Submitted:
05 June 2024
Posted:
07 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. History of CRISPR/Cas System
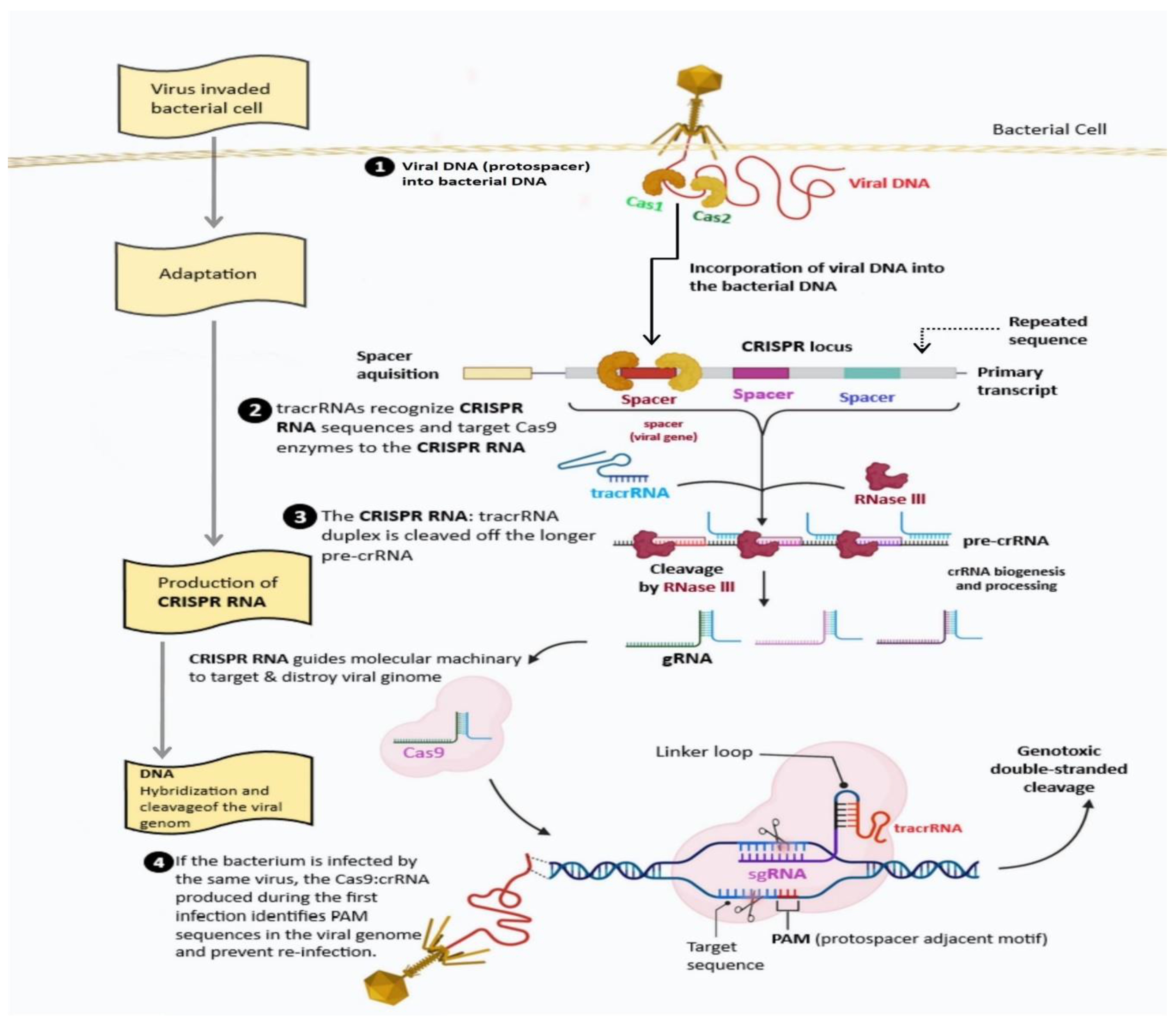
3. Nomenclature and Mechanism of CRISPR/Cas System
4. Cas proteins of the CRISPR System
4.1. Cas 1 and Cas 2 Proteins
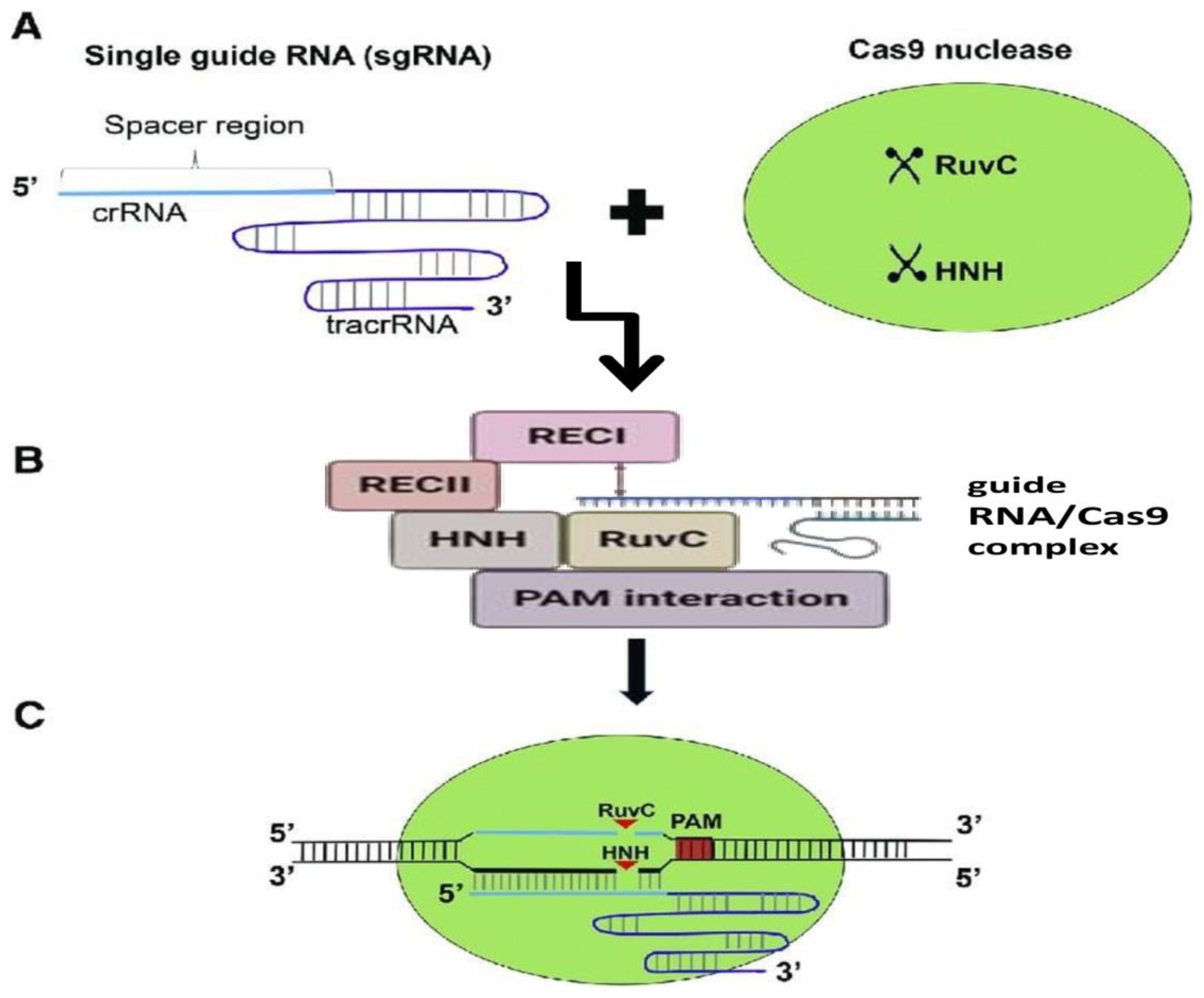
4.2. Cas9 Protein
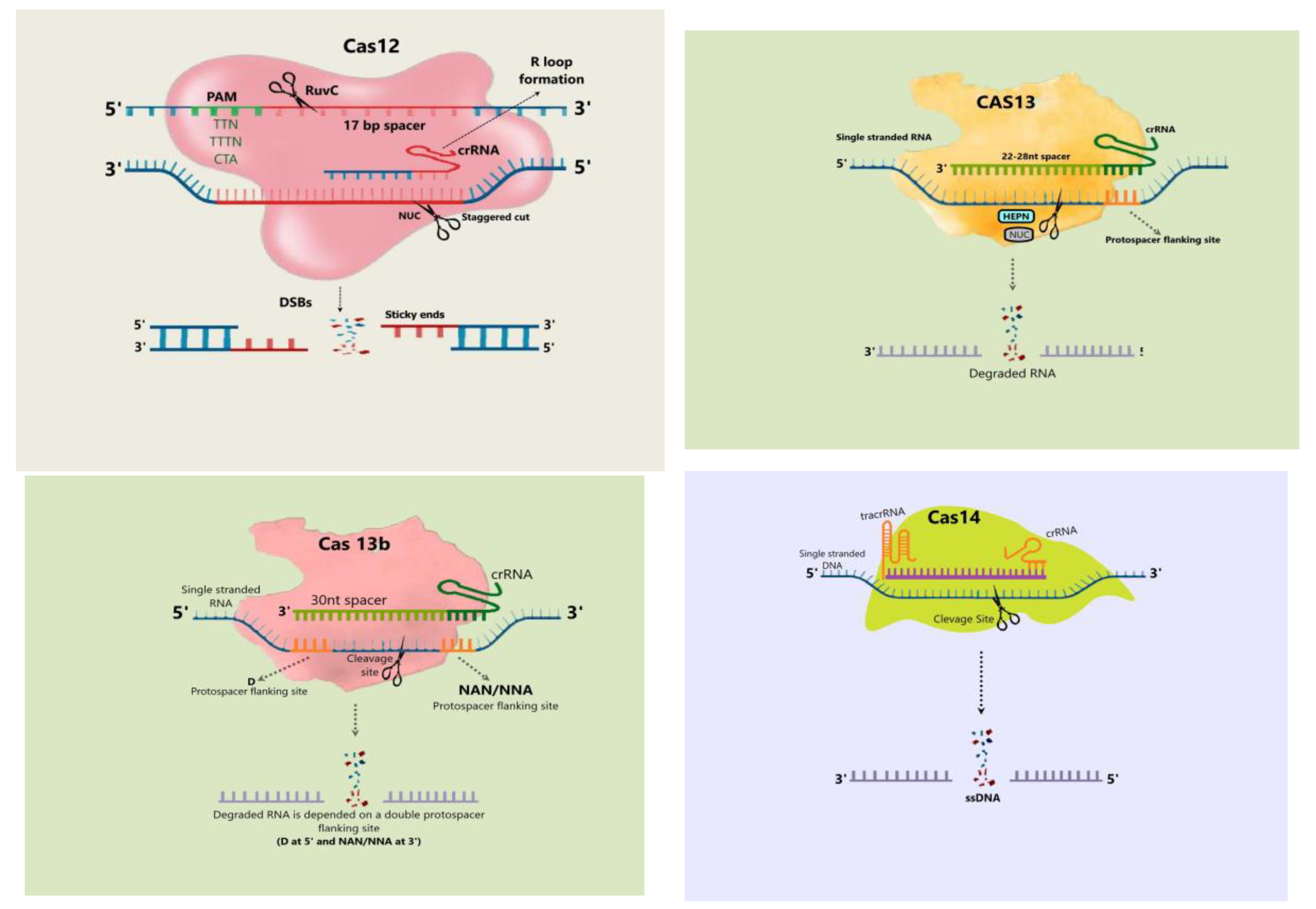
4.3. Cas12 Protein
4.4. Cas13 Protein
4.5. Cas14 Protein
5. Prime Editing
6. Application of CRISPR in Plant Abiotic and Biotic Stress Tolerance
6.1. CRISPR in Abiotic Stresses Tolerance
6.2. CRISPR in Biotic Stress Tolerance
| Stress | Pathogen Factor | Crop | The name of the target gene | References |
|---|---|---|---|---|
|
Insect disease |
Plant hopper | Rice (Oryza sativa) | CYTOCHROME P450 71A1 (OsCYP71A1) | [109] |
| Stem borer | Rice (Oryza sativa) | OsCYP71A1 | [109] | |
| Common cutworm | Soybean (Glycine max) | CALCIUM-DEPENDENT PROTEIN KINASE 38 (GmCDPK38) |
[110] |
|
|
Virus disease |
Rice tungro spherical virus | Rice (Oryza sativa) | eIF4G | [111] |
| Cucumber vein yellowing virus | Cucumber (Cucumis sativus) | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E(eIF4E) | [112] | |
| Zucchini yellow mosaic (Cucumis virus) | Cucumber (Cucumis sativus) | eIF4E | [112] |
|
| Papaya ring spot mosaic (Cucumis virus) | Cucumber (Cucumis sativus) | eIF4E | [112] | |
| Tomato mosaic virus | Tomato (Solanum lycopersicum) |
DICER-LIKE 2b (SlDCL2b) | [113] | |
| Potato virus X | Tomato (Solanum lycopersicum) | SlDCL2a and SlDCL2b | [113] | |
|
Fungus disease |
Rice Blast | Rice (Oryza sativa) | OsERF922 SUBUNIT OF THE |
[113] |
| Rice (Oryza sativa) | EXOCYST COMPLEX 3A (OsSEC3A) | [70] |
||
| Rice (Oryza sativa) | Pi21 and Bsr-d1 | [71] ,[50] |
||
| Powdery mildew | Tomato (Solanum lycopersicum) |
MILDEW RESISTANT LOCUS O (SlMLO) | [114] | |
| Wheat (Triticum aestivum) | TaMLO-A1, TaMLO-B1 and TaMLO-D1 | [115] | ||
| Grapevine (Vitis vinifera) | VvMOL3 | [115] | ||
| Tomato (Solanum lycopersicum) | POWDERY MILDEW RESISTANCE 4 (SlPMR4) | [67] | ||
|
Late blight |
Tomato (Solanum lycopersicum) | miR482b and miR482c | [113] | |
| Gray mould | Tomato (Solanum lycopersicum) | PECTATE LYASE (SlPL) | [116] | |
|
Bacterial disease |
Bacterial blight | Rice (Oryza sativa) | SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER 13 (OsSWEET13) | [117] |
| Citrus bacterial canker | Orange (Citrus sinensis) | LATERAL ORGAN BOUNDARY 1 (CsLOB1) | [118] |
|
| Bacterial leaf spot disease | Tomato (Solanum lycopersicum) | JASMONATE ZIM-DOMAIN 2 (SlJAZ2) | [119] |
| Crop Type | Monocotyledon Crops | Dicotyledon Crops |
|---|---|---|
| Prime Technologies | Examples | Examples |
|
Genomic Selection |
Rice (Oryza sativa) | Soybean (Glycine max) |
| Wheat (Triticum aestivum) | Cotton (Gossypium hirsutum) | |
| Maize (Zea mays) | Sunflower (Helianthus annuus) | |
|
CRISPR-Cas9 |
Barley (Hordeum vulgare) | Tomato (Solanum lycopersicum) |
| Sugarcane (Saccharum officinarum) | Potato (Solanum tuberosum) | |
| Sorghum (Sorghum bicolor) | Canola (Brassica napus) | |
|
Marker-Assisted Breeding |
Barley (Hordeum vulgare) | Common bean (Phaseolus vulgaris) |
| Sugarcane (Saccharum officinarum) | Pea (Pisum sativum) | |
|
High-Throughput Sequencing (NGS) |
Maize (Zea mays) | Soybean (Glycine max) |
| Rice (Oryza sativa) | Cotton (Gossypium hirsutum) | |
|
Biotechnology |
Sugarcane (Saccharum officinarum) | Cotton (Gossypium hirsutum) |
| Maize (Zea mays) | Soybean (Glycine max) | |
| Rice (Oryza sativa) | Canola (Brassica napus) | |
|
Precision Agriculture and Data Analytics |
Wheat (Triticum aestivum) | Sunflower (Helianthus annuus) |
| Rice (Oryza sativa) | Tomato (Solanum lycopersicum) | |
|
Traditional Breeding Methods |
Barley (Hordeum vulgare) | Pea (Pisum sativum) |
| Sugarcane (Saccharum officinarum) | Potato (Solanum tuberosum) | |
|
Grafting and Hybridization |
Bamboo (Various species) | Apple (Malus domestica) |
| Banana (Musa spp.) | Grape (Vitis vinifera) | |
|
Organic and Sustainable Farming Practices |
Bamboo (Various species) | Common bean (Phaseolus vulgaris) |
| Rice (Oryza sativa) | Pea (Pisum sativum) | |
|
Seed Enhancement Technologies |
Maize (Zea mays) | Cotton (Gossypium hirsutum) |
| Rice (Oryza sativa) | Soybean (Glycine max) |
7. Crop Improvement, Possible Risks and Ethical Concerns of CRISPR-Cas Based Genome Editing
8. Conclusion and Future Perspectives
Author Contributions
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- Clarke, J. L., & Zhang, P. (2013). Plant biotechnology for food security and bioeconomy. Plant molecular biology, 83(1-2), 1–3. [CrossRef]
- Gill, S. S., & Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant physiology and biochemistry: PPB, 48(12), 909–930. [CrossRef]
- Akram, F., Sahreen, S., Aamir, F., Haq, I. U., Malik, K., Imtiaz, M., Naseem, W., Nasir, N., & Waheed, H. M. (2023). An Insight into Modern Targeted Genome-Editing Technologies with a Special Focus on CRISPR/Cas9 and its Applications. Molecular biotechnology, 65(2), 227–242. [CrossRef]
- Sun, Y., Li, J., & Xia, L. (2016). Precise Genome Modification via Sequence-Specific Nucleases-Mediated Gene Targeting for Crop Improvement. Frontiers in plant science, 7, 1928. [CrossRef]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019b) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697. [CrossRef]
- Xu, Y., & Li, Z. (2020). CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Computational and structural biotechnology journal, 18, 2401–2415. [CrossRef]
- Rasheed, A., Gill, R. A., Hassan, M. U., Mahmood, A., Qari, S., Zaman, Q. U., Ilyas, M., Aamer, M., Batool, M., Li, H., & Wu, Z. (2021). A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises. Current issues in molecular biology, 43(3), 1950–1976. [CrossRef]
- Yang B. (2020). Grand Challenges in Genome Editing in Plants. Frontiers in genome editing, 2, 2. [CrossRef]
- Ishino, Y., Krupovic, M., & Forterre, P. (2018). History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. Journal of bacteriology, 200(7), e00580-17. [CrossRef]
- Gostimskaya I. (2022). CRISPR-Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochemistry. Biokhimiia, 87(8), 777–788. [CrossRef]
- Makarova, K.S., Wolf, Y.I., Iranzo, J. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83 (2020). Makarova, K.S., Wolf, Y.I., Iranzo, J. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83 (2020). [CrossRef]
- Makarova, K., Haft, D., Barrangou, R. et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol 9, 467–477 (2011). [CrossRef]
- Kim, E., Koo, T., Park, S. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8, 14500 (2017). [CrossRef]
- Wang, J.Y., Tuck, O.T., Skopintsev, P. et al. Genome expansion by a CRISPR trimmer-integrase. Nature 618, 855–861 (2023). [CrossRef]
- Liu, Z., Dong, H., Cui, Y., Cong, L., & Zhang, D. (2020). Application of different types of CRISPR/Cas-based systems in bacteria. Microbial cell factories, 19(1), 172. [CrossRef]
- Dangquan Zhang, Zhiyong Zhang, Turgay Unver, Baohong Zhang, CRISPR/Cas: A powerful tool for gene function study and crop improvement, Journal of Advanced Research, Volume 29, 2021, Pages 207-221. [CrossRef]
- Ceasar, S. A., Maharajan, T., Hillary, V. E., & Ajeesh Krishna, T. P. (2022). Insights to improve the plant nutrient transport by CRISPR/Cas system. Biotechnology advances, 59, 107963. [CrossRef]
- Bernabé-Orts, J. M., Casas-Rodrigo, I., Minguet, E. G., Landolfi, V., Garcia-Carpintero, V., Gianoglio, S., Vázquez-Vilar, M., Granell, A., & Orzaez, D. (2019). Assessment of Cas12a-mediated gene editing efficiency in plants. Plant biotechnology journal, 17(10), 1971–1984. [CrossRef]
- Rodríguez-Rodríguez, D. R., Ramírez-Solís, R., Garza-Elizondo, M. A., Garza-Rodríguez, M. L., & Barrera-Saldaña, H. A. (2019). Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases (Review). International journal of molecular medicine, 43(4), 1559–1574. [CrossRef]
- Moradpour, M., & Abdulah, S. N. A. (2020). CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant biotechnology journal, 18(1), 32–44. [CrossRef]
- Zhang, B., Ye, Y., Ye, W., Perčulija, V., Jiang, H., Chen, Y., Li, Y., Chen, J., Lin, J., Wang, S., Chen, Q., Han, Y. S., & Ouyang, S. (2019). Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nature communications, 10(1), 2544. [CrossRef]
- Wolter, F., & Puchta, H. (2019). In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. The Plant journal : for cell and molecular biology, 100(5), 1083–1094. [CrossRef]
- Safari, F., Zare, K., Negahdaripour, M., Barekati-Mowahed, M., & Ghasemi, Y. (2019). CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell & bioscience, 9, 36. [CrossRef]
- Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E., Joung, J., van der Oost, J., Regev, A., Koonin, E. V., & Zhang, F. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163(3), 759–771. [CrossRef]
- Li, G., Zhang, Y., Dailey, M., & Qi, Y. (2023). Hs1Cas12a and Ev1Cas12a confer efficient genome editing in plants. Frontiers in genome editing, 5, 1251903. [CrossRef]
- Bandyopadhyay, A., Kancharla, N., Javalkote, V. S., Dasgupta, S., & Brutnell, T. P. (2020). CRISPR-Cas12a (Cpf1): A Versatile Tool in the Plant Genome Editing Tool Box for Agricultural Advancement. Frontiers in plant science, 11, 584151. [CrossRef]
- Gong, Z., Previtera, D.A., Wang, Y. et al. Geminiviral-induced genome editing using miniature CRISPR/Cas12j (CasΦ) and Cas12f variants in plants. Plant Cell Rep 43, 71 (2024). [CrossRef]
- Gao, L., Cox, D. B. T., Yan, W. X., Manteiga, J. C., Schneider, M. W., Yamano, T., Nishimasu, H., Nureki, O., Crosetto, N., & Zhang, F. (2017). Engineered Cpf1 variants with altered PAM specificities. Nature biotechnology, 35(8), 789–792. [CrossRef]
- Tóth, E., Varga, É., Kulcsár, P. I., Kocsis-Jutka, V., Krausz, S. L., Nyeste, A., Welker, Z., Huszár, K., Ligeti, Z., Tálas, A., & Welker, E. (2020). Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic acids research, 48(7), 3722–3733. [CrossRef]
- Schindele, P., & Puchta, H. (2020). Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant biotechnology journal, 18(5), 1118–1120. [CrossRef]
- Zhang, F., Neik, T. X., Thomas, W. J. W., & Batley, J. (2023). CRISPR-Based Genome Editing Tools: An Accelerator in Crop Breeding for a Changing Future. International journal of molecular sciences, 24(10), 8623. [CrossRef]
- Kordyś, M., Sen, R., & Warkocki, Z. (2022). Applications of the versatile CRISPR-Cas13 RNA targeting system. Wiley interdisciplinary reviews. RNA, 13(3), e1694. [CrossRef]
- Sharma, V. K., Marla, S., Zheng, W., Mishra, D., Huang, J., Zhang, W., Morris, G. P., & Cook, D. E. (2022). CRISPR guides induce gene silencing in plants in the absence of Cas. Genome biology, 23(1), 6. [CrossRef]
- Lotfi, M., & Rezaei, N. (2020). CRISPR/Cas13: A potential therapeutic option of COVID-19. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 131, 110738. [CrossRef]
- Dong, G., Fan, Z. CRISPR/Cas-mediated germplasm improvement and new strategies for crop protection. Crop Health 2, 2 (2024). [CrossRef]
- Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., Cofsky, J. C., Kyrpides, N. C., Banfield, J. F., & Doudna, J. A. (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science (New York, N.Y.), 362(6416), 839–842. [CrossRef]
- Khan, M. Z., Haider, S., Mansoor, S., & Amin, I. (2019). Targeting Plant ssDNA Viruses with Engineered Miniature CRISPR-Cas14a. Trends in biotechnology, 37(8), 800–804. [CrossRef]
- Bin Zhou, Runlin Yang, Muhammad Sohail, Xiaoxue Kong, Xing Zhang, Ninghua Fu, Bingzhi Li. CRISPR/Cas14 provides a promising platform in facile and versatile aptasensing with improved sensitivity, Talenta, Volume 254, 2023, 124120. [CrossRef]
- Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L., and Corn, J. E. (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34 (3), 339–344. [CrossRef]
- Quadros, R. M., Miura, H., Harms, D. W., Akatsuka, H., Sato, T., Aida, T., et al. (2017). Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18 (1), 92. [CrossRef]
- Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. [CrossRef]
- Marzec, M., Brąszewska-Zalewska, A., and Hensel, G. (2020). Prime editing: A new way for genome editing. Trends Cell Biol. 30 (4), 257–259. [CrossRef]
- Liu Tingting, Zou Jinpeng, Yang Xi, Wang Kejian, Rao Yuchun, Wang Chun. Development and Application of Prime Editing in Plants, Rice Science, Volume 30, Issue 6, 2023, Pages 509-522. [CrossRef]
- Gupta, A., Liu, B., Chen, Q. J., & Yang, B. (2023). High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice. Plant biotechnology journal, 21(7), 1454–1464. [CrossRef]
- Huang, S., Weigel, D., Beachy, R. N., & Li, J. (2016). A proposed regulatory framework for genome-edited crops. Nature genetics, 48(2), 109–111. [CrossRef]
- Lu, Y., Tian, Y., Shen, R., Yao, Q., Zhong, D., Zhang, X., et al. (2021). Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 19 (3), 415–417. [CrossRef]
- Liu, Z., Ma, C., Hou, L., Wu, X., Wang, D., Zhang, L., et al. (2022). Exogenous SA affects rice seed germination under salt stress by regulating Na(+)/K(+) balance and endogenous GAs and ABA homeostasis. Int. J. Mol. Sci. 23 (6), 3293. [CrossRef]
- Al Murad, Musa, Abdul Latif Khan, and Sowbiya Muneer. 2020. „Silicon in Horticultural Crops: Cross-talk, Signaling, and Tolerance Mechanism under Salinity Stress” Plants 9, no. 4: 460. [CrossRef]
- Alam, M. S., Kong, J., Tao, R., Ahmed, T., Alamin, M., Alotaibi, S. S., Abdelsalam, N. R., & Xu, J. H. (2022). CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants (Basel, Switzerland), 11(9), 1184. [CrossRef]
- Nawaz, G., Usman, B., Peng, H., Zhao, N., Yuan, R., Liu, Y., & Li, R. (2020). Knockout of Pi21 by CRISPR/Cas9 and iTRAQ-Based Proteomic Analysis of Mutants Revealed New Insights into M. oryzae Resistance in Elite Rice Line. Genes, 11(7), 735. [CrossRef]
- Han, X., Chen, Z., Li, P., Xu, H.; Liu, K., Zha, W., Li, S., Chen, J., Yang, G., Huang, J., et al. Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability 2022, 14, 2621. [CrossRef]
- Santosh Kumar, V. V., Verma, R. K., Yadav, S. K., Yadav, P., Watts, A., Rao, M. V., & Chinnusamy, V. (2020). CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiology and molecular biology of plants : an international journal of functional plant biology, 26(6), 1099–1110. [CrossRef]
- Yue, E., Cao, H., & Liu, B. (2020). OsmiR535, a Potential Genetic Editing Target for Drought and Salinity Stress Tolerance in Oryza sativa. Plants (Basel, Switzerland), 9(10), 1337. [CrossRef]
- Vlčko, T., & Ohnoutková, L. (2020). Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants (Basel, Switzerland), 9(2), 195. [CrossRef]
- Yang B. (2020). Grand Challenges in Genome Editing in Plants. Frontiers in genome editing, 2, 2. [CrossRef]
- Du Y.T., Zhao M.J., Wang C.T., Gao Y., Wang Y.X., Liu Y.W., Ma Y.Z. Identification and Characterization of GmMYB118 Responses to Drought and Salt Stress. BMC Plant Biol. 2018;18:320. [CrossRef]
- Ogata, T., Ishizaki, T., Fujita, M., & Fujita, Y. (2020). CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PloS one, 15(12), e0243376. [CrossRef]
- Usman, B., Nawaz, G., Zhao, N., Liao, S., Liu, Y., & Li, R. (2020). Precise Editing of the OsPYL9 Gene by RNA-Guided Cas9 Nuclease Confers Enhanced Drought Tolerance and Grain Yield in Rice (Oryza sativa L.) by Regulating Circadian Rhythm and Abiotic Stress Responsive Proteins. International journal of molecular sciences, 21(21), 7854. [CrossRef]
- Lou D., Wang H., Liang G., Yu D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017;8:993. [CrossRef]
- Erdoğan İ, Cevher-Keskin B, Bilir Ö, Hong Y, Tör M. Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology (Basel). 2023 Jul 22;12(7):1037. [CrossRef]
- Nascimento FDS, Rocha AJ, Soares JMDS, Mascarenhas MS, Ferreira MDS, Morais Lino LS, Ramos APS, Diniz LEC, Mendes TAO, Ferreira CF, Santos-Serejo JAD, Amorim EP. Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review. Plants (Basel). 2023 Jan 9;12(2):305 . [CrossRef]
- Ding, Y., Shi, Y., & Yang, S. (2020). Molecular Regulation of Plant Responses to Environmental Temperatures. Molecular plant, 13(4), 544–564. [CrossRef]
- Zeng, Y., Wen, J., Zhao, W., Wang, Q., & Huang, W. (2020). Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR-Cas9 System. Frontiers in plant science, 10, 1663. [CrossRef]
- Li, J., Ding, J., Zhu, J., Xu, R., Gu, D., Liu, X., Liang, J., Qiu, C., Wang, H., Li, M., Qin, R., & Wei, P. (2023). Prime editing-mediated precise knockin of protein tag sequences in the rice genome. Plant communications, 4(3), 100572. [CrossRef]
- Savary S., Ficke A., Aubertot J.N., Hollier C. Crop Losses Due to Diseases and Their Implications for Global Food Production Losses and Food Security. Food Secur. 2012;4:519–537. [CrossRef]
- Wan, D. Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y., & Wen, Y. Q. (2020). CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Horticulture research, 7, 116. [CrossRef]
- Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., & Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nature biotechnology, 31(8), 691–693. [CrossRef]
- Santillán Martínez, M.I., Bracuto, V., Koseoglou, E. et al. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol 20, 284 (2020). [CrossRef]
- Zhang, M., Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., Ren, R., Xu, J., & Lou, L. (2020). CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant cell reports, 39(5), 589–595. [CrossRef]
- Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., Liu, Y. G., & Zhao, K. (2016). Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PloS one, 11(4), e0154027. [CrossRef]
- Ma, J., Chen, J., Wang, M., Ren, Y., Wang, S., Lei, C., Cheng, Z., & Sodmergen (2018). Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. Journal of experimental botany, 69(5), 1051–1064. [CrossRef]
- Thomazella, D. P. T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., Qi, T., Pham, J., Giuseppe, P., Lee, C. Y., Ortega, A., Cho, M. J., Hutton, S. F., & Staskawicz, B. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proceedings of the National Academy of Sciences of the United States of America, 118(27), e2026152118. [CrossRef]
- Chu, C., Huang, R., Liu, L., Tang, G., Xiao, J., Yoo, H., & Yuan, M. (2022). The rice heavy-metal transporter OsNRAMP1 regulates disease resistance by modulating ROS homeostasis. Plant, cell & environment, 45(4), 1109–1126. [CrossRef]
- Pixley, K.V., Falck-Zepeda, J.B., Paarlberg, R.L. et al. Genome-edited crops for improved food security of smallholder farmers. Nat Genet 54, 364–367 (2022). [CrossRef]
- Menz, J., Modrzejewski, D., Hartung, F., Wilhelm, R., and Sprink, T. (2020). Genome edited crops touch the market: a view on the global development and regulatory environment. Front. Plant Sci. 11:586027. [CrossRef]
- Schmidt, S. M., Belisle, M., & Frommer, W. B. (2020). The evolving landscape around genome editing in agriculture: Many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO reports, 21(6), e50680. [CrossRef]
- Zess, E., Begemann, M. CRISPR-Cas9 and beyond: what’s next in plant genome engineering. In Vitro Cell.Dev.Biol.-Plant 57, 584–594 (2021). [CrossRef]
- Podevin, N., Davies, H. V., Hartung, F., Nogué, F., and Casacuberta, J. M. (2013). Site-directed nucleases: a paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol. 31, 375–383. [CrossRef]
- Agricultural Chief Scientists (G20) Communique: G20 Italy 2021, 10th Meeting of Agricultural Chief Scientists (MACS) (2021); https://www.macs-g20.org/fileadmin/macs/Annual_Meetings/2021_Italy/Documents/Communique.pdf.
- Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., Dickman, M. J., Makarova, K. S., Koonin, E. V., & van der Oost, J. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science (New York, N.Y.), 321(5891), 960–964. [CrossRef]
- Marraffini, L. A., & Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science (New York, N.Y.), 322(5909), 1843–1845. [CrossRef]
- Koonin, E. V., & Makarova, K. S. (2017). Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back. Genome biology and evolution, 9(10), 2812–2825. [CrossRef]
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.), 337(6096), 816–821. [CrossRef]
- Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E., Joung, J., van der Oost, J., Regev, A., Koonin, E. V., & Zhang, F. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163(3), 759–771. [CrossRef]
- Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., Verdine, V., Cox, D. B. T., Kellner, M. J., Regev, A., Lander, E. S., Voytas, D. F., Ting, A. Y., & Zhang, F. (2017). RNA targeting with CRISPR-Cas13. Nature, 550(7675), 280–284. [CrossRef]
- Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., & Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution, 60(2), 174–182. [CrossRef]
- Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P. D., Wu, X., Jiang, W., Marraffini, L. A., & Zhang, F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.), 339(6121), 819–823. [CrossRef]
- Esvelt, K. M., Mali, P., Braff, J. L., Moosburner, M., Yaung, S. J., & Church, G. M. (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature methods, 10(11), 1116–1121. [CrossRef]
- Horvath, P., Romero, D. A., Coûté-Monvoisin, A. C., Richards, M., Deveau, H., Moineau, S., Boyaval, P., Fremaux, C., & Barrangou, R. (2008). Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. Journal of bacteriology, 190(4), 1401–1412. [CrossRef]
- Yang, H., Gao, P., Rajashankar, K. R., & Patel, D. J. (2016). PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell, 167(7), 1814–1828.e12. [CrossRef]
- Yamano, T., Nishimasu, H., Zetsche, B., Hirano, H., Slaymaker, I. M., Li, Y., Fedorova, I., Nakane, T., Makarova, K. S., Koonin, E. V., Ishitani, R., Zhang, F., & Nureki, O. (2016). Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell, 165(4), 949–962. [CrossRef]
- Walton, R. T., Christie, K. A., Whittaker, M. N., & Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science (New York, N.Y.), 368(6488), 290–296. [CrossRef]
- Miller, S. M., Wang, T., Randolph, P. B., Arbab, M., Shen, M. W., Huang, T. P., Matuszek, Z., Newby, G. A., Rees, H. A., & Liu, D. R. (2020). Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nature biotechnology, 38(4), 471–481. [CrossRef]
- Chen, J. S., Dagdas, Y. S., Kleinstiver, B. P., Welch, M. M., Sousa, A. A., Harrington, L. B., Sternberg, S. H., Joung, J. K., Yildiz, A., & Doudna, J. A. (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature, 550(7676), 407–410. [CrossRef]
- Casini, A., Olivieri, M., Petris, G., Montagna, C., Reginato, G., Maule, G., Lorenzin, F., Prandi, D., Romanel, A., Demichelis, F., Inga, A., & Cereseto, A. (2018). A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nature biotechnology, 36(3), 265–271. [CrossRef]
- Lee, J. K., Jeong, E., Lee, J., Jung, M., Shin, E., Kim, Y. H., Lee, K., Jung, I., Kim, D., Kim, S., & Kim, J. S. (2018). Directed evolution of CRISPR-Cas9 to increase its specificity. Nature communications, 9(1), 3048. [CrossRef]
- Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., Zeina, C. M., Gao, X., Rees, H. A., Lin, Z., & Liu, D. R. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature, 556(7699), 57–63. [CrossRef]
- Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., Noda, T., Abudayyeh, O. O., Gootenberg, J. S., Mori, H., Oura, S., Holmes, B., Tanaka, M., Seki, M., Hirano, H., Aburatani, H., Ishitani, R., Ikawa, M., Yachie, N., Zhang, F., … Nureki, O. (2018). Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science (New York, N.Y.), 361(6408), 1259–1262. [CrossRef]
- Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., & Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science (New York, N.Y.), 351(6268), 84–88. [CrossRef]
- Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., & Joung, J. K. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529(7587), 490–495. [CrossRef]
- Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Topkar, V. V., Zheng, Z., & Joung, J. K. (2015). Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nature biotechnology, 33(12), 1293–1298. [CrossRef]
- Hirano, H., Gootenberg, J. S., Horii, T., Abudayyeh, O. O., Kimura, M., Hsu, P. D., Nakane, T., Ishitani, R., Hatada, I., Zhang, F., Nishimasu, H., & Nureki, O. (2016). Structure and Engineering of Francisella novicida Cas9. Cell, 164(5), 950–961. [CrossRef]
- Ran, F. A., Cong, L., Yan, W. X., Scott, D. A., Gootenberg, J. S., Kriz, A. J., Zetsche, B., Shalem, O., Wu, X., Makarova, K. S., Koonin, E. V., Sharp, P. A., & Zhang, F. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520(7546), 186–191. [CrossRef]
- Hou, Z., Zhang, Y., Propson, N. E., Howden, S. E., Chu, L. F., Sontheimer, E. J., & Thomson, J. A. (2013). Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences of the United States of America, 110(39), 15644–15649. [CrossRef]
- Magadán, A. H., Dupuis, M. È., Villion, M., & Moineau, S. (2012). Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PloS one, 7(7), e40913. [CrossRef]
- Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A., & Charpentier, E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature, 532(7600), 517–521. [CrossRef]
- Li, SY., Cheng, QX., Liu, JK. et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res 28, 491–493 (2018). [CrossRef]
- Hillary, V. E., & Ceasar, S. A. (2023). A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Molecular biotechnology, 65(3), 311–325. [CrossRef]
- Lu, H. P., Luo, T., Fu, H. W., Wang, L., Tan, Y. Y., Huang, J. Z., Wang, Q., Ye, G. Y., Gatehouse, A. M. R., Lou, Y. G., & Shu, Q. Y. (2018). Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nature plants, 4(6), 338–344. [CrossRef]
- Li, R., Char, S.N., Yang, B. (2019). Creating Large Chromosomal Deletions in Rice Using CRISPR/Cas9. In: Qi, Y. (eds) Plant Genome Editing with CRISPR Systems. Methods in Molecular Biology, vol 1917. Humana, New York, NY. [CrossRef]
- Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., Voytas, D. F., Choi, I. R., & Chadha-Mohanty, P. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant biotechnology journal, 16(11), 1918–1927. [CrossRef]
- Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., Sherman, A., Arazi, T., & Gal-On, A. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Molecular plant pathology, 17(7), 1140–1153. [CrossRef]
- Wang, T., Deng, Z., Zhang, X., Wang, H., Wang, Y., Liu, X., Liu, S., Xu, F., Li, T., Fu, D., Zhu, B., Luo, Y., & Zhu, H. (2018). Tomato DCL2b is required for the biosynthesis of 22-nt small RNAs, the resulting secondary siRNAs, and the host defense against ToMV. Horticulture research, 5, 62. [CrossRef]
- Wan, D. Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y., & Wen, Y. Q. (2020). CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Horticulture research, 7, 116. [CrossRef]
- Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., & Qiu, J. L. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature biotechnology, 32(9), 947–951. [CrossRef]
- Silva, C. J., van den Abeele, C., Ortega-Salazar, I., Papin, V., Adaskaveg, J. A., Wang, D., Casteel, C. L., Seymour, G. B., & Blanco-Ulate, B. (2021). Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. Journal of experimental botany, 72(7), 2696–2709. [CrossRef]
- Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., Huang, S., Liu, S., Vera Cruz, C., Frommer, W. B., White, F. F., & Yang, B. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. The Plant journal : for cell and molecular biology, 82(4), 632–643. [CrossRef]
- Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., Yao, L., & Zou, X. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant biotechnology journal, 15(12), 1509–1519. [CrossRef]
- Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., & Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant biotechnology journal, 17(3), 665–673. [CrossRef]
| CRISPR/Cas System Used |
Host Organism | Name of the Effector | Types of Protein | Target Molecule | Reference |
|---|---|---|---|---|---|
| Cas7, Cas5, Cas8, and Cas3 |
E. coli |
Cas3, Cascade, and crRNA | Cas 3 | ssDNA | [80] |
| Cas7, Cas5, and Cas1 |
S. epidermics |
Cmr/Csm, crRNA, and Cas10 | Cas 6 | ssDNA | [81] |
| Cas7, Cas5, and Csf1 | - | - | Csf1 | - | [82] |
|
Cas9 |
S. thermophilus and S. pyogenes | Cas9, tracrRNA, and crRNA | Cas9 | dsDNA | [83] |
|
Cas12 |
F. novicida |
Cpf1, crRNA and tracrRNA | Cpf1 | ssDNA and ds DNA | [84] |
|
Cas13 |
- | C2c1, and crRNA | C2c2 | ssRNA | [85] |
| Host Organism | Target Molecule | Protein Name | References |
|---|---|---|---|
| C.bacterjejuna | DNA | Cas9 | [13] |
| S. thermophilus | DNA | Cas9 | [89] |
| S. thermophilus | DNA | Cas9 | - |
| N.meningitidis | DNA | Cas9 | [88] |
| S. aureus | DNA | Cas9 | [88] |
| F. novicida | DNA | Cas9 | - |
| S. pyogenes | DNA | Cas9 | [87] |
| S. pyogenes | dsDNA | Cas9 | [86] |
| Acidaminococcus sp. | DNA | Cpf1 | [91] |
| P.Francisella | DNA | Cpf1 | [84] |
| A.bacillusacidoterrestris | DNA | C2c1 | [90] |
| Acidaminococcus sp. | DNA | Cas12a | [13] |
| L. bacterium | ssRNA | Cas13 | [85] |
| Uarchaea | ssDNA | Cas14 | [36] |
| Name | Cas | CRISPR/Cas | PAM | PAM location | Resources | Reference |
|---|---|---|---|---|---|---|
| SpRY SpG Cas9-NRNH HypaCas9 evoCas9 Sniper-Cas9 xCas9 SpCas9-NG eSpCas9 SpCas9-HF SaCas9-KKH Modified SpCas9 FnCas9variant SpCas9 SaCas9 FnCas9 NmCas9 CjCas9 St1Cas9 St1Cas9 FnCas12a LbCas12a AsCas12a LsCas13# Cas14 |
Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas9 Cas12a(cpf1) Cas12a(cpf1) Cas12a(cpf1) Cas13(C2c2) Cas14 |
Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II Type II TypeVI NA |
NRN or NYN NGN NRNH NGG NGG NGG NG NG NGG NGG NNNRRT NGA or NAG YG NGG NNGRRT NGG NNNNGATT NNNNRYAC NNAGAAW NGGNG TTTN or YTN TTTV TTTV NA NA |
3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 3′ 5′ 5′ 5′ NA NA. |
Engineered SpCas9 Engineered SpCas9 Engineered SpCas9 Mutated SpCas9-HF Mutated SpCas9 Engineered SpCas9 Engineered SpCas9 Engineered SpCas9 Engineered SpCas9 Engineered SpCas9 Engineered SaCas9 Engineered SpCas9 Modified FnCas9 S. pyogenes S. aureus F. Novicida N. meningitidis C. jejuni S. thermophilus S. thermophilus F. novicida L. bacterium Acidaminococcus sp. L. shahii Archaea |
[92] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103] [83] [103] [102] [104] [13] [105] [105] [84] [84] [106] [85] [36] |
| Parameter | Cas9 | Cas12 | Cas13 | Cas14 |
|---|---|---|---|---|
|
Size of Protein (Amino Acid) |
~1000-1600 |
~1300 |
~1400 |
~400-700 |
|
Target |
DNA |
DNA |
RNA |
DNA |
|
RNA |
Two RNA molecules |
Single RNA molecules |
Two RNA molecules |
Single RNA molecules |
|
Nuclease Site |
2 nuclease domains HNH and RuvC | Single nuclease RuvC-Nuc |
Target RNA domain HEPN |
DNA binding domain RuvC |
|
Pattern of cut |
Blunt |
Sticky-ended |
Degraded |
NA |
|
Spacer Size |
16-20nt |
16-25nt |
25-35nt |
NA |
|
Protospacer restriction |
PAM |
PAM |
PFS |
PAM |
|
Single guide molecular size (Nucleotides, nt) |
17-24nt |
42-44nt |
-64nt |
-140nt |
|
Non-specifically cut nucleic acids (DNA or RNA) |
DNA(SS) | DNA(SS) | RNA(SS) | DNA(SS) |
| Cas Proteins | Mechanism | Applications | Merits | Demerits | Reference |
|---|---|---|---|---|---|
| Cas 9 |
|
|
|
|
[83] |
| Cas 12 |
|
|
|
|
[107] |
| Cas 13 |
|
|
|
|
[85] |
| Cas 14 |
|
|
|
|
[108] |
| Non-specifically cut nucleic acids (DNA or RNA) | DNA(SS) | DNA(SS) | RNA(SS) | DNA(SS) |
| Stress | Cas Enzmyes | Crop | The Name of the Target Gene | References |
|---|---|---|---|---|
|
Salinity |
Cas9 | Rice (Oryza sativa) | BASIC HELIX-LOOP-HELIX 024 (OsbHLH024) | [49] |
| Rice (Oryza sativa) | RESPONSE REGULAT ORS 22 (OsRR22) | [51] | ||
| Rice (Oryza sativa) | RELATED TO ABI3/VP1 2 (OsRAV2) | [43] | ||
| Rice (Oryza sativa) | DROUGHT AND SALT TOLERANCE (OsDST) | [43] | ||
| Rice (Oryza sativa) | NAM, ATAF and CUC 041 (OsNAC041) | [70] | ||
| Rice (Oryza sativa) | OsmiR535 | [53] | ||
| Barley (Hordeum vulgare) | INOSITOLTRISPHOSPHATE 5/6 KINASES 1 (HvITPK1) | [54] | ||
| Tomato (Solanum lycopersicum) | HYBRID PROLINE-RICH PROTEIN 1 (SlHyPRP1) | [43] |
||
| Tomato (Solanum lycopersicum) | Auxin Response Factor 4 (SlARF4) | [43] | ||
|
Drought |
Rice (Oryza sativa) | ENHANCED RESPONSE TO ABA1 (OsERA1) | [57] | |
| Rice (Oryza sativa) | OsDST | [52] | ||
| Rice (Oryza sativa) | PYRABACTIN RESISTANCE-LIKE 9 (OsPYL9) | [58] | ||
| Rice (Oryza sativa) | SEMI-ROLLED LEAF 1 (SRL1) and SEMI-ROLLED LEAF 2 (SRL2) | [58] | ||
| Maize (Zea mays) | AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE 8 (ZmARGOS8) | [43] | ||
| Cas9 | Wheat (Triticum aestivum) | DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN 2 (TaDREB2) | [13] |
|
| Wheat (Triticum aestivum) | ETHYLENE-RESPONSE FACTOR 3 (TaERF3) | [13] | ||
| Tomato (Solanum lycopersicum) | GA-INSENSITIVE DWARF1 1 (SlGID1) | [43] | ||
| Tomato (Solanum lycopersicum) | LATERAL ORGAN BOUNDARIES DOMAIN 40 (SlLBD40) | [15] | ||
| Arsenic Caesium | Cas 9 | Rice (Oryza sativa) | HIGH-AFFINITY POTASSIUM TRANSPORTER 1 (OSHAK1) | [43] |
| Rice (Oryza sativa) | ARSENITE-RESPONSIVE MYB1 (OsARM1) | [43] | ||
|
Low temperature |
Rice (Oryza sativa) | PIN-FORMED 5b (OsPIN5b) | [43] | |
| Cas9 | Rice (Oryza sativa) | GRAIN SIZE (GS3) | [43] | |
| Rice (Oryza sativa) | V-MYB AVIAN MYELOBLASTOSIS VIRAL ONCOGENE HOMOLOG 30 (OsMYB30) |
[43] | ||
|
High temperature |
Rice (Oryza sativa) | PYRABACTIN RESISTANCE-LIKE 1/4/6 (OsPYL1/4/6) | [43] | |
| Cas9 | Tomato (Solanum lycopersicum) | MITOGEN-ACTIVATED PROTEIN KINASES 3 (SlMAPK3) | [43] | |
|
Cadmium |
Rice (Oryza sativa) | NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 5 (OsNRAMP5) |
[73] | |
| Cas9 | Rice (Oryza sativa) | LOW-AFFINITY CATION TRANSPORTER 1 (OsLCT1) | [43] | |
| Rice (Oryza sativa) | NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 1 (OsNRAMP1) |
[73] |
| Crop Species | Trait | Gene Targeted | Achievement made | Reference |
|---|---|---|---|---|
|
O. sativa |
|
|
|
[79] |
| M. balbisiana | Enhanced resistance | Mutation in downy mildew resistance 6 (DMR6) | Musa dmr6 transgenic mutants of banana showed enhanced resistance to BXW, and did not show any detrimental effect on plant growth | [80] |
| M. domestica | Disease resistance | Mutation in apple DIPM-1, DIPM-2 and DIPM-4 | Resistance to fire blast disease in non-transgenic but mutant apple lines | [81] |
| S. lycopersicum | Disease resistance | Loss of function mutation in SlDMR6-1 gene | Mutants do not have detrimental effects on growth and had multiple disease resistance P. syringae, P. capsici and Xanthomonas spp. | [82] |
| C. sinensis Osbeck | Canker resistance | Mutation and loss of function in CsWRKY22 | Mutant orange plants showed decreased susceptibility to citrus canker | [83] |
| C. sinensis Osbeck | Canker resistance | CRISPR/Cas9-targeted mutation in CsLOB1 promoter in citrus | Promoter editing of CsLOB1 alone was sufficient to enhance citrus canker resistance in citrus. | [84] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





