Submitted:
10 April 2024
Posted:
10 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
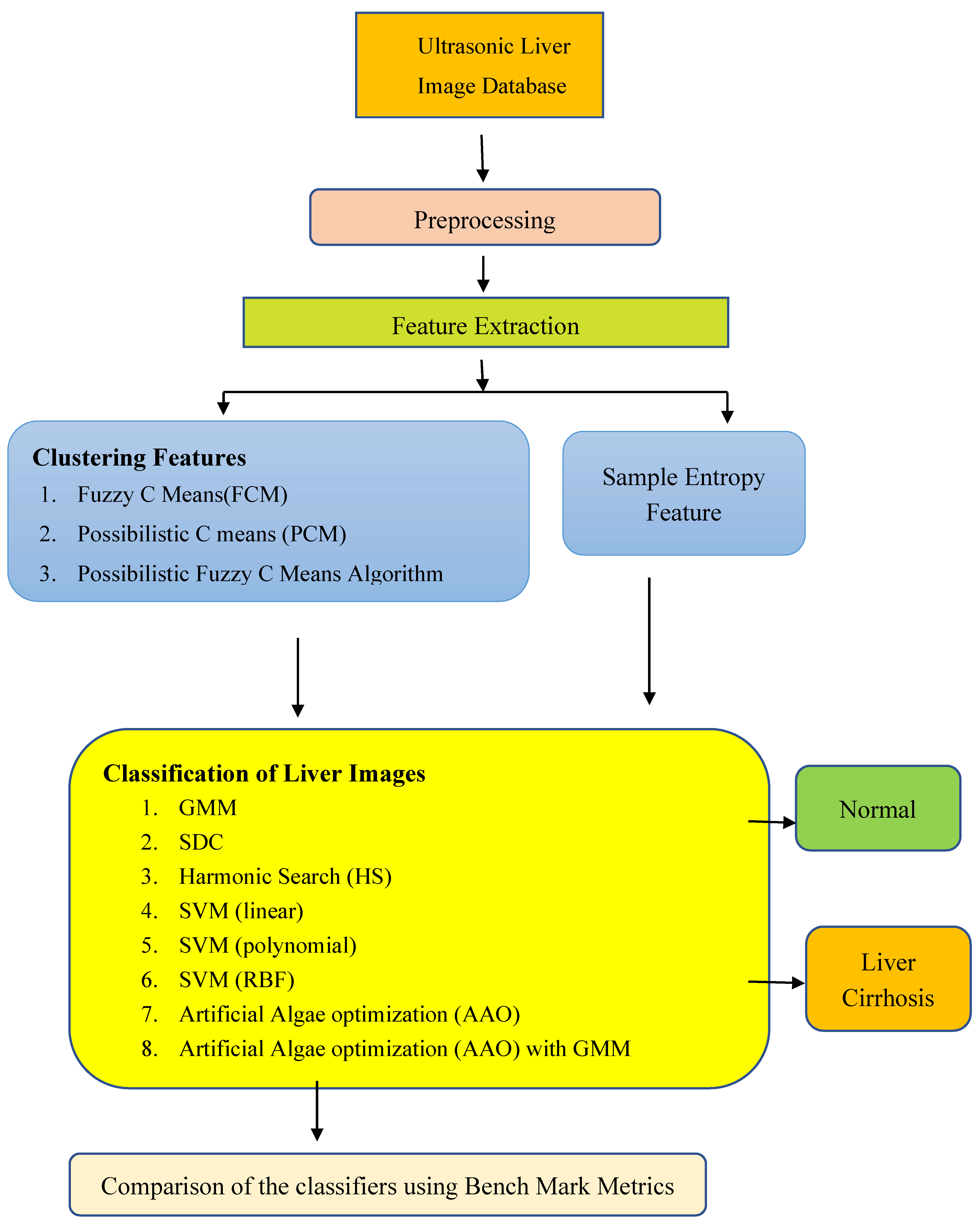
2. Materials and Methods
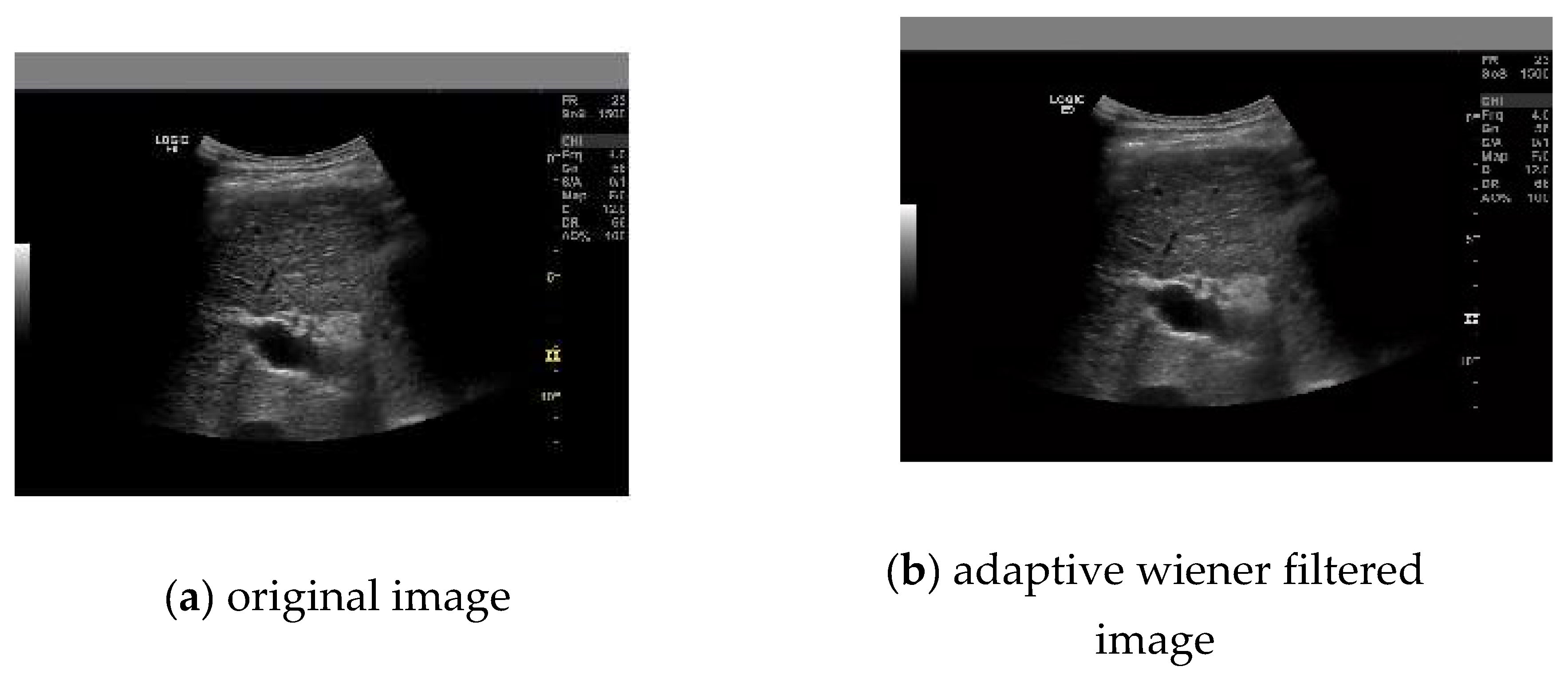
2.1. Preprocessing
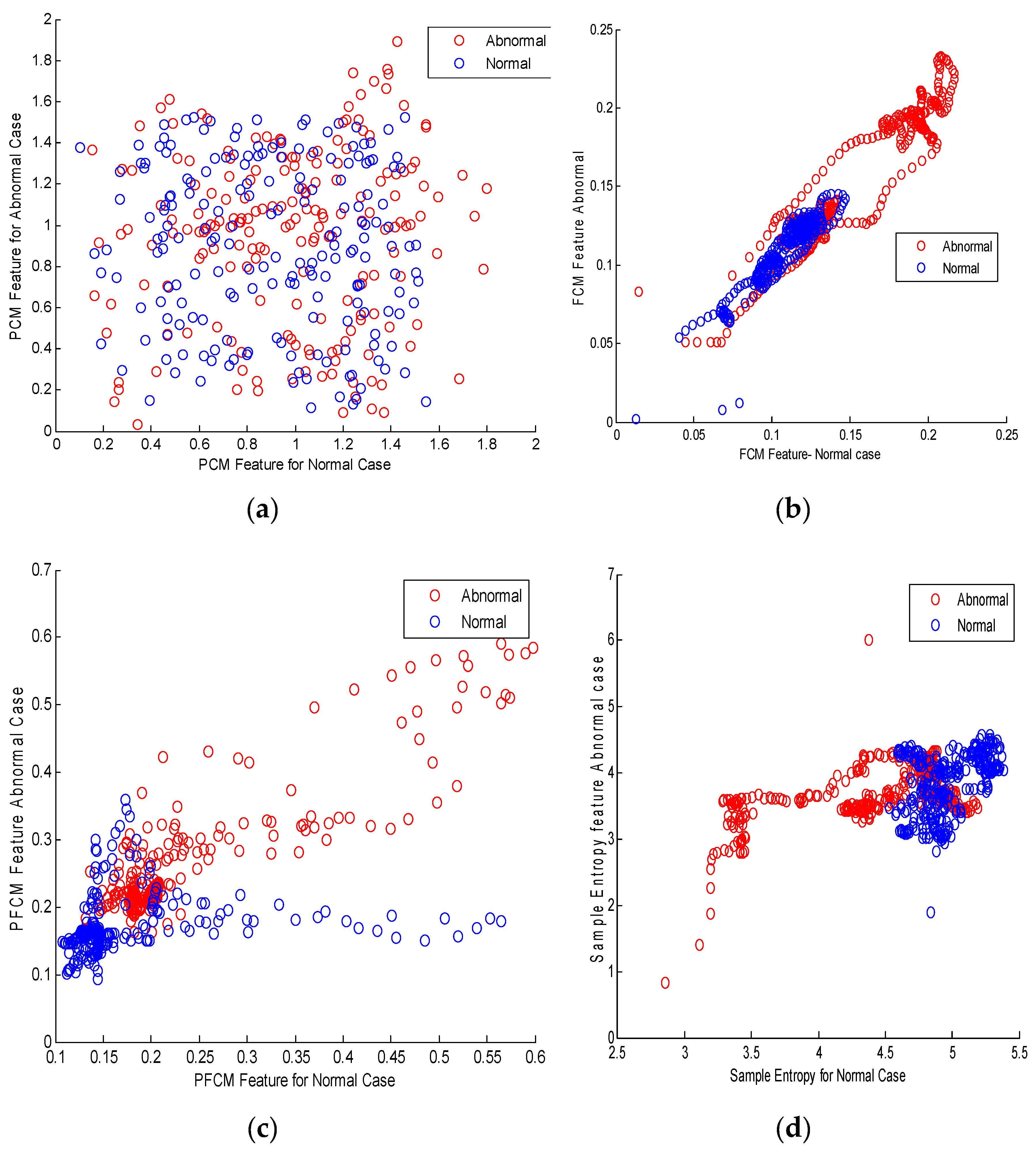
2.2. Clustering Methods for Feature Extraction
2.2.1. Fuzzy C Means Clustering (FCM)
2.2.2. Possibilistic C means (PCM) Algorithm
2.2.3. Possibilistic Fuzzy C Means Algorithm (PFCM)
2.2.4. Sample Entropy as a Feature
3. Bio Inspired Classifiers for Classification of Liver Cirrhosis from Extracted Features
3.1. Gaussian Mixture Model (GMM) as a Classifier
3.2. Softmax Discriminant Classifier (SDC)
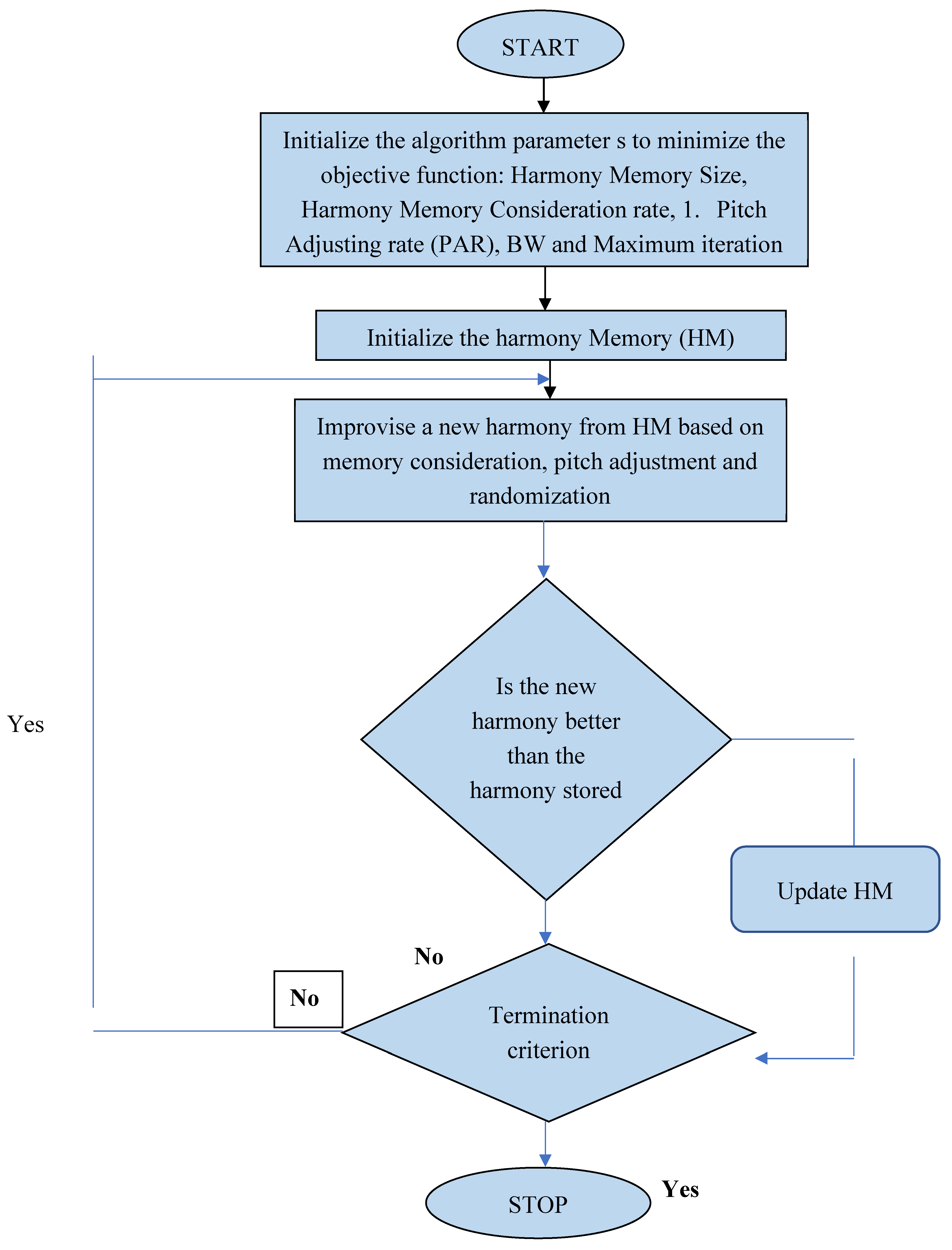
3.3. Harmonic Search Algorithm (HSA) as a Classifier
- Harmony Memory size (SHM)
- Harmony memory consideration rate (HMCR)
- Pitch Adjusting rate(PAR)
- Bandwidh(BW)
- Maximum number of Iteration(Maxitr)
- Consideration of memory
- Pitch change
- Random selection
3.4. Support Vector Machine as a Classifier
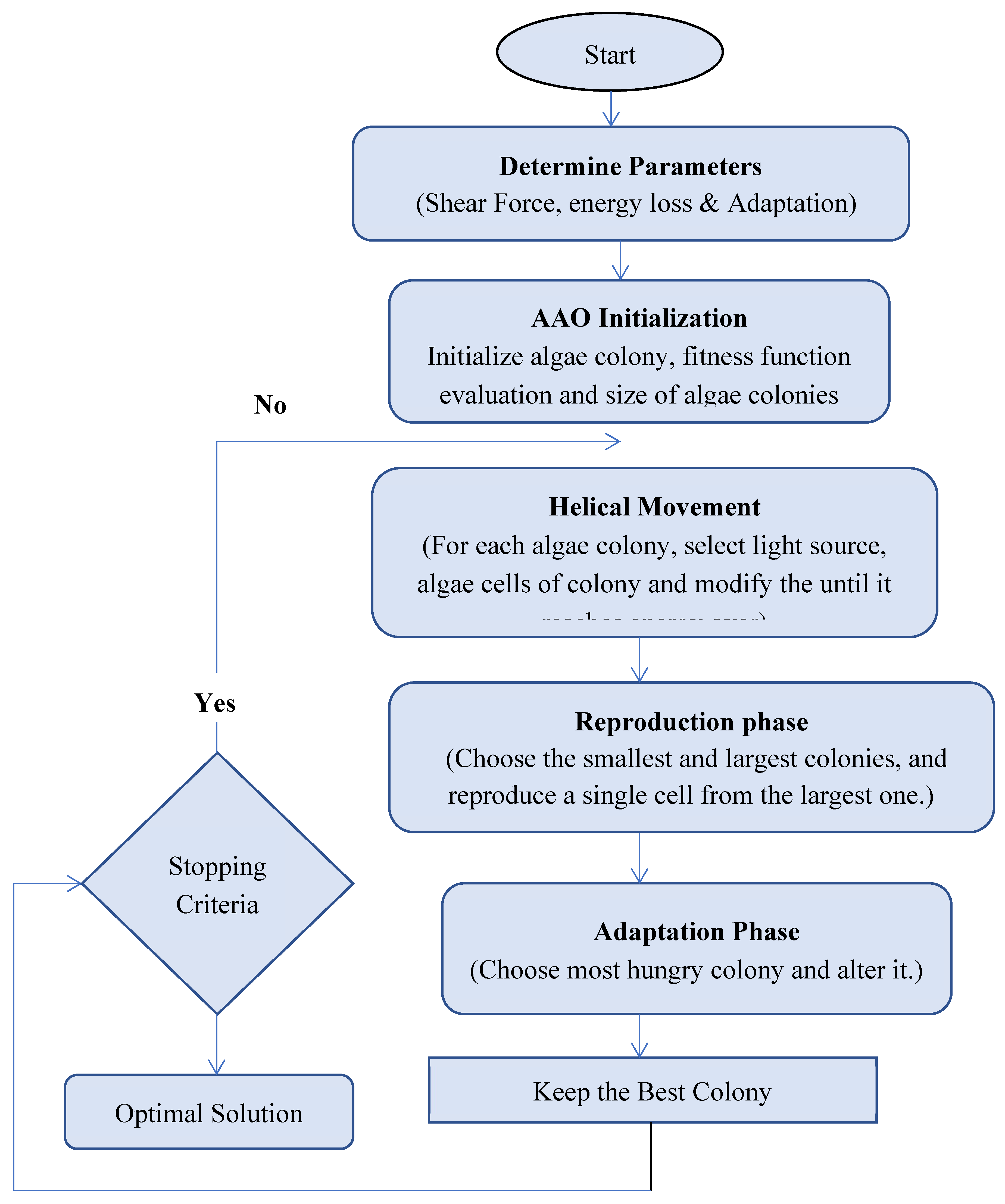
3.5. Artificial Algae Optimization Algorithm (AAO)
3.6. AAO with GMM
4. Results and Discussion
4.1. Selection of Classifier Parameters
4.2. Classifier Performance Analysis
5. Conclusions
References
- S. K. Randhawa, R. K. Sunkaria and A. K. Bedi, "Prediction of Liver Cirrhosis Using Weighted Fisher Discriminant Ratio Algorithm," 2018 First International Conference on Secure Cyber Computing and Communication (ICSCCC), 2018, pp. 184-187. [CrossRef]
- R.Karthikamani,Harikumar Rajaguru Analysis Of K Means Clustering And Classifiers In Diagnosing Abnormality Of The Ultrasonic Liver Images International Journal of Aquatic Science ISSN: 2008-8019,Vol 12, Issue 03, 2021pp1589-1595.
- Web reference, https://www.mana.md/different-uses-for-ultrasound/.
- Jagdeep Singh, SachinBagga, RanjodhKaur, Software-based Prediction of Liver Disease with Feature Selection and Classification Techniques, Procedia Computer Science,Volume 167,2020,Pages 1970-1980,ISSN 1877-0509.
- M. Tsiplakidou, M. G. Tsipouras, P. Manousou, N. Giannakeas and A. T. Tzallas, "Automated Hepatic Steatosis Assessment through Liver Biopsy Image Processing," 2016 IEEE 18th Conference on Business Informatics (CBI), 2016, pp. 61-67. [CrossRef]
- https://gco.iarc.fr/today/data/factsheets/cancers/39-All-cancers-fact-sheet.pdf.
- R. Karthikamani; Harikumar Rajaguru, Detection of Liver Cirrhosis in Ultrasonic Images from GLCM Features and Classifiers, S. Vlad and N. M. Roman(Eds.): MEDITECH 2020, IFMBE Proceedings 88, Springer, pp. 161–172, 2022. [Google Scholar] [CrossRef]
- 8. Harikumar Rajaguru, R.Karthikamani, Detection Of Abnormal Liver In Ultrasonic Images From FCM Features International Journal of Aquatic Science ISSN: 2008-8019,Vol 12, Issue 03,2021pp1581-1588.
- R. Suganya and S. Rajaram, "Feature extraction and classification of ultrasound liver images using haralick texture-primitive features: Application of SVM classifier," 2013 International Conference on Recent Trends in Information Technology (ICRTIT), 2013.
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011 Mar-Apr;61(2):69-90. Epub 2011 Feb 4. Erratum in: CA Cancer J Clin. 2011 Mar-Apr;61(2):134. [CrossRef] [PubMed]
- Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019 Jan;70(1):151-171. Epub 2018 Sep 26. [CrossRef] [PubMed]
- Nasrul Humaimi Mahmood and NoraishikinZulkarnain and Nor Saradatul and AkmarZulkifli, Ultrasound Liver Image Enhancement Using Watershed Segmentation Method, International Journal of Engineering Research and Applications (IJERA) 2012.
- Jain N, Kumar V. IFCM Based Segmentation Method for Liver Ultrasound Images. J Med Syst. 2016 Nov;40(11):249. Epub 2016 Oct 4. [CrossRef] [PubMed]
- Bharath, Ramkrishna& Mishra, Pradeep&Pachamuthu, Rajalakshmi. (2017). Automated quantification of ultrasonic fatty liver texture based on curvelet transform and SVD. Biocybernetics and Biomedical Engineering. 38. 10.1016/j.bbe.2017.12.004.
- Acharya UR, Raghavendra U, Fujita H, Hagiwara Y, Koh JE, Jen Hong T, Sudarshan VK, Vijayananthan A, Yeong CH, Gudigar A, Ng KH. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. ComputBiol Med. 2016 Dec 1;79:250-258. Epub 2016 Oct 29. [CrossRef] [PubMed]
- U RajendraAcharya, Hamido Fujita, Vidya K Sudarshan, Muthu Rama Krishnan Mookiah, Joel EW Koh, Jen Hong Tan, Yuki Hagiwara, Chua Kuang Chua, Sameer Padmakumar Junnarkar, Anushya Vijayananthan, and Kwan Hoong Ng. 2016. An integrated index for identification of fatty liver disease using radon transform and discrete cosine transform features in ultrasound images. Inf. Fusion 31, C (September 2016), 43–53.
- Wang, Q. , et al. (2017). "Liver cirrhosis detection using texture analysis and ensemble clustering. " Computer Methods and Programs in Biomedicine, 151, 15-22.
- Saranya, K. , & Kanagalakshmi, K. (2019). "Liver cirrhosis diagnosis using K-means clustering and SVM classification. " Journal of King Saud University-Computer and Information Sciences, 31(1), 32-38.
- Binish Khan, Piyush Kumar Shukla, Manish Kumar Ahirwar, “Strategic Analysis in Prediction of Liver Disease Using Different Classification Algorithms,” International Journal of Computer Sciences and Engineering, Vol.7, Issue.7, pp.71-76, 2019.
- Fahnun, Budi &Mutiara, Achmad&Prasetyo, Eri& Harlan, Johan & Abdullah, Apriyadi&Latief, Muhammad. (2018). Filtering Techniques for Noise Reduction in Liver Ultrasound Images. 261-266. 10.1109/EIConCIT.2018.8878547.
- Wu, F.; Yang, W.; Xiao, L.; Zhu, J. Adaptive Wiener Filter and Natural Noise to Eliminate Adversarial Perturbation. Electronics 2020, 9, 1634. [Google Scholar] [CrossRef]
- R. Xu, D. Wunsch II, ―Survey of clustering algorithms‖, IEEE Transactions on Neural Networks, 16, 2005, 645-678.
- SoumiGhosh and Sanjay Kumar Dubey, “Comparative Analysis of K-Means and Fuzzy C-Means Algorithms” International Journal of Advanced Computer Science and Applications(IJACSA), 4(4), 2013.
- M. A. Balafar, A. R. Ramli, S. Mashohor and A. Farzan, "Compare different spatial based fuzzy-C_mean (FCM) extensions for MRI image segmentation," 2010 The 2nd International Conference on Computer and Automation Engineering (ICCAE), 2010, pp .609-611. [CrossRef]
- R. Krishnapuram and J. M. Keller, "The possibilistic C-means algorithm: insights and recommendations," in IEEE Transactions on Fuzzy Systems, vol. 4, no. 3, pp. 385-393, Aug. 1996. [CrossRef]
- S. Askari, N. Montazerin, M.H. FazelZarandi, Generalized Possibilistic Fuzzy C-Means with novel cluster validity indices for clustering noisy data, Applied Soft Computing, Volume 53,2017.
- X. Wu, "A Possibilistic C-Means Clustering Algorithm Based on Kernel Methods," 2006 International Conference on Communications, Circuits and Systems, 2006, pp. 2062-2066. [CrossRef]
- N.R. Pal, K. Pal, J.C. Bezdek A mixed c-means clustering model Proceedings of the Sixth IEEE International Conference on Fuzzy Systems, vol. 1 (1997), pp. 11-21.
- N. R. Pal, K. Pal, J. M. Keller and J. C. Bezdek, "A possibilistic fuzzy c-means clustering algorithm," in IEEE Transactions on Fuzzy Systems, vol. 13, no. 4, pp. 517-530, Aug. 2005. [CrossRef]
- Lio, Pietro& Song, Yuedong. (2010). A new approach for epileptic seizure detection: sample entropy based feature extraction and extreme learning machine. Journal of Biomedical Science and Engineering. 3. 556-567. 10.4236/jbise.2010.36078.
- H. Wan, H. Wang, B. Scotney and J. Liu, "A Novel Gaussian Mixture Model for Classification," 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC), 2019, pp.3298-3303. [CrossRef]
- Prabhakar, S.K. , Rajaguru H. (2017) Performance Analysis of GMM Classifier for Classification of Normal and Abnormal Segments in PPG Signals. In: Goh J., Lim C., Leo H. (eds) The 16th International Conference on Biomedical Engineering. IFMBE Proceedings, vol 61. Springer, Singapore. [CrossRef]
- F. Zang and J. Zhang, "Softmax Discriminant Classifier," 2011 Third International Conference on Multimedia Information Networking and Security, 2011, pp.16-19. [CrossRef]
- H. Rajaguru and S. K. Prabhakar, "Softmax discriminant classifier for detection of risk levels in alcoholic EEG signals," 2017 International Conference on Computing Methodologies and Communication (ICCMC), 2017, pp. 989-991. [CrossRef]
- MahimaDubey, A Systematic Review on Harmony Search Algorithm: Theory, Literature, and Applications, Mathematical Problems in Engineering Volume 2021, Article ID 5594267.
- JoongHoon Kim, Harmony Search Algorithm: A Unique Music-inspired Algorithm, ProcediaEngineering,Volume 154,2016,Pages 1401-1405,ISSN 1877-7058.
- H. Rajaguru and S. K. Prabhakar, "A comprehensive analysis of support vector machine and Gaussian mixture model for classification of epilepsy from EEG signals," 2017 International conference of Electronics, Communication and Aerospace Technology (ICECA), 2017, pp. 585-593.
- Rajaguru, Harikumar, Ganesan, Karthick, Bojan, Vinoth Kumar, Earlier detection of cancer regions from MR image features and SVM classifiers, International Journal of Imaging Systems and Technology, 2016.
- Korkmaz, S. , Babalik, A. &Kiran, M.S. An artificial algae algorithm for solving binary optimization problems. Int. J. Mach. Learn. & Cyber. 9, 1233–1247 (2018).
- Sait Ali Uymaz, Gulay Tezel, EsraYel,Artificial algae algorithm (AAA) for nonlinear global optimization,Applied Soft Computing,Volume 31,2015,Pages 153-171,ISSN 1568-4946.
- Sait Ali Uymaz, Gulay Tezel, EsraYel,Artificial algae algorithm with multi-light source for numerical optimization and applications, Biosystems,Volume 138,2015,Pages 25-38,ISSN 0303-2647.
- Kumar, M. , Thakur, A.R., & Singh, S. (2018). Optimization of Some Standard Functions using Artificial Algae Algorithm. International journal of engineering research and technology.

| Feature Extraction Method |
PCM | FCM | PFCM | Sample Entropy | ||||
|---|---|---|---|---|---|---|---|---|
| Statistical Parameters | Cirrhosis | Normal | Cirrhosis | Normal | Cirrhosis | Normal | Cirrhosis | Normal |
| Mean | 0.5054 | 0.4968 | 0.1509 | 0.1569 | 0.2828 | 0.2833 | 5.339727 | 5.2504 |
| Variance | 0.0468 | 0.0456 | 0.0010 | 0.0014 | 0.0079 | 0.0089 | 0.137262 | 0.2105 |
| Skewness | -0.2156 | -0.1960 | -0.6516 | -0.764 | 1.6003 | 1.7111 | -1.75338 | -1.6610 |
| Kurtosis | -0.8087 | -0.8578 | 0.7095 | 0.9353 | 3.1021 | 3.1411 | 10.75434 | 7.3273 |
| Pearson Correlation coefficient (PCC) | 0.0092 | -0.0212 | 0.5839 | 0.5453 | 0.3721 | 0.4374 | 0.5030 | 0.4914 |
| Canonical Correlation Analysis (CCA) | 0.4190 | 0.7655 | 0.7569 | 0.6046 | ||||
| Feature Extraction | Classifiers | TP | TN | FP | FN | MSE |
|---|---|---|---|---|---|---|
| PCM | GMM | 509 | 531 | 398 | 421 | 1.09E-04 |
| SDC | 576 | 597 | 332 | 354 | 5.19E-05 | |
| Harmonic Search (HS) | 642 | 509 | 420 | 288 | 1.17E-04 | |
| SVM (linear) | 509 | 465 | 464 | 421 | 3.33E-04 | |
| SVM (polynomial) | 531 | 597 | 332 | 399 | 7.38E-05 | |
| SVM (RBF) | 700 | 640 | 289 | 160 | 2.53E-05 | |
| Artificial Algae optimization (AAO) | 772 | 641 | 288 | 158 | 2.31E-05 | |
| AAO GMM | 797 | 619 | 310 | 133 | 2.22E-05 | |
| FCM | GMM | 619 | 752 | 177 | 311 | 3E-05 |
| SDC | 664 | 575 | 354 | 266 | 4.09E-05 | |
| Harmonic Search (HS) | 774 | 663 | 266 | 156 | 1.62E-05 | |
| SVM (linear) | 907 | 486 | 443 | 23 | 2.01E-04 | |
| SVM (polynomial) | 752 | 752 | 177 | 178 | 1.22E-05 | |
| SVM (RBF) | 818 | 796 | 133 | 112 | 4.57E-06 | |
| Artificial Algae optimization (AAO) | 870 | 860 | 69 | 60 | 1.22E-06 | |
| Artificial Algae optimization (AAO) with GMM | 841 | 840 | 89 | 89 | 1.39E-06 | |
| PFCM | GMM | 819 | 885 | 44 | 111 | 2.05E-06 |
| SDC | 554 | 774 | 155 | 377 | 3.701E-05 | |
| Harmonic Search (HS) | 819 | 686 | 243 | 111 | 1.249E-05 | |
| SVM (linear) | 797 | 708 | 221 | 133 | 1.306E-05 | |
| SVM (polynomial) | 797 | 752 | 177 | 133 | 8.545E-06 | |
| SVM (RBF) | 841 | 796 | 133 | 89 | 3.145E-06 | |
| Artificial Algae optimization(AAO) | 863 | 774 | 155 | 67 | 5.125E-06 | |
| AAO GMM | 886 | 885 | 44 | 44 | 2.6E-07 | |
| Sample Entropy | GMM | 686 | 818 | 111 | 244 | 1.331E-05 |
| SDC | 753 | 774 | 155 | 177 | 1.265E-05 | |
| Harmonic Search(HS) | 664 | 553 | 376 | 266 | 4.42E-05 | |
| SVM (linear) | 664 | 531 | 398 | 266 | 5.545E-05 | |
| SVM (polynomial) | 797 | 664 | 265 | 133 | 1.702E-05 | |
| SVM (RBF) | 841 | 818 | 111 | 89 | 1.945E-06 | |
| Artificial Algae optimization(AAO) | 886 | 863 | 66 | 44 | 6.5E-07 | |
| AAO GMM | 819 | 841 | 88 | 111 | 2.745E-06 |
| Classifier | Optimal Parameter of the Classifier |
|---|---|
| Gaussian Mixture Model (GMM) | The mean and covariance of the input samples, as well as the tuning parameter, are estimated using the Expectation-Maximization (EM) algorithm. Criterion: MSE |
| Softmax Discriminant Classifier( SDC) | The value of λ is 0.5, and the mean of the target values for each class is 0.1 and 0.85, respectively. |
| Harmonic Search Algorithm | Class harmony will always be maintained at the predetermined target values of 0.85 and 0.1. Adjustments are made to the upper and lower bounds using a step size of = 0.005 for each. The final harmony aggregation is achieved when the MSE is less than 10^-5 or when the maximum iteration count reaches 1000, depending on which comes first. Criterion: MSE |
| SVM (linear) | C (Regularization Parameter): 0.85, Class weight: 0.4, Convergence Criterion: MSE |
| SVM (polynomial) | C=0.8, kernel function Coefficient γ: 10, Class weight: 0.5, Convergence Criterion: MSE |
| SVM (RBF) | C=0.8, kernel function coefficient γ: 100, Class weight: 0.87, Convergence Criterion: MSE |
| Artificial Algae optimization(AAO) | Share force: 3, Energy Loss: 0.4, Adaptation: 0.3, Convergence Criterion: MSE |
| AAO with GMM | Mean , Covariance of the input samples and tuning parameter is EM steps, Share force: 3, Energy Loss: 0.4, Adaptation: 0.3, Convergence Criterion: MSE |
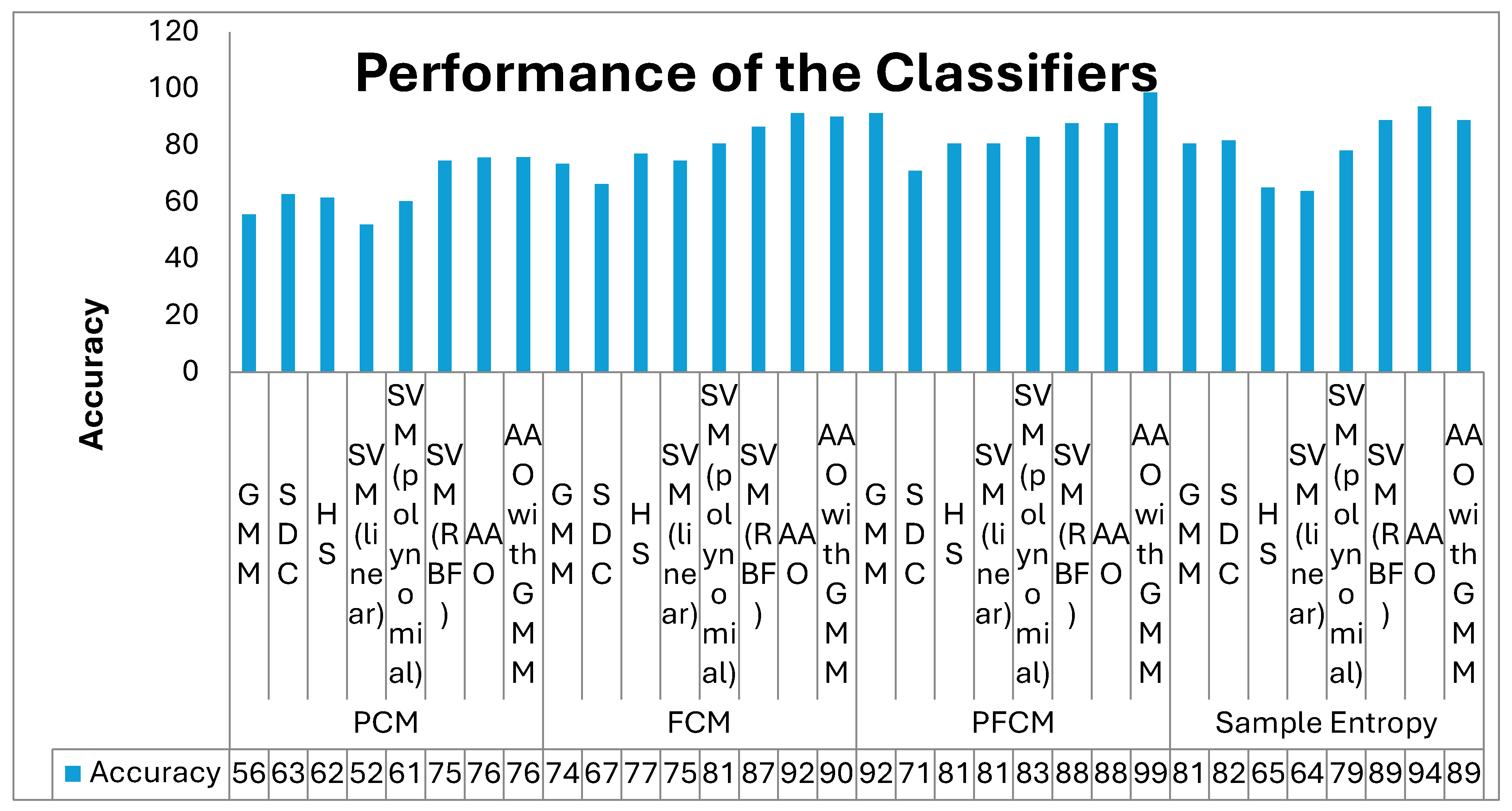
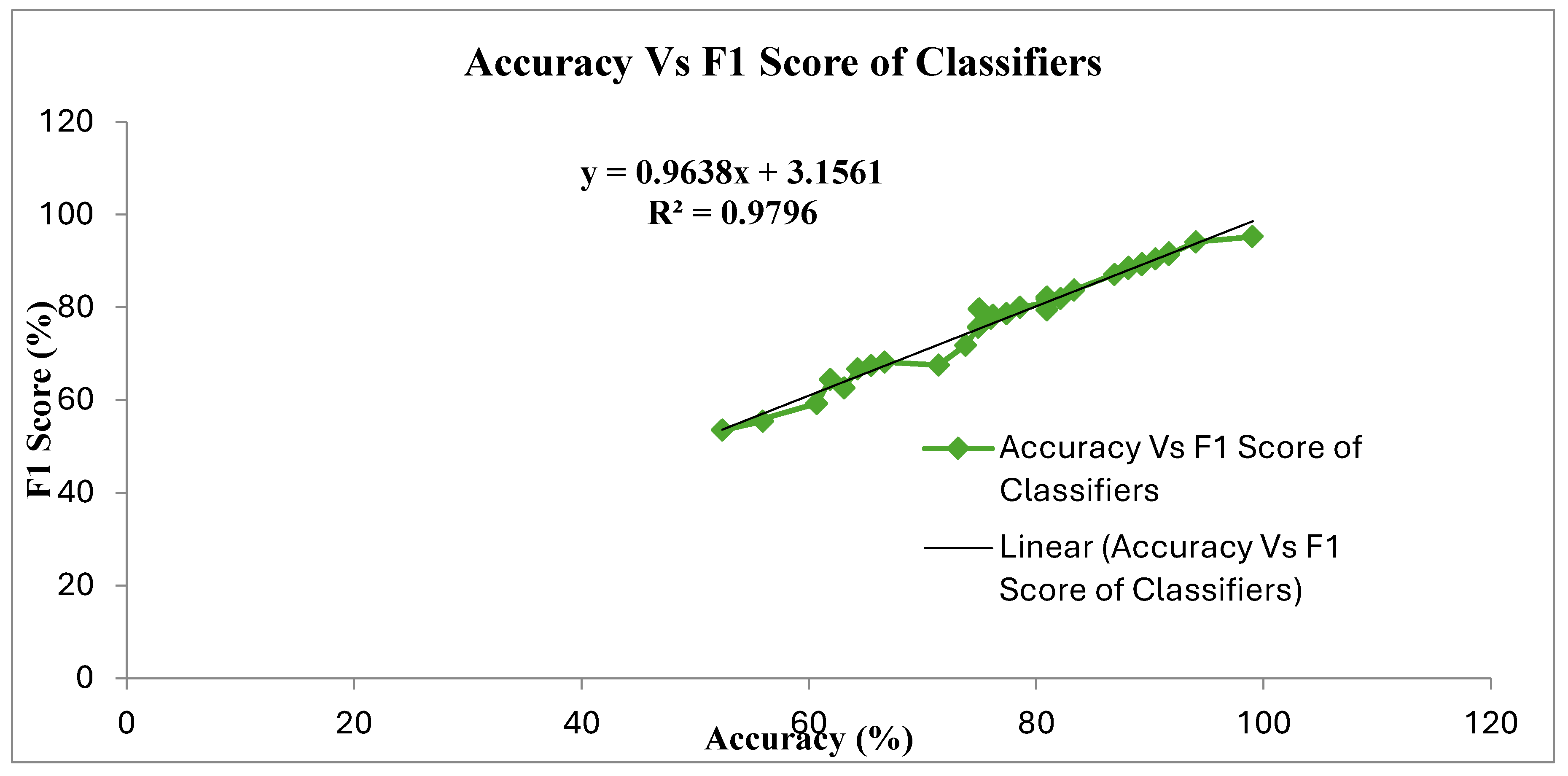
| Feature Extraction | Classifiers | Accuracy | F1 Score | MCC | F Measure | ER | JM |
|---|---|---|---|---|---|---|---|
| PCM | GMM | 55.95 | 55.42 | 0.12 | 0.55 | 44.05 | 38.33 |
| SDC | 63.10 | 62.65 | 0.26 | 0.63 | 36.90 | 45.61 | |
| Harmonic Search | 61.90 | 64.44 | 0.24 | 0.65 | 38.10 | 47.54 | |
| SVM (linear) | 52.38 | 53.49 | 0.05 | 0.54 | 47.62 | 36.51 | |
| SVM (polynomial) | 60.71 | 59.26 | 0.21 | 0.59 | 39.29 | 42.11 | |
| SVM (RBF) | 74.90 | 75.72 | 0.51 | 0.76 | 25.10 | 60.92 | |
| AAO | 76.01 | 77.59 | 0.53 | 0.78 | 23.99 | 63.38 | |
| AAO with GMM | 76.19 | 78.26 | 0.53 | 0.79 | 23.81 | 64.29 | |
| FCM | GMM | 73.81 | 71.79 | 0.48 | 0.72 | 26.19 | 56.00 |
| SDC | 66.67 | 68.18 | 0.33 | 0.68 | 33.33 | 51.72 | |
| Harmonic Search | 77.38 | 78.65 | 0.55 | 0.79 | 22.62 | 64.81 | |
| SVM (linear) | 75.00 | 79.61 | 0.56 | 0.81 | 25.00 | 66.13 | |
| SVM (polynomial) | 80.95 | 80.95 | 0.62 | 0.81 | 19.05 | 68.00 | |
| SVM (RBF) | 86.90 | 87.06 | 0.74 | 0.87 | 13.10 | 77.08 | |
| AAO | 91.67 | 91.76 | 0.83 | 0.92 | 8.33 | 84.78 | |
| AAO with GMM | 90.48 | 90.48 | 0.81 | 0.90 | 9.52 | 82.61 | |
| PFCM | GMM | 91.67 | 91.36 | 0.84 | 0.91 | 8.33 | 84.09 |
| SDC | 71.43 | 67.57 | 0.44 | 0.68 | 28.57 | 51.02 | |
| Harmonic Search | 80.95 | 82.22 | 0.63 | 0.82 | 19.05 | 69.81 | |
| SVM (linear) | 80.95 | 81.82 | 0.62 | 0.82 | 19.05 | 69.23 | |
| SVM (polynomial) | 83.33 | 83.72 | 0.67 | 0.84 | 16.67 | 72.00 | |
| SVM (RBF) | 88.10 | 88.37 | 0.76 | 0.88 | 11.90 | 79.17 | |
| AAO | 88.10 | 88.64 | 0.77 | 0.89 | 11.90 | 79.59 | |
| AAO with GMM | 99.03 | 95.24 | 0.90 | 0.95 | 0.97 | 90.91 | |
| Sample Entropy | GMM | 80.95 | 79.49 | 0.63 | 0.80 | 19.05 | 65.96 |
| SDC | 82.14 | 81.93 | 0.64 | 0.82 | 17.86 | 69.39 | |
| Harmonic Search | 65.48 | 67.42 | 0.31 | 0.68 | 34.52 | 50.85 | |
| SVM (linear) | 64.29 | 66.67 | 0.29 | 0.67 | 35.71 | 50.00 | |
| SVM (polynomial) | 78.57 | 80.00 | 0.58 | 0.80 | 21.43 | 66.67 | |
| SVM (RBF) | 89.29 | 89.41 | 0.79 | 0.89 | 10.71 | 80.85 | |
| AAO | 94.05 | 94.12 | 0.88 | 0.94 | 5.95 | 88.89 | |
| AAO with GMM | 89.29 | 89.16 | 0.79 | 0.89 | 10.71 | 80.43 |
| Feature Extraction Technique | Classifiers | Accuracy | F1 Score | JM |
|---|---|---|---|---|
| PCM | SVM (linear) | 52.38 | 53.49 | 36.51 |
| AAO with GMM | 76.19 | 78.26 | 64.29 | |
| FCM | SDC | 66.67 | 68.18 | 51.72 |
| AAO | 91.67 | 91.76 | 84.78 | |
| PFCM | SDC | 71.43 | 67.57 | 51.02 |
| AAO with GMM | 99.03 | 95.24 | 90.91 | |
| Sample Entropy | SVM (linear) | 64.29 | 66.67 | 50.00 |
| AAO | 94.05 | 94.12 | 88.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




