Submitted:
25 April 2024
Posted:
26 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
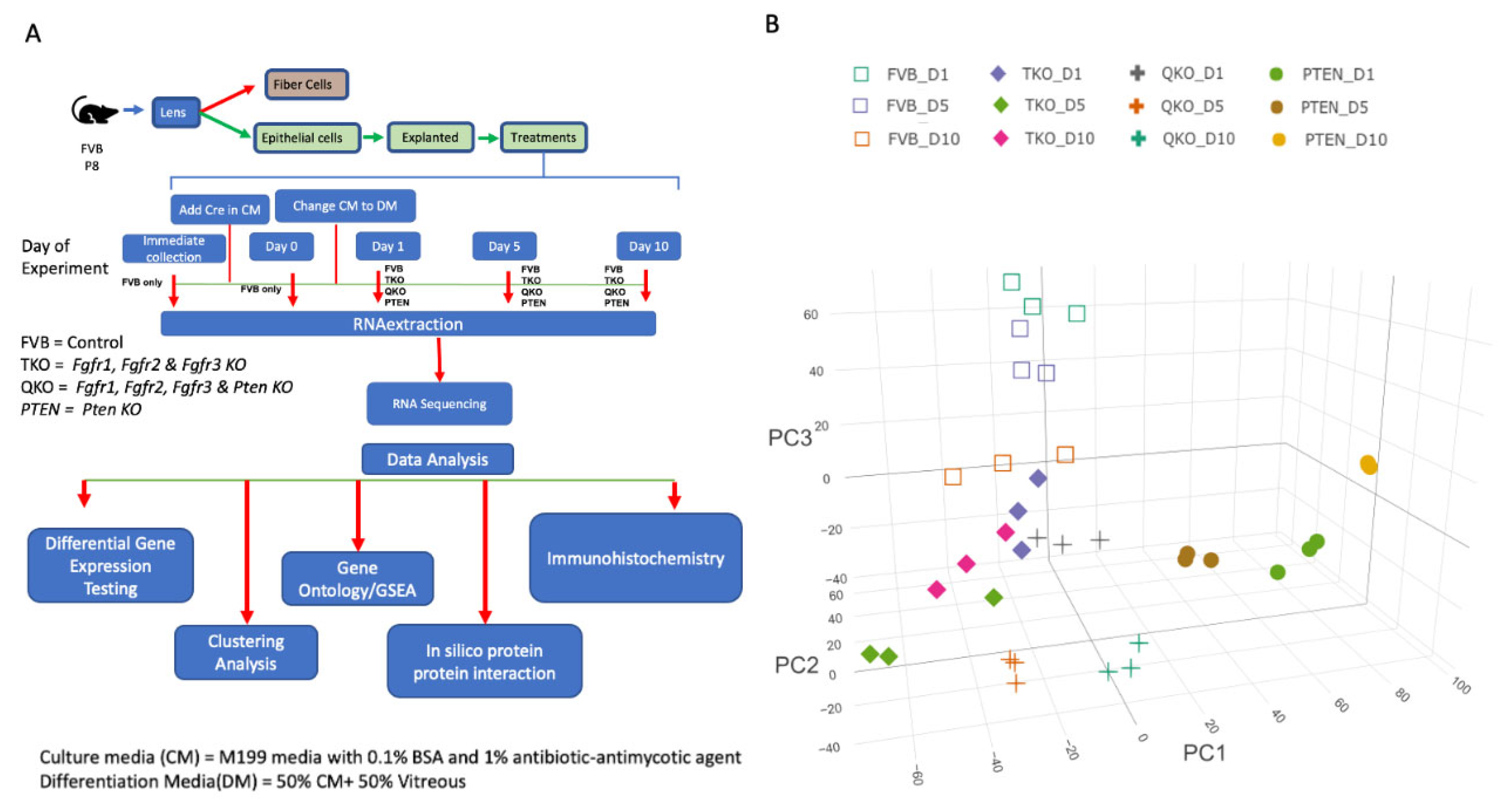
2. Materials and Methods
2.1. Animals
2.2. Lens Epithelial Explant Generation and Culture
2.3. RNA-seq Library Preparation and Sequencing
2.4. RNA seq Data Analysis
2.5. Clustering of DEGs
2.6. Gene Set Enrichment Analysis (GSEA) and Protein Protein Interaction
2.7. Lens Explant Immunofluorescence
3. Results
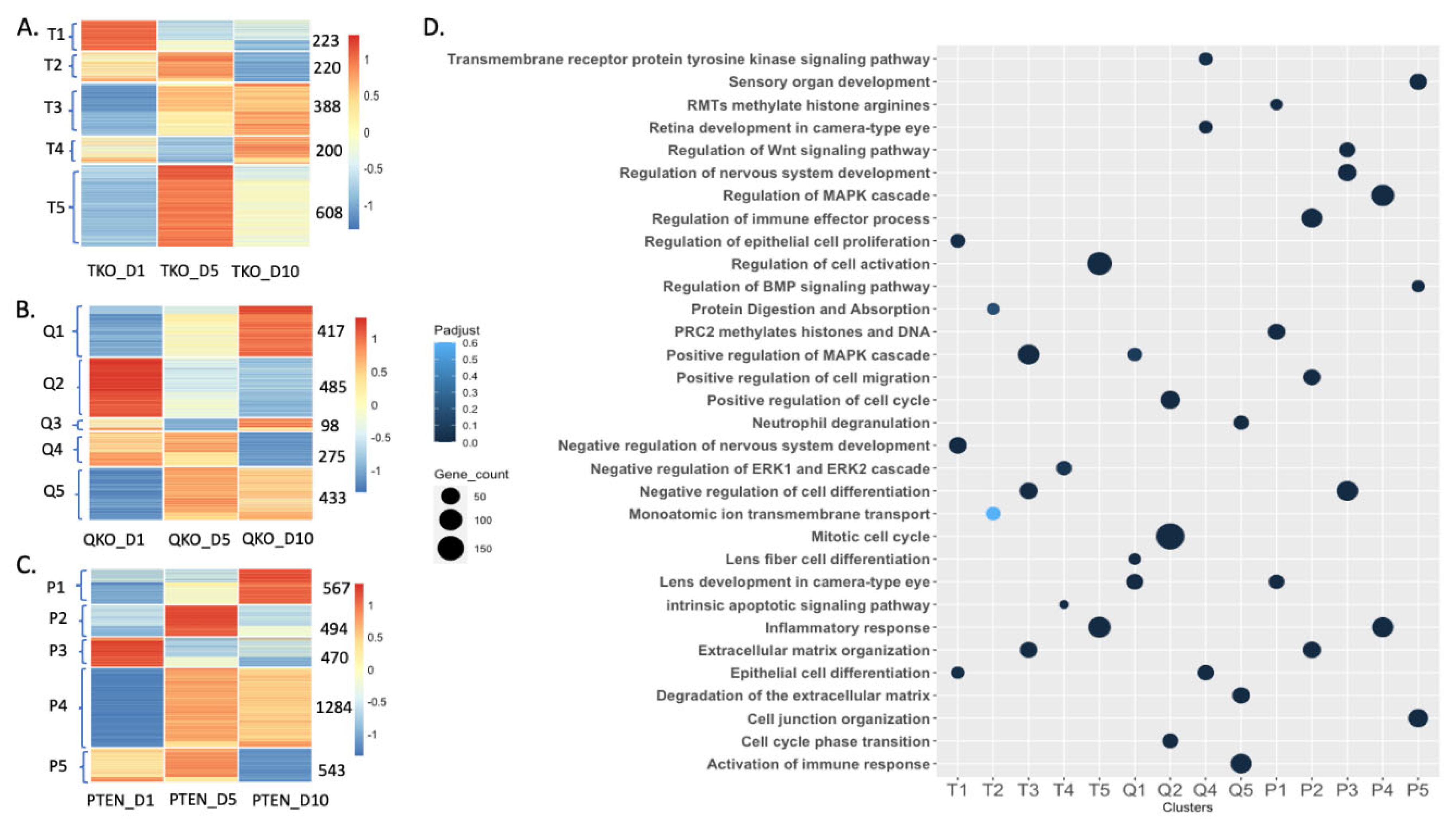
3.1. Overall Transcriptomic Changes in Lens Epithelial Explants Correlated Both to Genotype and Duration of Exposure to Vitreous Humor
3.2. Loss of Fgfrs, Pten or Both Fgfrs and Pten Induced Different Patterns of Differential Gene Expression Following Vitreous Exposure
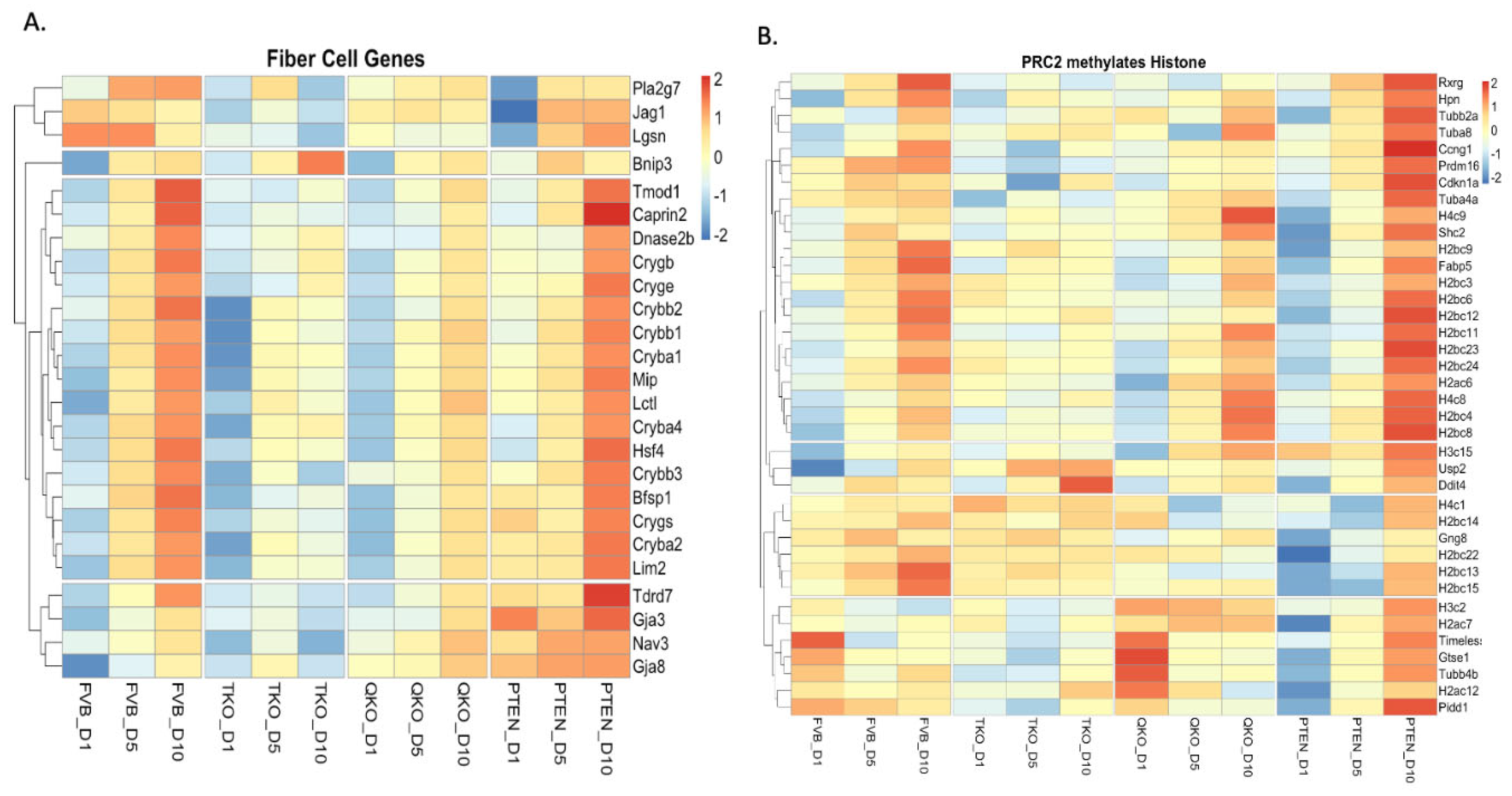
3.3. Fgfrs and Pten Influence Gene Expression in Opposite Directions with Respect to Fiber Cell Differentiation and Chromatin Remodeling Following DM Exposure
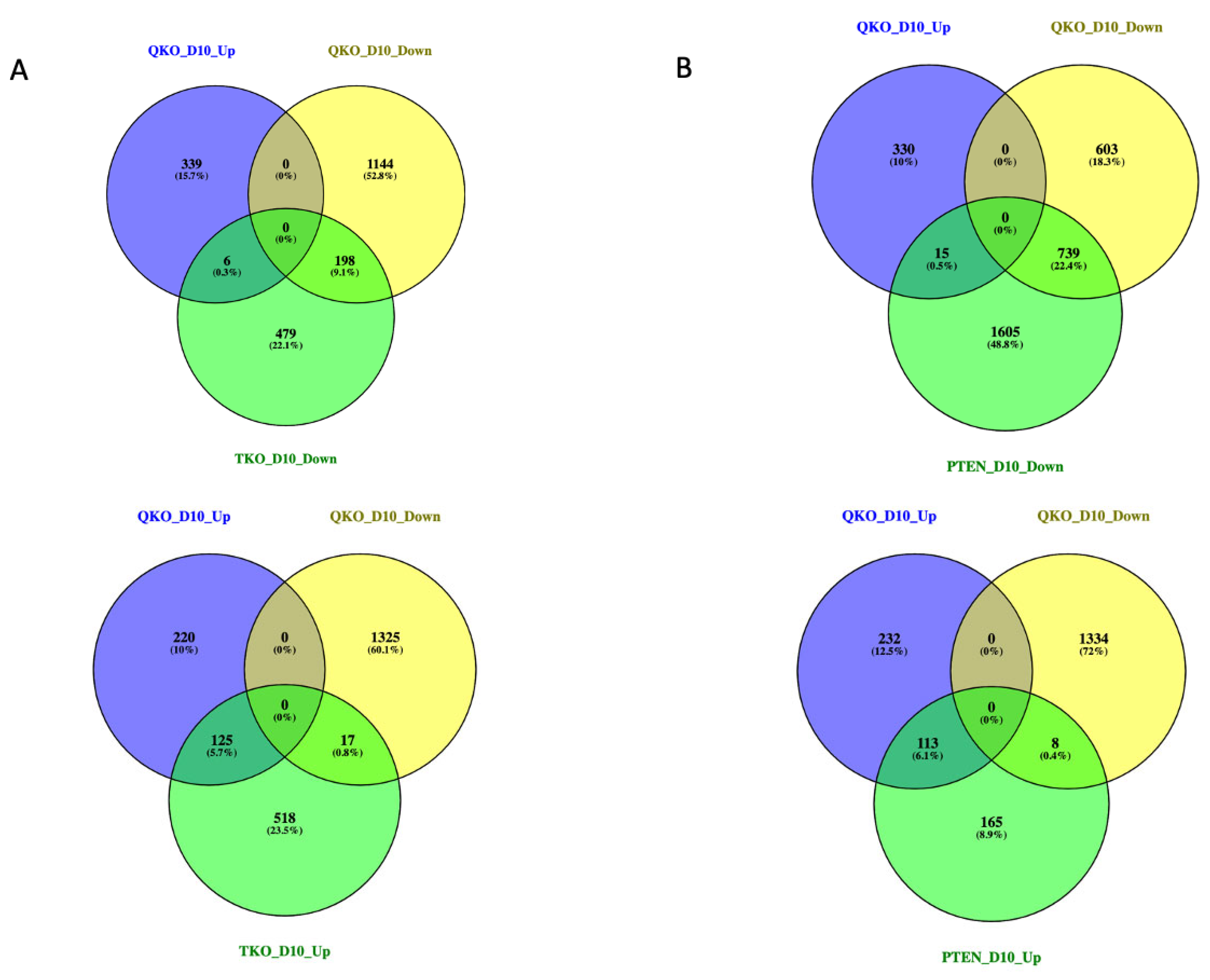
3.4. Epistasis between FGFR-Signaling and PTEN in Lens Epithelial Explants in Response to DM
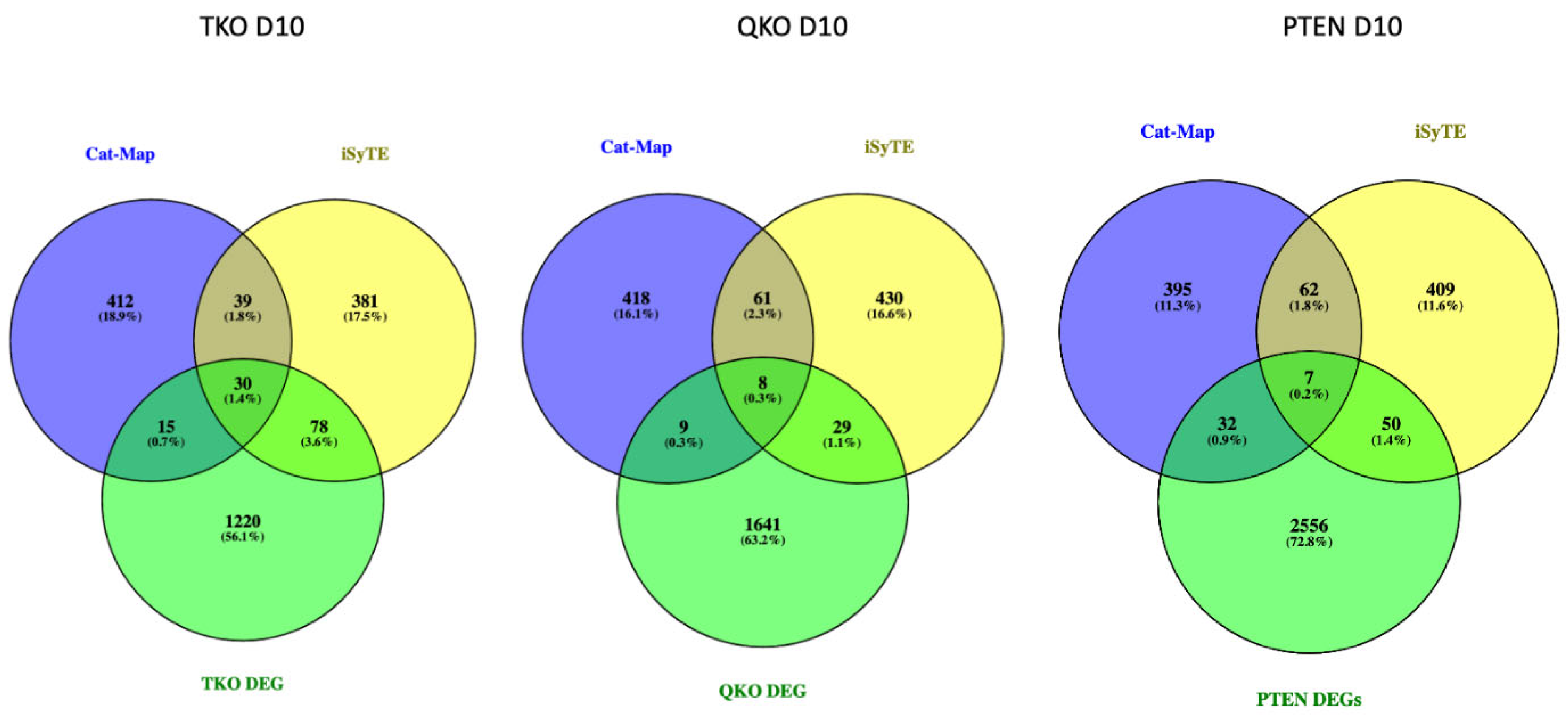
3.5. The Effect of Fgfr/Pten Epistasis on the Expression of Genes Listed in iSyTE and Cat-Map
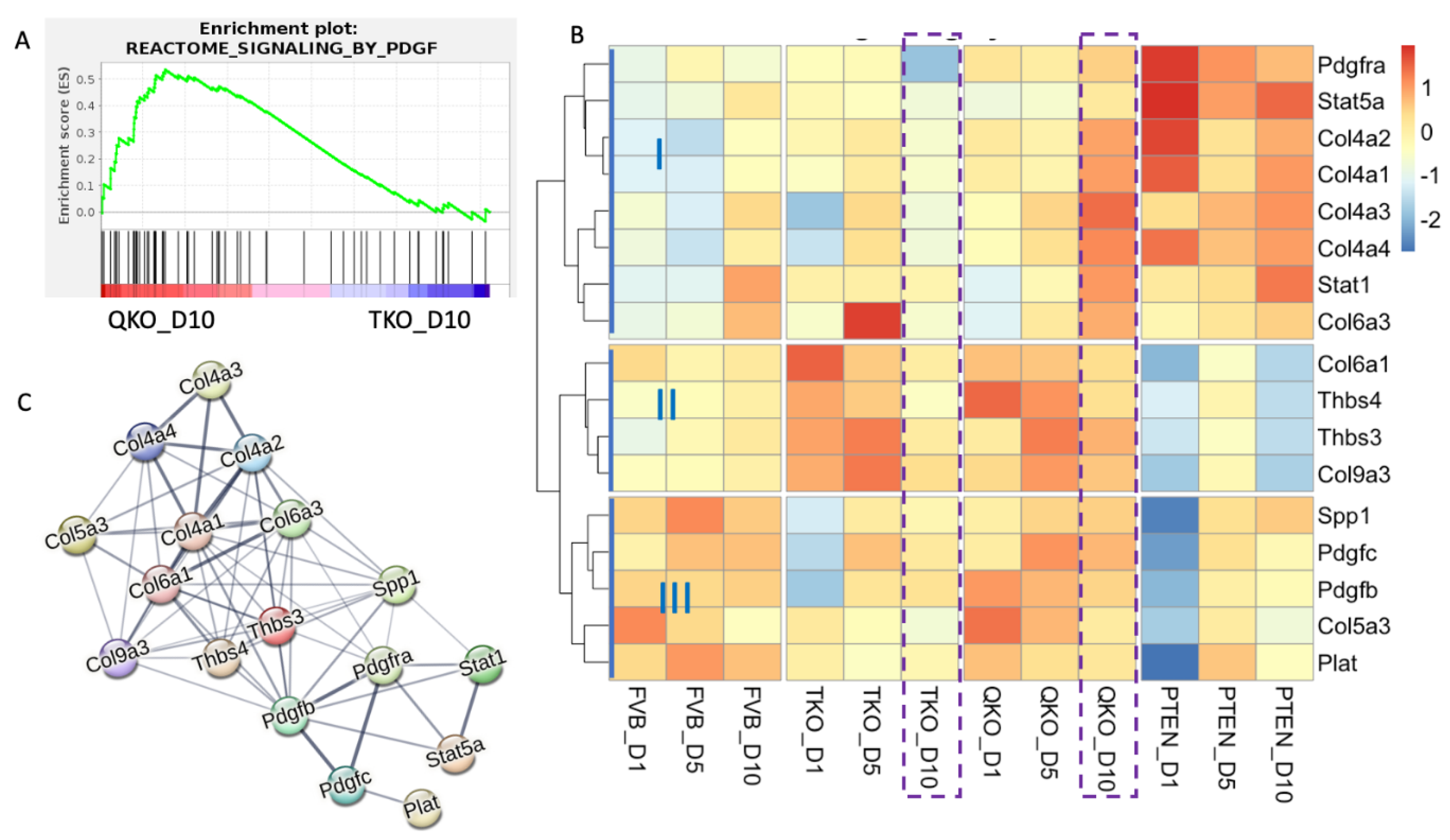
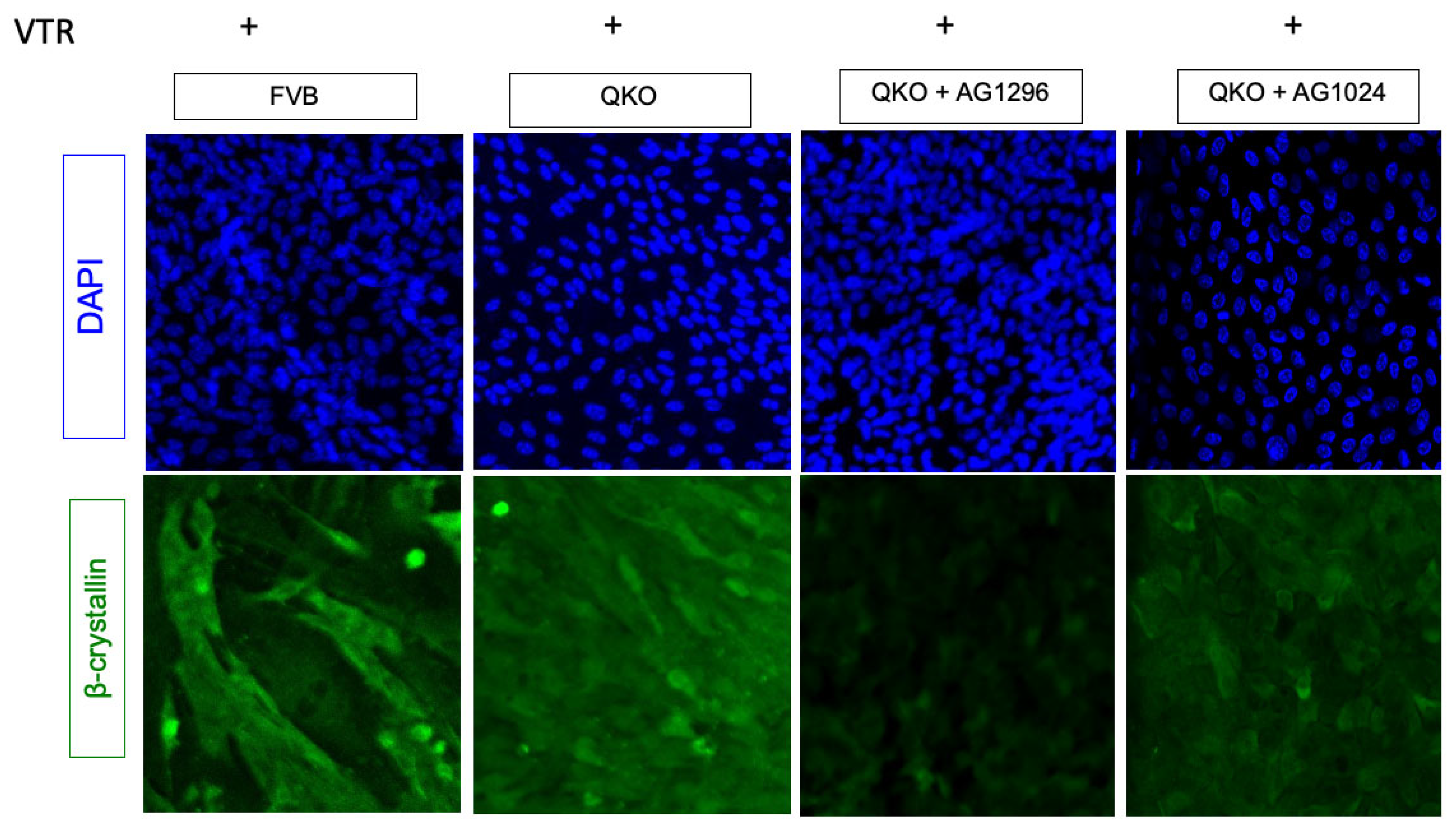
3.6. PDGFRa: A Potential Alternative Pathway for Fiber Cell Differentiation in Absence of FGFRs and PTEN
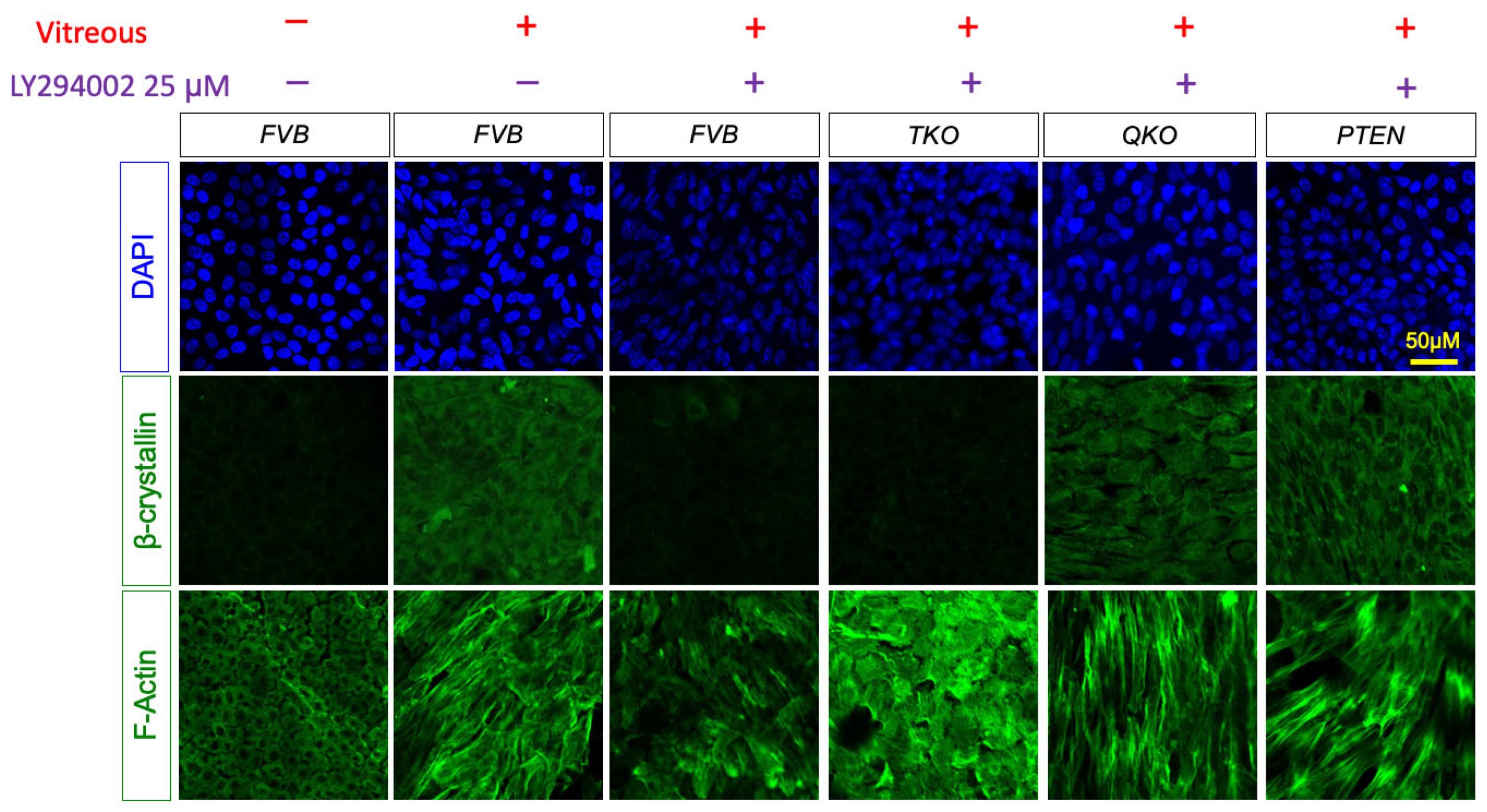
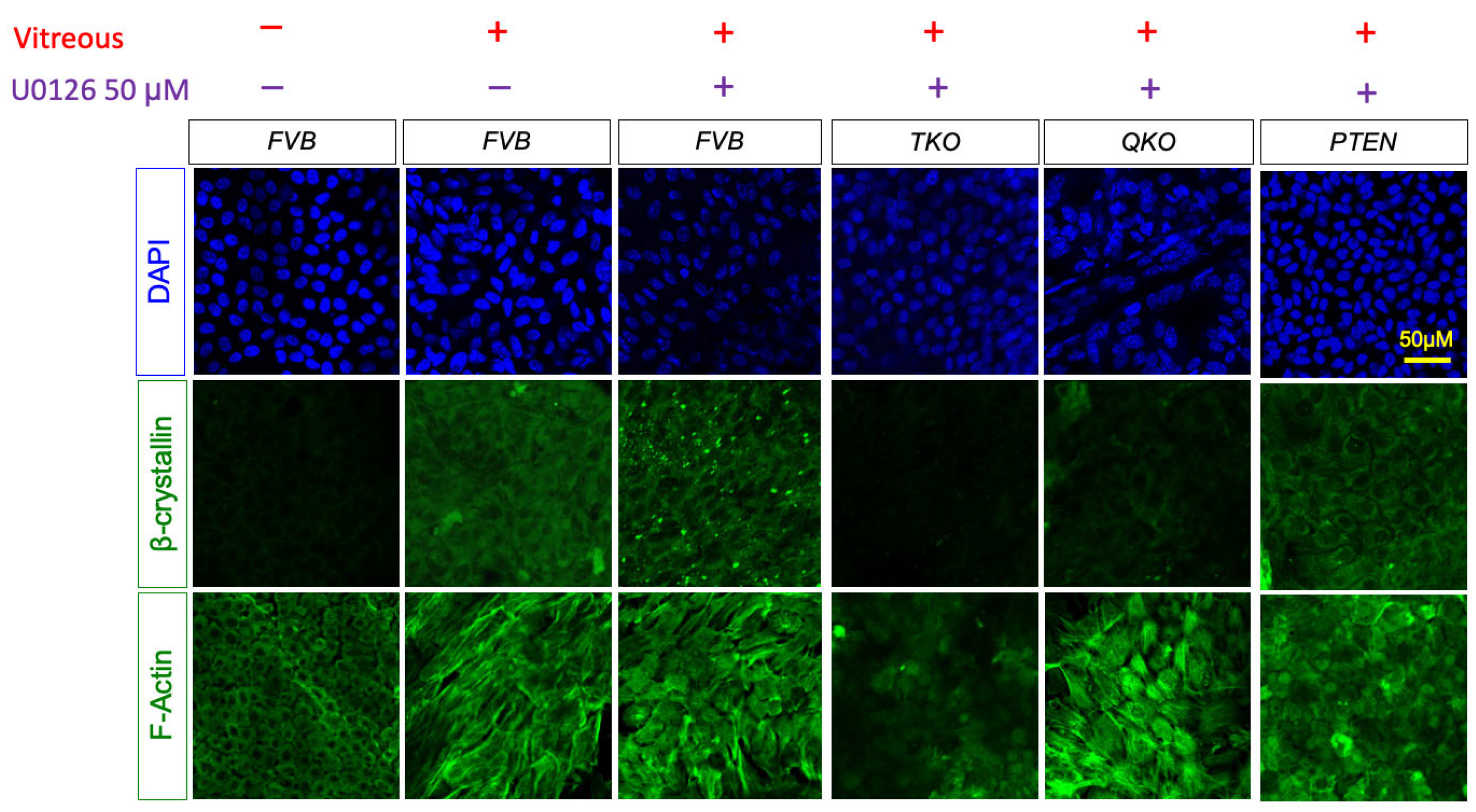
3.7. ERK Signaling is Essential for Fiber Cell Differentiation in Absence of FGFRs and PTEN
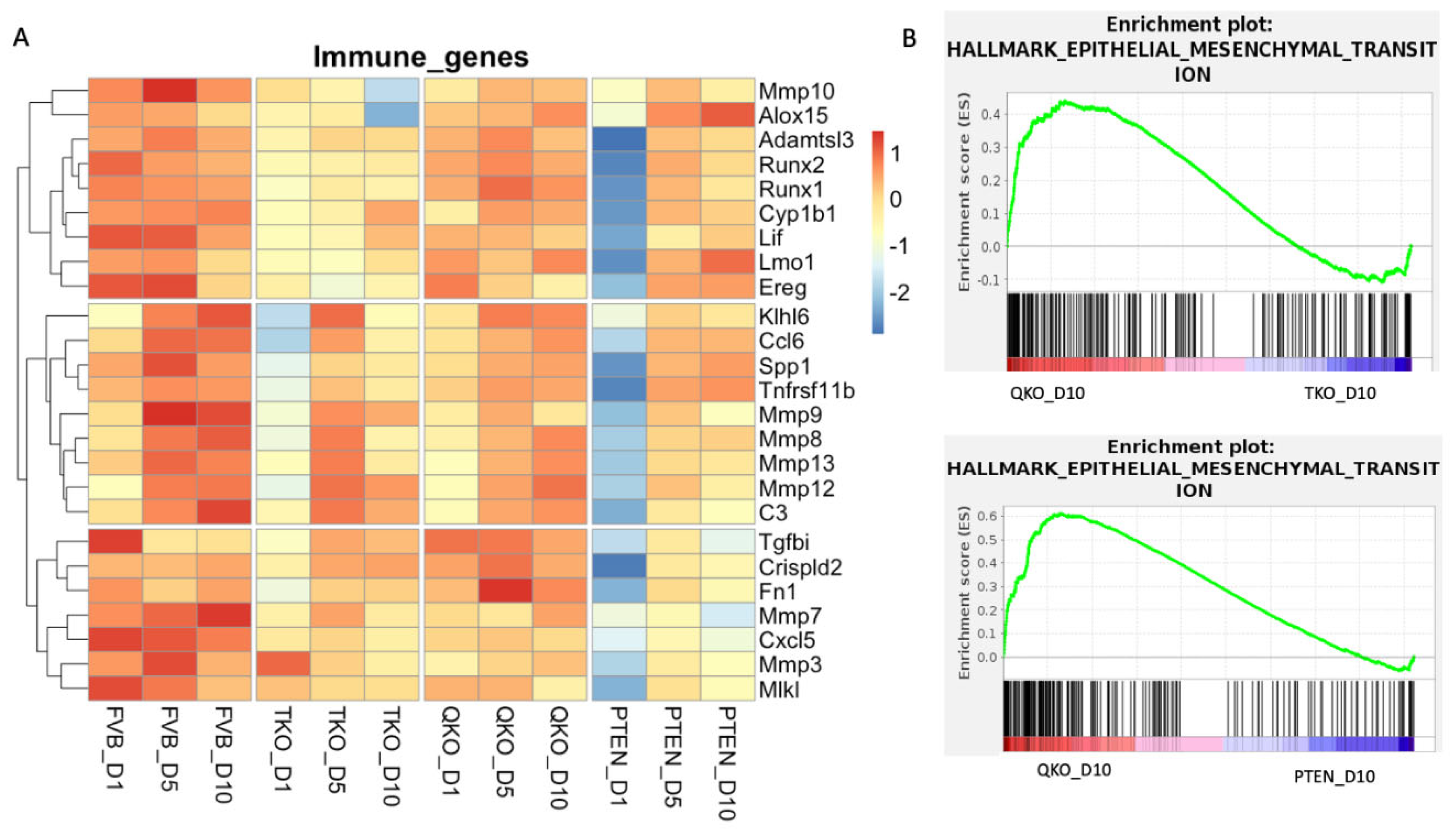
3.8. The Removal of Pten Increased the Expression of Genes Related to Immune Response and Epithelial to Mesenchymal Transition in Explants Lacking Fgfrs
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Q.; McAvoy, J.W.; Lovicu, F.J. Growth Factor Signaling in Vitreous Humor-Induced Lens Fiber Differentiation. Invest. Ophthalmol. Vis. Sci. 2010, 51, 3599–3610. [CrossRef]
- Garcia, C.M.; Shui, Y.-B.; Kamath, M.; DeVillar, J.; Johnson, R.S.; Gerber, H.-P.; Ferrara, N.; Robinson, M.L.; Beebe, D.C. The Function of VEGF-A in Lens Development: Formation of the Hyaloid Capillary Network and Protection against Transient Nuclear Cataracts. Exp. Eye Res. 2009, 88, 270–276. [CrossRef]
- Orr-Urtreger, A.; Givol, D.; Yayon, A.; Yarden, Y.; Lonai, P. Developmental Expression of Two Murine Fibroblast Growth Factor Receptors, Flg and Bek. Development 1991, 113, 1419–1434. [CrossRef]
- Peters, K.; Ornitz, D.; Werner, S.; Williams, L. Unique Expression Pattern of the FGF Receptor 3 Gene during Mouse Organogenesis. Dev. Biol. 1993, 155, 423–430. [CrossRef]
- Garcia, C.M.; Yu, K.; Zhao, H.; Ashery-Padan, R.; Ornitz, D.M.; Robinson, M.L.; Beebe, D.C. Signaling through FGF Receptor-2 Is Required for Lens Cell Survival and for Withdrawal from the Cell Cycle during Lens Fiber Cell Differentiation. Dev. Dyn. 2005, 233, 516–527. [CrossRef]
- Potts, J.D.; Bassnett, S.; Kornacker, S.; Beebe, D.C. Expression of Platelet-Derived Growth Factor Receptors in the Developing Chicken Lens. Invest. Ophthalmol. Vis. Sci. 1994, 35, 3413–3421.
- Reneker, L.W.; Silversides, D.W.; Patel, K.; Overbeek, P.A. TGF Alpha Can Act as a Chemoattractant to Perioptic Mesenchymal Cells in Developing Mouse Eyes. Development 1995, 121, 1669–1680. [CrossRef]
- Reneker, L.W.; Silversides, D.W.; Xu, L.; Overbeek, P.A. Formation of Corneal Endothelium Is Essential for Anterior Segment Development - a Transgenic Mouse Model of Anterior Segment Dysgenesis. Development 2000, 127, 533–542. [CrossRef]
- Shirke, S.; Faber, S.C.; Hallem, E.; Makarenkova, H.P.; Robinson, M.L.; Overbeek, P.A.; Lang, R.A. Misexpression of IGF-I in the Mouse Lens Expands the Transitional Zone and Perturbs Lens Polarization. Mech. Dev. 2001, 101, 167–174. [CrossRef]
- Ash, J.D.; Overbeek, P.A. Lens-Specific VEGF-A Expression Induces Angioblast Migration and Proliferation and Stimulates Angiogenic Remodeling. Dev. Biol. 2000, 223, 383–398. [CrossRef]
- Reneker, L.W.; Overbeek, P.A. Lens-Specific Expression of PDGF-A Alters Lens Growth and Development. Dev. Biol. 1996, 180, 554–565. [CrossRef]
- Robinson, M.L.; Overbeek, P.A.; Verran, D.J.; Grizzle, W.E.; Stockard, C.R.; Friesel, R.; Maciag, T.; Thompson, J.A. Extracellular FGF-1 Acts as a Lens Differentiation Factor in Transgenic Mice. Development 1995, 121, 505–514. [CrossRef]
- Robinson, M.L.; Ohtaka-Maruyama, C.; Chan, C.C.; Jamieson, S.; Dickson, C.; Overbeek, P.A.; Chepelinsky, A.B. Disregulation of Ocular Morphogenesis by Lens-Specific Expression of FGF-3/int-2 in Transgenic Mice. Dev. Biol. 1998, 198, 13–31. [CrossRef]
- Lovicu, F.J.; Overbeek, P.A. Overlapping Effects of Different Members of the FGF Family on Lens Fiber Differentiation in Transgenic Mice. Development 1998, 125, 3365–3377. [CrossRef]
- Gotoh, N. Regulation of Growth Factor Signaling by FRS2 Family Docking/scaffold Adaptor Proteins. Cancer Sci. 2008, 99, 1319–1325. [CrossRef]
- Madakashira, B.P.; Kobrinski, D.A.; Hancher, A.D.; Arneman, E.C.; Wagner, B.D.; Wang, F.; Shin, H.; Lovicu, F.J.; Reneker, L.W.; Robinson, M.L. Frs2α Enhances Fibroblast Growth Factor-Mediated Survival and Differentiation in Lens Development. Development 2012, 139, 4601–4612. [CrossRef]
- Zhao, H.; Yang, T.; Madakashira, B.P.; Thiels, C.A.; Bechtle, C.A.; Garcia, C.M.; Zhang, H.; Yu, K.; Ornitz, D.M.; Beebe, D.C.; et al. Fibroblast Growth Factor Receptor Signaling Is Essential for Lens Fiber Cell Differentiation. Dev. Biol. 2008, 318, 276–288. [CrossRef]
- Garcia, C.M.; Huang, J.; Madakashira, B.P.; Liu, Y.; Rajagopal, R.; Dattilo, L.; Robinson, M.L.; Beebe, D.C. The Function of FGF Signaling in the Lens Placode. Dev. Biol. 2011, 351, 176–185. [CrossRef]
- Jerde, T.J. Phosphatase and Tensin Homologue: Novel Regulation by Developmental Signaling. J. Signal Transduct. 2015, 2015, 282567. [CrossRef]
- Chaffee, B.R.; Hoang, T.V.; Leonard, M.R.; Bruney, D.G.; Wagner, B.D.; Dowd, J.R.; Leone, G.; Ostrowski, M.C.; Robinson, M.L. FGFR and PTEN Signaling Interact during Lens Development to Regulate Cell Survival. Dev. Biol. 2016, 410, 150–163. [CrossRef]
- Guntur, A.R.; Reinhold, M.I.; Cuellar, J., Jr; Naski, M.C. Conditional Ablation of Pten in Osteoprogenitors Stimulates FGF Signaling. Development 2011, 138, 1433–1444. [CrossRef]
- Hertzler-Schaefer, K.; Mathew, G.; Somani, A.-K.; Tholpady, S.; Kadakia, M.P.; Chen, Y.; Spandau, D.F.; Zhang, X. Pten Loss Induces Autocrine FGF Signaling to Promote Skin Tumorigenesis. Cell Rep. 2014, 6, 818–826. [CrossRef]
- Padula, S.L.; Anand, D.; Hoang, T.V.; Chaffee, B.R.; Liu, L.; Liang, C.; Lachke, S.A.; Robinson, M.L. High-Throughput Transcriptome Analysis Reveals That the Loss of Pten Activates a Novel NKX6-1/RASGRP1 Regulatory Module to Rescue Microphthalmia Caused by Fgfr2-Deficient Lenses. Hum. Genet. 2019, 138, 1391–1407. [CrossRef]
- Upreti, A.; Padula, S.L.; Tangeman, J.A.; Wagner, B.D.; O’Connell, M.J.; Jaquish, T.J.; Palko, R.K.; Mantz, C.J.; Anand, D.; Lovicu, F.J.; et al. Lens Epithelial Explants Treated with Vitreous Humor Undergo Alterations in Chromatin Landscape with Concurrent Activation of Genes Associated with Fiber Cell Differentiation and Innate Immune Response. Cells 2023, 12. [CrossRef]
- Padula, S.L.; Sidler, E.P.; Wagner, B.D.; Manz, C.J.; Lovicu, F.J.; Robinson, M.L. Lens Fiber Cell Differentiation Occurs Independently of Fibroblast Growth Factor Receptor Signaling in the Absence of Pten. Dev. Biol. 2020, 467, 1–13. [CrossRef]
- West-Mays, J.A.; Pino, G.; Lovicu, F.J. Development and Use of the Lens Epithelial Explant System to Study Lens Differentiation and Cataractogenesis. Prog. Retin. Eye Res. 2010, 29, 135–143. [CrossRef]
- Zelenka, P.S.; Gao, C.Y.; Saravanamuthu, S.S. Preparation and Culture of Rat Lens Epithelial Explants for Studying Terminal Differentiation. J. Vis. Exp. 2009, doi:10.3791/1519. [CrossRef]
- Schulz, M.W.; Chamberlain, C.G.; de Iongh, R.U.; McAvoy, J.W. Acidic and Basic FGF in Ocular Media and Lens: Implications for Lens Polarity and Growth Patterns. Development 1993, 118, 117–126. [CrossRef]
- Davis, M.P.; van Dongen, S.; Abreu-Goodger, C.; Bartonicek, N.; Enright, A.J. Kraken: A Set of Tools for Quality Control and Analysis of High-Throughput Sequence Data☆. Methods 2013, 63, 41–49. [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17, 10–12. [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [CrossRef]
- Adams, R.P. K-Means Clustering and Related Algorithms. Princeton University 2018.
- Website Available online: Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. J. R. Stat. Soc. Ser. C. Appl. Stat. 1979, 28, 100 R Core Team R: A Language and Envirronment for Statistical Computing. Available online: https//www.R-project.org/.
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 15545–15550. [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-Responsive Genes Involved in Oxidative Phosphorylation Are Coordinately Downregulated in Human Diabetes. Nat. Genet. 2003, 34, 267–273. [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [CrossRef]
- Hoang, T.V.; Kumar, P.K.R.; Sutharzan, S.; Tsonis, P.A.; Liang, C.; Robinson, M.L. Comparative Transcriptome Analysis of Epithelial and Fiber Cells in Newborn Mouse Lenses with RNA Sequencing. Mol. Vis. 2014, 20, 1491–1517.
- van Zyl, T.; Yan, W.; McAdams, A.M.; Monavarfeshani, A.; Hageman, G.S.; Sanes, J.R. Cell Atlas of the Human Ocular Anterior Segment: Tissue-Specific and Shared Cell Types. Proc. Natl. Acad. Sci. U. S. A. 2022, 119, e2200914119. [CrossRef]
- Tangeman, J.A.; Rebull, S.M.; Grajales-Esquivel, E.; Weaver, J.M.; Bendezu-Sayas, S.; Robinson, M.L.; Lachke, S.A.; Del Rio-Tsonis, K. Integrated Single-Cell Multiomics Uncovers Foundational Regulatory Mechanisms of Lens Development and Pathology. Development 2024, 151. [CrossRef]
- Maddala, R.; Gao, J.; Mathias, R.T.; Lewis, T.R.; Arshavsky, V.Y.; Levine, A.; Backer, J.M.; Bresnick, A.R.; Rao, P.V. Absence of S100A4 in the Mouse Lens Induces an Aberrant Retina-Specific Differentiation Program and Cataract. Sci. Rep. 2021, 11, 2203. [CrossRef]
- Kakrana, A.; Yang, A.; Anand, D.; Djordjevic, D.; Ramachandruni, D.; Singh, A.; Huang, H.; Ho, J.W.K.; Lachke, S.A. iSyTE 2.0: A Database for Expression-Based Gene Discovery in the Eye. Nucleic Acids Res. 2018, 46, D875–D885. [CrossRef]
- Shiels, A.; Bennett, T.M.; Hejtmancik, J.F. Cat-Map: Putting Cataract on the Map. Mol. Vis. 2010, 16, 2007–2015.
- Faber, S.C.; Dimanlig, P.; Makarenkova, H.P.; Shirke, S.; Ko, K.; Lang, R.A. Fgf Receptor Signaling Plays a Role in Lens Induction. Development 2001, 128, 4425–4438. [CrossRef]
- Kim, Y.-J.; Bahn, M.; Kim, Y.H.; Shin, J.-Y.; Cheong, S.-W.; Ju, B.-G.; Kim, W.-S.; Yeo, C.-Y. Xenopus Laevis FGF Receptor Substrate 3 (XFrs3) Is Important for Eye Development and Mediates Pax6 Expression in Lens Placode through Its Shp2-Binding Sites. Dev. Biol. 2015, 397, 129–139. [CrossRef]
- Anchan, R.M.; Lachke, S.A.; Gerami-Naini, B.; Lindsey, J.; Ng, N.; Naber, C.; Nickerson, M.; Cavallesco, R.; Rowan, S.; Eaton, J.L.; et al. Pax6- and Six3-Mediated Induction of Lens Cell Fate in Mouse and Human ES Cells. PLoS One 2014, 9, e115106. [CrossRef]
- Sun, J.; Rockowitz, S.; Xie, Q.; Ashery-Padan, R.; Zheng, D.; Cvekl, A. Identification of in Vivo DNA-Binding Mechanisms of Pax6 and Reconstruction of Pax6-Dependent Gene Regulatory Networks during Forebrain and Lens Development. Nucleic Acids Res. 2015, 43, 6827–6846. [CrossRef]
- Huang, J.; Rajagopal, R.; Liu, Y.; Dattilo, L.K.; Shaham, O.; Ashery-Padan, R.; Beebe, D.C. The Mechanism of Lens Placode Formation: A Case of Matrix-Mediated Morphogenesis. Dev. Biol. 2011, 355, 32–42. [CrossRef]
- Wenzel, P.L.; Chong, J.-L.; Sáenz-Robles, M.T.; Ferrey, A.; Hagan, J.P.; Gomez, Y.M.; Rajmohan, R.; Sharma, N.; Chen, H.-Z.; Pipas, J.M.; et al. Cell Proliferation in the Absence of E2F1-3. Dev. Biol. 2011, 351, 35–45. [CrossRef]
- Saravanamuthu, S.S.; Le, T.T.; Gao, C.Y.; Cojocaru, R.I.; Pandiyan, P.; Liu, C.; Zhang, J.; Zelenka, P.S.; Brown, N.L. Conditional Ablation of the Notch2 Receptor in the Ocular Lens. Dev. Biol. 2012, 362, 219–229. [CrossRef]
- Coulombre, J.L.; Coulombre, A.J. LENS DEVELOPMENT: FIBER ELONGATION AND LENS ORIENTATION. Science 1963, 142, 1489–1490. [CrossRef]
- Chamberlain, C.G.; McAvoy, J.W. Induction of Lens Fibre Differentiation by Acidic and Basic Fibroblast Growth Factor (FGF). Growth Factors 1989, 1, 125–134. [CrossRef]
- O’Connor, M.D.; Wederell, E.D.; de Iongh, R.; Lovicu, F.J.; McAvoy, J.W. Generation of Transparency and Cellular Organization in Lens Explants. Exp. Eye Res. 2008, 86, 734–745. [CrossRef]
- Iyengar, L.; Wang, Q.; Rasko, J.E.J.; McAvoy, J.W.; Lovicu, F.J. Duration of ERK1/2 Phosphorylation Induced by FGF or Ocular Media Determines Lens Cell Fate. Differentiation 2007, 75, 662–668. [CrossRef]
- Sellitto, C.; Li, L.; Gao, J.; Robinson, M.L.; Lin, R.Z.; Mathias, R.T.; White, T.W. AKT Activation Promotes PTEN Hamartoma Tumor Syndrome-Associated Cataract Development. J. Clin. Invest. 2013, 123, 5401–5409. [CrossRef]
- Ito, S.; Umehara, T.; Koseki, H. Polycomb-Mediated Histone Modifications and Gene Regulation. Biochem. Soc. Trans. 2024, 52, 151–161. [CrossRef]
- Dong, N.; Xu, B.; Xu, J. EGF-Mediated Overexpression of Myc Attenuates miR-26b by Recruiting HDAC3 to Induce Epithelial-Mesenchymal Transition of Lens Epithelial Cells. Biomed Res. Int. 2018, 2018, 7148023. [CrossRef]
- Zhang, L.; Wang, L.; Hu, X.-B.; Hou, M.; Xiao, Y.; Xiang, J.-W.; Xie, J.; Chen, Z.-G.; Yang, T.-H.; Nie, Q.; et al. MYPT1/PP1-Mediated EZH2 Dephosphorylation at S21 Promotes Epithelial-Mesenchymal Transition in Fibrosis through Control of Multiple Families of Genes. Adv. Sci. 2022, 9, e2105539. [CrossRef]
- Cui, X.; Du, C.; Wan, S.; Wu, D.; Yan, L.; Zhang, J.; Li, J.; Li, H.; Yang, Z.; Zhang, H.; et al. Deficiency of Heat Shock Factor 4 Promotes Lens Epithelial Cell Senescence through Upregulating p21 Expression. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166233. [CrossRef]
- Yang, J.; Yin, Y. PTEN in Chromatin Remodeling. Cold Spring Harb. Perspect. Med. 2020, 10. [CrossRef]
- Chen, Z.H.; Zhu, M.; Yang, J.; Liang, H.; He, J.; He, S.; Wang, P.; Kang, X.; McNutt, M.A.; Yin, Y.; et al. PTEN Interacts with Histone H1 and Controls Chromatin Condensation. Cell Rep. 2014, 8, 2003–2014. [CrossRef]
- Fan, X.; Kraynak, J.; Knisely, J.P.S.; Formenti, S.C.; Shen, W.H. PTEN as a Guardian of the Genome: Pathways and Targets. Cold Spring Harb. Perspect. Med. 2020, 10. [CrossRef]
- Tuzon, C.T.; Rigueur, D.; Merrill, A.E. Nuclear Fibroblast Growth Factor Receptor Signaling in Skeletal Development and Disease. Curr. Osteoporos. Rep. 2019, 17, 138–146. [CrossRef]
- Ma, J.; Benitez, J.A.; Li, J.; Miki, S.; Ponte de Albuquerque, C.; Galatro, T.; Orellana, L.; Zanca, C.; Reed, R.; Boyer, A.; et al. Inhibition of Nuclear PTEN Tyrosine Phosphorylation Enhances Glioma Radiation Sensitivity through Attenuated DNA Repair. Cancer Cell 2019, 35, 504–518.e7. [CrossRef]
- Li, H.; Mao, Y.; Bouaziz, M.; Yu, H.; Qu, X.; Wang, F.; Feng, G.-S.; Shawber, C.; Zhang, X. Lens Differentiation Is Controlled by the Balance between PDGF and FGF Signaling. PLoS Biol. 2019, 17, e3000133. [CrossRef]
- Li, Q.; Li, Z.; Luo, T.; Shi, H. Targeting the PI3K/AKT/mTOR and RAF/MEK/ERK Pathways for Cancer Therapy. Mol Biomed 2022, 3, 47. [CrossRef]
- Gu, J.; Tamura, M.; Yamada, K.M. Tumor Suppressor PTEN Inhibits Integrin- and Growth Factor-Mediated Mitogen-Activated Protein (MAP) Kinase Signaling Pathways. J. Cell Biol. 1998, 143, 1375–1383. [CrossRef]
- Jechlinger, M.; Sommer, A.; Moriggl, R.; Seither, P.; Kraut, N.; Capodiecci, P.; Donovan, M.; Cordon-Cardo, C.; Beug, H.; Grünert, S. Autocrine PDGFR Signaling Promotes Mammary Cancer Metastasis. J. Clin. Invest. 2006, 116, 1561–1570. [CrossRef]
- Zheng, H.; Kang, Y. Multilayer Control of the EMT Master Regulators. Oncogene 2014, 33, 1755–1763. [CrossRef]
- Mongroo, P.S.; Rustgi, A.K. The Role of the miR-200 Family in Epithelial-Mesenchymal Transition. Cancer Biol. Ther. 2010, 10, 219–222. [CrossRef]
- Vu, N.D.; Chepko, G.; Zelenka, P. Decreased Turnover of Phosphatidylinositol Accompanies in Vitro Differentiation of Embryonic Chicken Lens Epithelial Cells into Lens Fibers. Biochim. Biophys. Acta 1983, 750, 105–111. [CrossRef]
- Nath, P.; Getzenberg, R.; Beebe, D.; Pallansch, L.; Zelenka, P. C-Myc mRNA Is Elevated as Differentiating Lens Cells Withdraw from the Cell Cycle. Exp. Cell Res. 1987, 169, 215–222. [CrossRef]
- Lovicu, F.J.; Chamberlain, C.G.; McAvoy, J.W. Differential Effects of Aqueous and Vitreous on Fiber Differentiation and Extracellular Matrix Accumulation in Lens Epithelial Explants. Invest. Ophthalmol. Vis. Sci. 1995, 36, 1459–1469.
- Wang, Q.; Stump, R.; McAvoy, J.W.; Lovicu, F.J. MAPK/ERK1/2 and PI3-Kinase Signalling Pathways Are Required for Vitreous-Induced Lens Fibre Cell Differentiation. Exp. Eye Res. 2009, 88, 293–306. [CrossRef]
| Mouse Model | Pax6cKO | Pax6cKO | E2F1:2:3cKO | E2F1:2:3cKO | Notch2cKO |
| Stage | E9.5 | E10.5 | E17.5 | P0 | E19.5 |
| Pdgfra (f.c.) | -1.6 | 6.6 | -1.9 | -6.4 | -4.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





