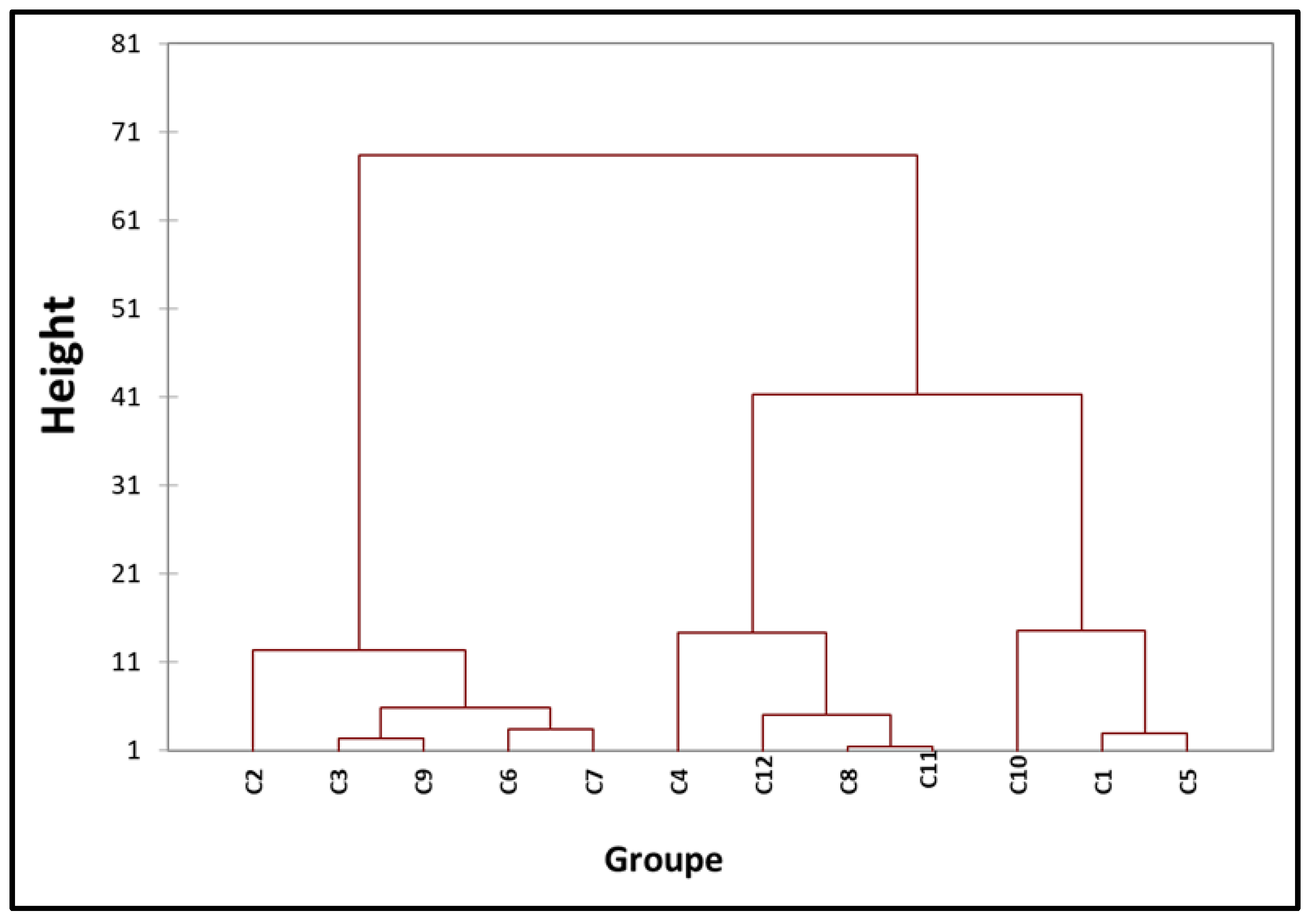
3.6. Hierarchical Ascending Classification
Multivariate analysis was employed to cluster strains based on their geographical origin, TAL and non-TAL type 3 effector content, and year of isolation. Considering these parameters for each strain, 12 distinct groups were identified, as depicted in the dendrogram in
Figure 5. The dendrogram facilitated the categorization of all
Xcc strains into 12 classes. Strains isolated in 2012 were dispersed among classes C5, C10, and C11, while those from 2020 were predominantly represented in class C7. In contrast, strains isolated in 2021 were distributed across the remaining classification classes. Among the three strains isolated in 2020 and 2021 with all tested TALEs, two (Xcc85 and Xcc104) were placed in class C7. Strains with three TALEs (Profile 2) isolated in 2012 were more prominent in class C11, while those from 2012 (Xcc92 and Xcc94) were grouped in class C4.
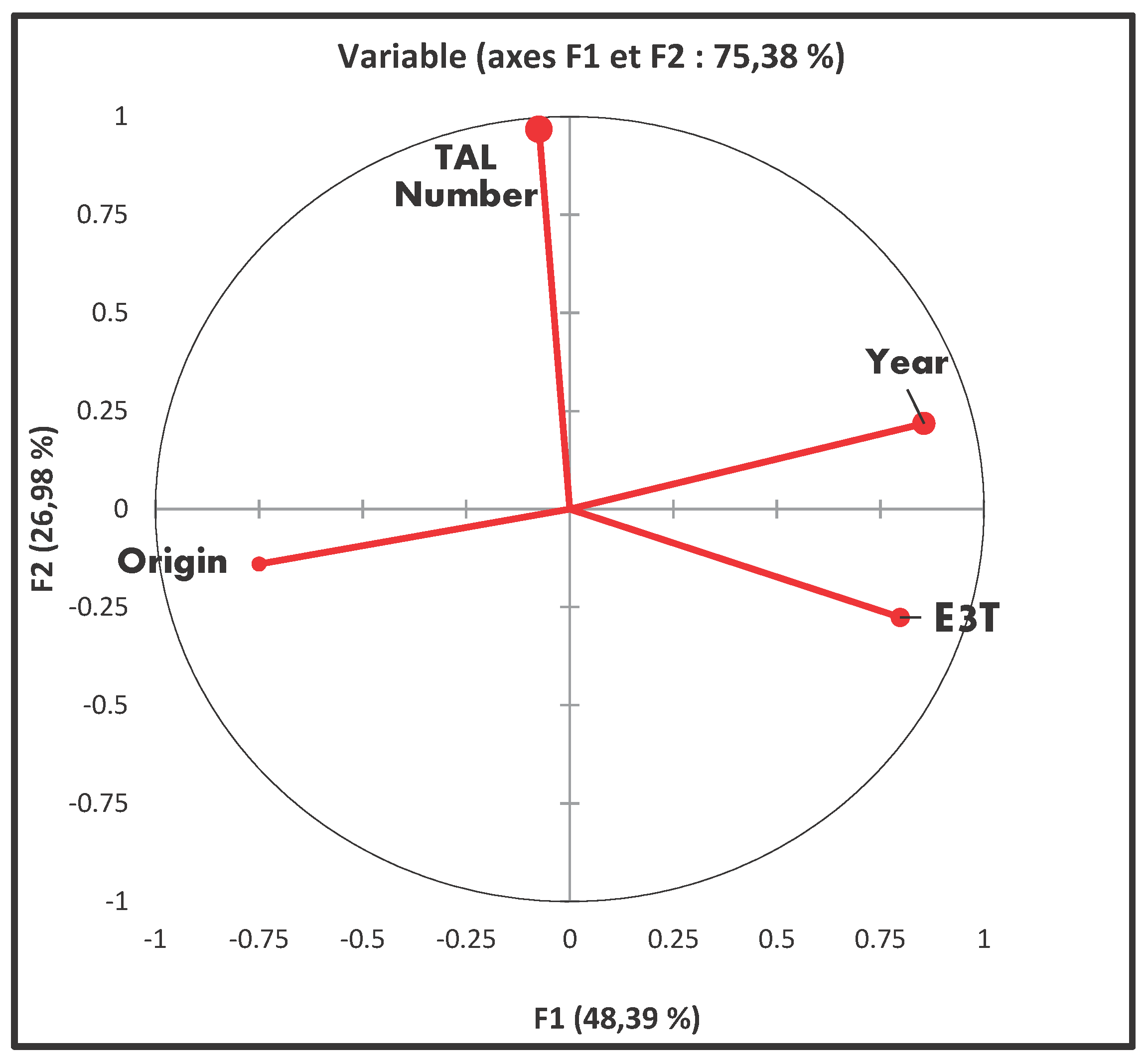
3.7. Principal Component Analysis
The graph depicting the correlation matrix between the geographical origin of the strains, their years of isolation, and their TALE and T3E content is presented in
Figure 7. Analysis of variance (p-value ˂ 0.0001) indicated correlations among the different factors. A positive correlation was observed between the T3E repertoire and the years of strain isolation. Conversely, a negative correlation existed between the years of strain collection and their geographical origins. Notably, no correlation was found between the TALE content of the strains and either their years of isolation or their geographical origins.
3. Discussion
The characterization T3E content in
Xcc strains unveiled the presence of T3E gene polymorphism. Specifically, three T3E genes XopE2, XopN, and AvrBs2 were universally present in all
Xcc strains. Notably, strains isolated in 2020 and 2021 exhibited a shared T3E repertoire and displayed greater diversity compared to strains isolated in 2012. The consistent presence of these genes aligns with the findings of [
15], suggesting their involvement in
Xcc pathogenicity. The observed differences in T3E content between 2012 and subsequent years could be attributed to the continued cultivation of susceptible varieties and the exchange of plant material within the country and neighboring regions such as Mali and Côte d'Ivoire, fostering more diverse and extensive
Xcc populations.
The analysis of TALE content revealed that all screened
Xcc strains carried TALEs. Out of the 48 tested strains, 11 had three TALEs, while 37 had two. This minimum presence of two TALEs aligns with [
8]. Strains with three TALEs, exhibiting characteristics of the A* pathotype, were observed, consistent with previous research by [
13] indicating the presence of the A* pathotype in Burkina Faso.
However, the weak structuring of T3E content based on geographical origin implies limited strain variability, potentially due to the restricted number of strains analyzed and the predominant sampling from the Orodara locality, a key area for seedling production and distribution. Indeed, [
6] showed that most
Xcc strains from Mauritius or Réunion were grouped in RCCs distributed throughout the island. Most of the haplotypes from Grande Comore, Anjouan and Mayotte were closely related genetically. The intra-island distribution of
Xcc haplotypes is often restricted to a single citrus block or to a spatially close block. This suggests the spread of contaminated citrus plant material from commercial nurseries. Diversification factors, including environmental conditions, may also contribute to the observed strain structuring, with lower variability in environmental factors across collection areas hindering significant strain evolution. [
13] showed that strains originating from Mali and Burkina Faso were genetically closer because of their geographical proximity and almost identical inter-zone environmental conditions. However, our results are in disagreement with the work of [
16] who showed that the geography and relative proximity of southern states, in particular Santa Catarina and Rio Grande do Sul, and neighbouring countries where citrus canker is endemic in South America (Argentina, Paraguay and Uruguay) may have facilitated the exchange of infected plant material, leading to the establishment of relatively different populations of
X.
citri subsp.
citri.
Analyzing the relationship between T3E distribution and the year of collection revealed a positive correlation, suggesting an evolution in strain distribution over time, independent of geographical origin. This underscores potential scenarios governing the epidemiology and evolutionary dynamics of
Xcc populations in Burkina Faso. Escalons' work suggests that the pathogenicity of
Xcc strains evolves by lineage through convergence. These results also illustrate the dynamic nature of a pathogen's host range dynamic nature of a pathogen's host range [
17].
Conclusion
The investigation into the T3E content of Xcc strains demonstrated a common repertoire of three identical T3E genes (XopE2, XopN, and AvrBs2) among all tested strains. However, variability in T3E content was evident in strains isolated in 2020 and 2021, contrasting with the limited diversity observed in strains from 2012. Characterization of TALEs indicated the presence of the 17.5-repeat TALE B in all tested strains, a key contributor to canker symptoms in pathotype A strains. The study also identified two distinct TALE profiles, with 11 strains belonging to profile 2 and 37 strains to profile 1. Geographical origin did not strongly structure strain distribution, but a positive correlation emerged between T3E content and years of isolation, highlighting the independent distribution of strains over geographical locations and revealing the evolutionary dynamics of Xcc populations in Burkina Faso.
Author Contributions
Conceptualization, I.W. and K.B.F.Z.; methodology, K.B.F.Z. and FY; software, K.B.F.Z. and FY; formal analysis, K.B.F.Z; writing original draft preparation, K.B.F.Z. and FY., supervision, I.W. and I.S All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are available contacting the authors.
Acknowledgments
The authors would like to thank the phytopathology/bacteriology laboratory of INERA Farako-Bâ, Bobo Dioulasso (Burkina Faso), LMI PathoBios, France, and the plant clinic of NAZI Boni University, Bobo Dioulasso (Burkina Faso).
Conflicts of Interest
The authors of this article declare that they have no conflicts of interest.
References
- Ference, C. M.; Gochez, A. M.; Behlau, F.; Wang, N.; Graham, J. H.; Jones, J. B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Wang, N. The citrus hanglongbing crisis and potential solutions. Mol. Plant 2019, 12, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, C. C.; Leduc, A.; Boyer, C.; Guérin, F.; Vernière, C.; Pruvost, O.; Wonni, I.; Ouedraogo, L. First report of Xanthomonas citri pv. citri Causing Asiatic Citrus Canker in Burkina Faso. Plant Dis. 2013, 97, 1653–1653. [Google Scholar] [CrossRef] [PubMed]
- Zerbo, K. B. F.; Wonni, I.; Yameogo, F.; Somda, I. , Antibacterial activity of some substances against citrus bacterial canker caused by Xanthomonas citri pv. citri (Hasse) in Burkina Faso. Afr. J. Plant Sci. 2022, 16, 138–147. [Google Scholar]
- Young, J. M.; Park, D.-C.; Shearman, H. M.; Fargier, E. A Multilocus Sequence Analysis of the Genus Xanthomonas. Syst. Appl. Microbiol. 2008, 31, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Pruvost, O.; Richard, D.; Boyer, K.; Javegny, S.; Boyer, C.; Chiroleu, F.; Grygiel, P.; Parvedy, E.; Robène, I.; Maillot-Lebon, V.; Hamza, A.; Lobin, K. K.; Naiken, M.; Vernière, C. Diversity and geographical structure of Xanthomonas citri pv. citri on Citrus in the South West Indian Ocean Region. Microorganisms 2021, 9. [Google Scholar] [CrossRef]
- Bui Thi Ngoc, L.; Vernière, C.; Belasque, J.; Vital, K.; Boutry, S.; Gagnevin, L.; Pruvost, O. Ligation-Mediated PCR, a fast and reliable technique for insertion sequence-based typing of Xanthomonas citri pv. citri. FEMS Microbiol. Lett. 2008, 288, 33–39. [Google Scholar] [CrossRef]
- 8. Aline Escalon. Evolution et spécialisation du pouvoir pathogène de Xanthomonas citri pv. citri : Rôle des effecteurs de Type 3. Agritop.
- Kay, S.; Hahn, S.; Marois, E.; Wieduwild, R.; Bonas, U. Detailed analysis of the DNA recognition motifs of the Xanthomonas Type III Effectors AvrBs3 and AvrBs3Δrep16. Plant J. 2009, 59, 859–871. [Google Scholar] [CrossRef]
- Da Silva, A. C. R.; Ferro, J. A.; Reinach, F. C.; Farah, C. S.; Furlan, L. R.; Quaggio, R. B.; Monteiro-Vitorello, C. B.; Sluys, M. A. V.; Almeida, N. F.; Alves, L. M. C.; Do Amaral, A. M.; Bertolini, M. C.; Camargo, L. E. A.; Camarotte, G.; Cannavan, F.; Cardozo, J.; Chambergo, F.; Ciapina, L. P.; Cicarelli, R. M. B.; Coutinho, L. L.; Cursino-Santos, J. R.; El-Dorry, H.; Faria, J. B.; Ferreira, A. J. S.; Ferreira, R. C. C.; Ferro, M. I. T.; Formighieri, E. F.; Franco, M. C.; Greggio, C. C.; Gruber, A.; Katsuyama, A. M.; Kishi, L. T.; Leite, R. P.; Lemos, E. G. M.; Lemos, M. V. F.; Locali, E. C.; Machado, M. A.; Madeira, A. M. B. N.; Martinez-Rossi, N. M.; Martins, E. C.; Meidanis, J.; Menck, C. F. M.; Miyaki, C. Y.; Moon, D. H.; Moreira, L. M.; Novo, M. T. M.; Okura, V. K.; Oliveira, M. C.; Oliveira, V. R.; Pereira, H. A.; Rossi, A.; Sena, J. A. D.; Silva, C.; De Souza, R. F.; Spinola, L. A. F.; Takita, M. A.; Tamura, R. E.; Teixeira, E. C.; Tezza, R. I. D.; Trindade Dos Santos, M.; Truffi, D.; Tsai, S. M.; White, F. F.; Setubal, J. C.; Kitajima, J. P. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Castiblanco, L. F.; Gil, J.; Rojas, A.; Osorio, D.; Gutiérrez, S.; Muñoz-Bodnar, A.; Perez-Quintero, A. L.; Koebnik, R.; Szurek, B.; López, C.; Restrepo, S.; Verdier, V.; Bernal, A. J. TALE 1 from Xanthomonas axonopodis pv. manihotis acts as a Transcriptional Activator in plant cells and is important for pathogenicity in cassava Plants. Mol. Plant Pathol. 2013, 14, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Bonas, U.; Lahaye, T. TAL Effectors – pathogen strategies and plant resistance engineering. New Phytol. 2014, 204, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Leduc, A.; Traoré, Y. N.; Boyer, K.; Magne, M.; Grygiel, P.; Juhasz, C. C.; Boyer, C.; Guerin, F.; Wonni, I.; Ouedraogo, L.; Vernière, C.; Ravigné, V.; Pruvost, O. Bridgehead invasion of a monomorphic plant pathogenic bacterium: Xanthomonas citri pv. citri, an emerging citrus pathogen in Mali and Burkina Faso. Environ. Microbiol. 2015, 17, 4429–4442. [Google Scholar] [CrossRef] [PubMed]
- Hajri, A.; Pothier, J. F.; Fischer-Le Saux, M.; Bonneau, S.; Poussier, S.; Boureau, T.; Duffy, B.; Manceau, C. Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola. Appl. Environ. Microbiol. 2012, 78, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Escalon, A.; Javegny, S.; Vernière, C.; Noël, L. D.; Vital, K.; Poussier, S.; Hajri, A.; Boureau, T.; Pruvost, O.; Arlat, M.; Gagnevin, L. Variations in Type III Effector repertoires, pthological phenotypes and host range of Xanthomonas citri pv. citri pathotypes. Mol. Plant Pathol. 2013, 14, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Jaciani, F. J.; Ferro, J. A.; Ferro, M. I. T.; Vernière, C.; Pruvost, O.; Belasque, J. Genetic Diversity of a Brazilian strain collection of Xanthomonas citri subsp. citri based on the Type III Effector Protein Genes. Plant Dis. 2012, 96, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P.; Panstruga, R. A Molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 2011, 16, 117–125. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Geographical location of the Xcc strains used in this study.
Figure 1.
Geographical location of the Xcc strains used in this study.
Figure 2.
Citrus bacterial canker symptoms on Citrus reticulata × Citrus paradisi (A) and Morphological appearance of isolated Xcc strains (B).
Figure 2.
Citrus bacterial canker symptoms on Citrus reticulata × Citrus paradisi (A) and Morphological appearance of isolated Xcc strains (B).
Figure 3.
Symptoms induced by Xcc strains after inoculation Symptoms on the lower surface, (A) and Symptoms on the upper surface (B).
Figure 3.
Symptoms induced by Xcc strains after inoculation Symptoms on the lower surface, (A) and Symptoms on the upper surface (B).
Figure 4.
Distribution of E3T by localities. Legend: Each colour represents a T3E. The size of the diagram for each site depends on the number and the variability of T3E present.
Figure 4.
Distribution of E3T by localities. Legend: Each colour represents a T3E. The size of the diagram for each site depends on the number and the variability of T3E present.
Figure 5.
PCR detection of TALEs.
Figure 5.
PCR detection of TALEs.
Figure 6.
Hierarchical ascending classification. C1 (876) ; C2 (84) ; C3 (102) ; C4 (92, 94, 106) ; C5 (888, 877, 878, 874, 875, 889) ; C6 (95) ; C7 (82, 85, 104, 98, 81, 83, 86, 107) ; C8 (897) ; C9 (99, 100) ; C10 (864, 865, 866, 867, 868, 869, 870, 872, 873, 880, 883, 884, 885, 886, 887, 890, 891, 895) ; C11 (871, 879, 881, 882, 896) ; C12 (80). Dissimilarity : Euclidean distance, aggregation method: Ward's method, interclass, percentage: 95 %.
Figure 6.
Hierarchical ascending classification. C1 (876) ; C2 (84) ; C3 (102) ; C4 (92, 94, 106) ; C5 (888, 877, 878, 874, 875, 889) ; C6 (95) ; C7 (82, 85, 104, 98, 81, 83, 86, 107) ; C8 (897) ; C9 (99, 100) ; C10 (864, 865, 866, 867, 868, 869, 870, 872, 873, 880, 883, 884, 885, 886, 887, 890, 891, 895) ; C11 (871, 879, 881, 882, 896) ; C12 (80). Dissimilarity : Euclidean distance, aggregation method: Ward's method, interclass, percentage: 95 %.
Figure 7.
Degree of correlation between the distribution of T3E and the origin and year of collection of the strains.
Figure 7.
Degree of correlation between the distribution of T3E and the origin and year of collection of the strains.
Table 1.
Characteristics of strains used.
Table 1.
Characteristics of strains used.
| Strain code |
Host plant |
Province |
Site |
Year of collection |
| 864 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 865 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 866 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 867 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 868 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 869 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 870 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 871 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 872 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 873 |
C. reticulata × C. paradisi |
Kénédougou |
Orodara |
2012 |
| 879 |
C. clementina |
Kénédougou |
Orodara |
2012 |
| 880 |
C. clementina |
Kénédougou |
Orodara |
2012 |
| 881 |
C. clementina |
Kénédougou |
Orodara |
2012 |
| 882 |
C. sinensis |
Kénédougou |
Orodara |
2012 |
| 883 |
C. sinensis |
Kénédougou |
Orodara |
2012 |
| 884 |
C. sinensis |
Kénédougou |
Orodara |
2012 |
| 885 |
C. sinensis |
Kénédougou |
Orodara |
2012 |
| 886 |
C. reticulata |
Kénédougou |
Orodara |
2012 |
| 890 |
C. volkameriana |
Kénédougou |
Orodara |
2012 |
| 891 |
C. volkameriana |
Kénédougou |
Orodara |
2012 |
| 895 |
C. volkameriana |
Kénédougou |
Orodara |
2012 |
| 896 |
C. volkameriana |
Kénédougou |
Orodara |
2012 |
| 897 |
C. volkameriana |
Kénédougou |
Orodara |
2012 |
| 887 |
C. sinensis |
Houet |
Tangoma |
2012 |
| 876 |
C. reticulata × C. paradisi |
Houet |
Badala |
2012 |
| 888 |
C. sinensis |
Houet |
Gnadga |
2012 |
| 889 |
C. reticulata × C. paradisi |
Kénédougou |
Koloko |
2012 |
| 874 |
C. reticulata × C. paradisi |
Kénédougou |
Koloko |
2012 |
| 875 |
C. reticulata × C. paradisi |
Kénédougou |
Koloko |
2012 |
| 877 |
C. sinensis |
Kénédougou |
Koloko |
2012 |
| 878 |
C. sinensis |
Kénédougou |
Koloko |
2012 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).