Submitted:
29 April 2024
Posted:
01 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
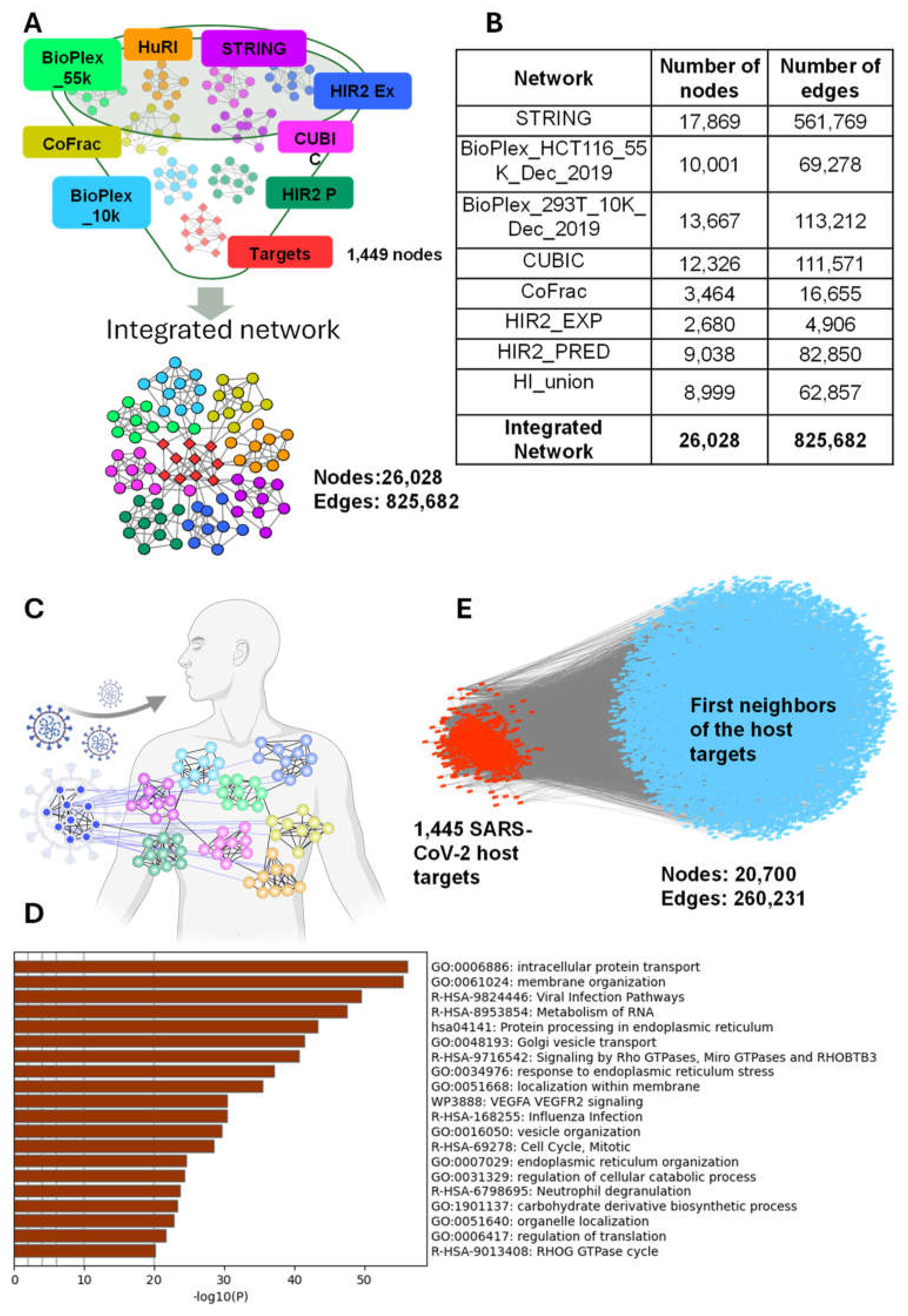
2.1. Construction of a Comprehensive Human Protein Interaction Network
2.2. Building a SARS-CoV-2 Host Proteins Specific Human Protein Interaction Network
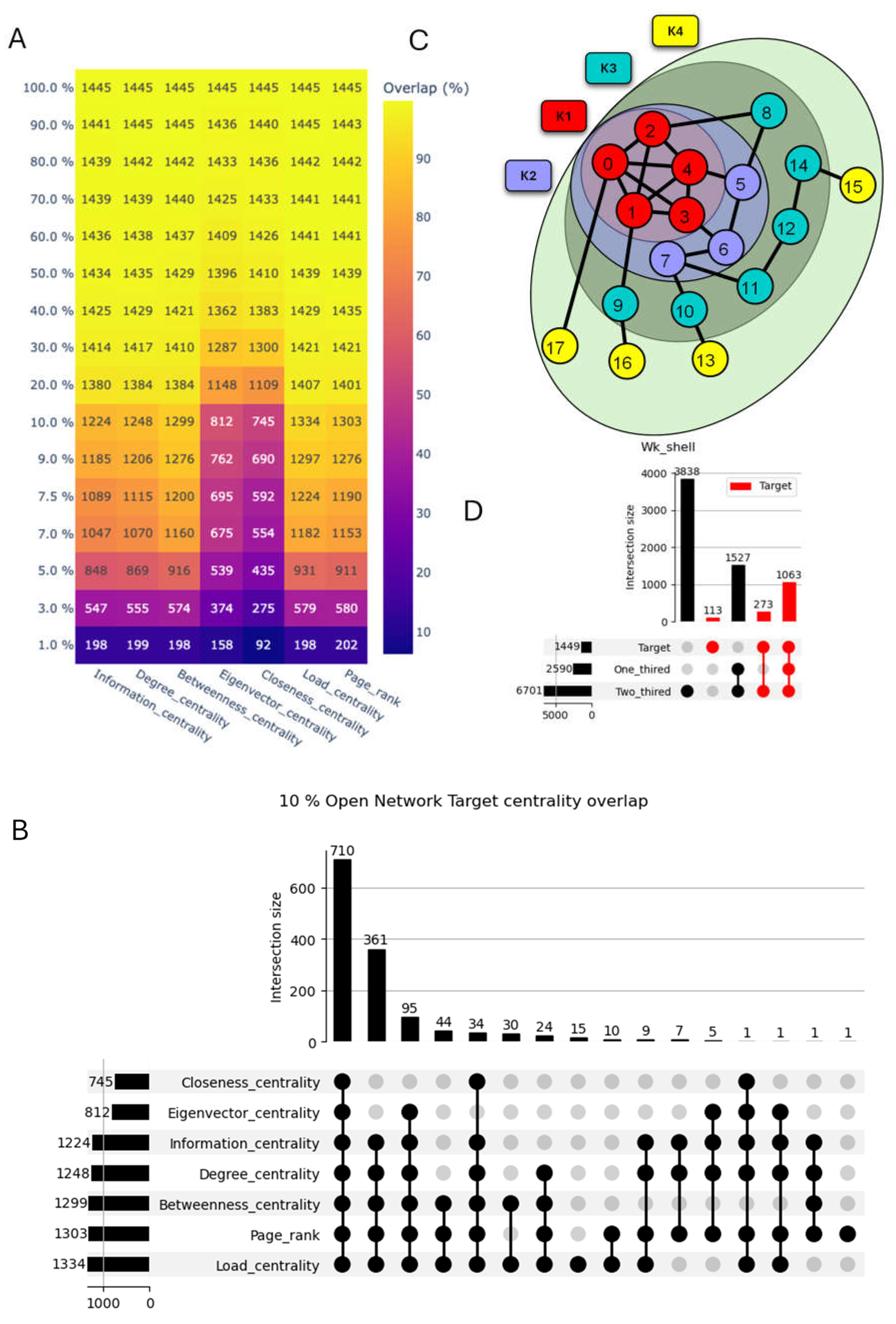
2.3. Network Centrality Analysis
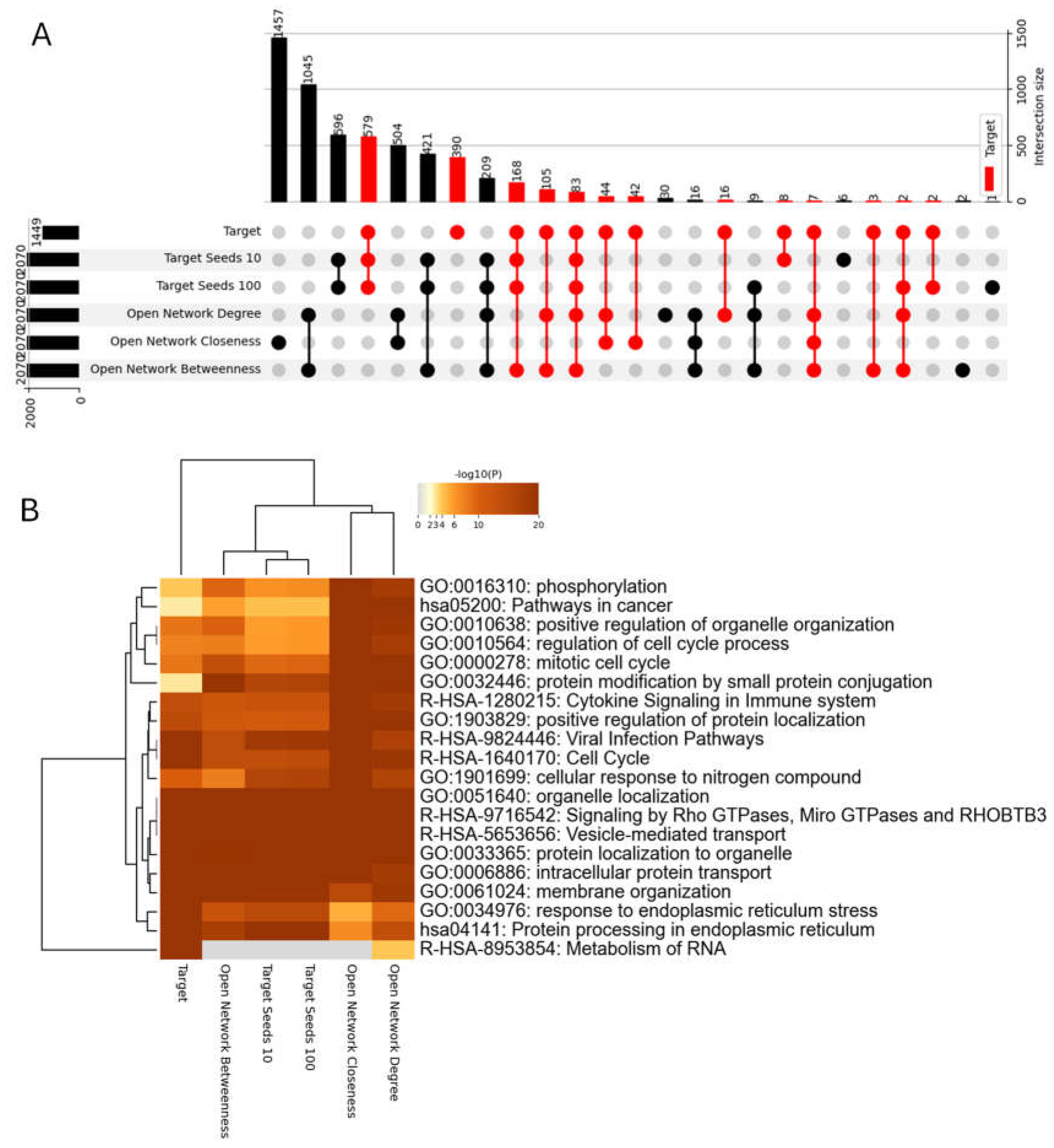
2.4. Ranking Nodes with Respect to the SARS-CoV-2 Host Proteins
- SARS-CoV-2 Host Protein Based Seeds: We utilized two variations. In the first, we selected 10% of the 1445 known SARS-CoV-2 host proteins. In the second variation, we included all 1445 known host proteins. This approach prioritizes nodes with established connections to the virus.
- Centrality Based Seeds: We identified the top 10% of nodes based on three centrality measures: Degree centrality, Betweenness centrality, and Closeness centrality. These nodes are inherently very interconnected within the network, making them prime candidates for further investigation.
3. Results and Discussion
3.1. A comprehensive Human Protein–Protein Interactome and SARS-CoV-2 Host Targets
3.2. Diverse Centrality Measures to Reveal the Enrichment of SARS-CoV-2 Targets in Human Interactome
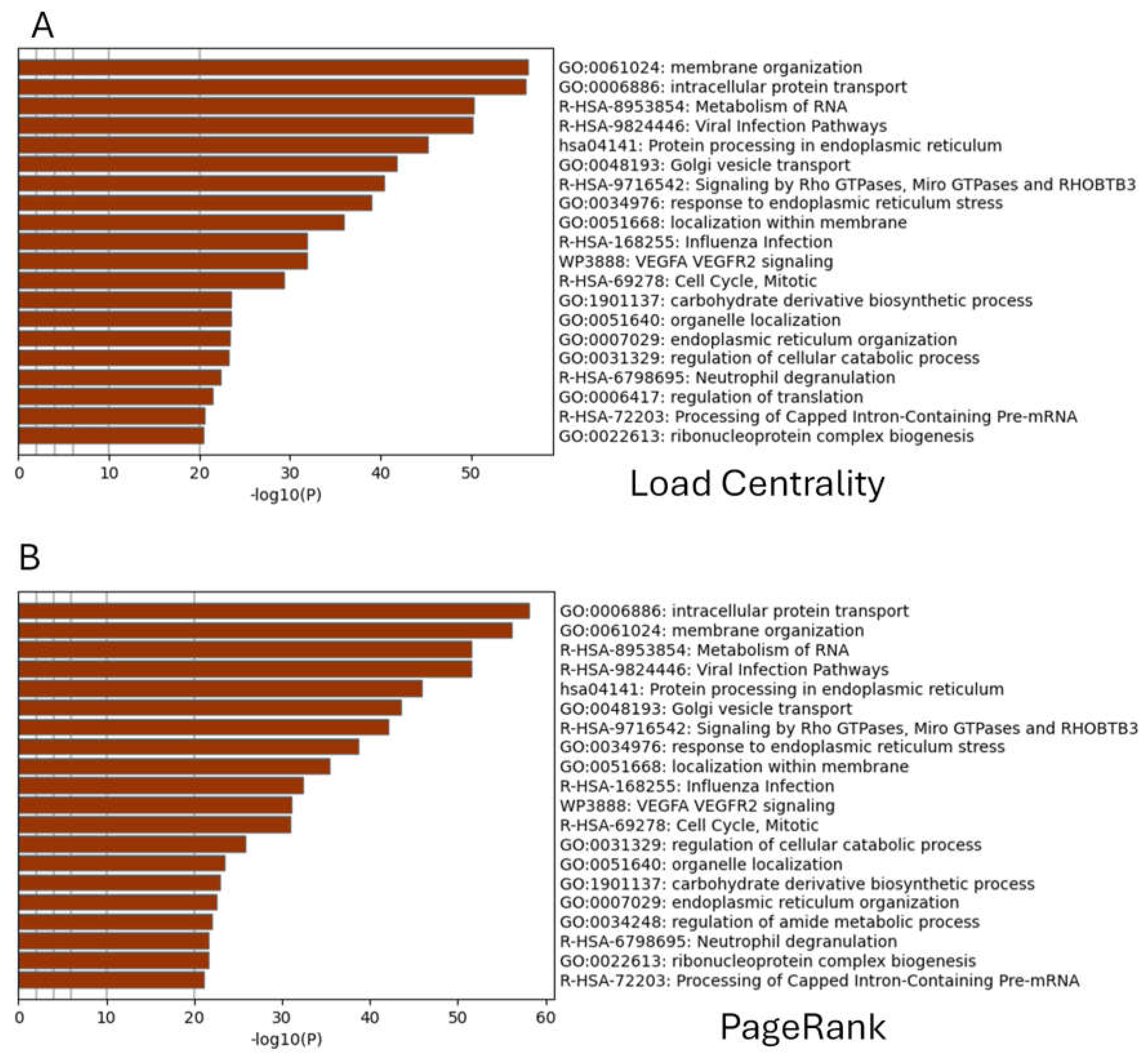
3.3. CentralityCosDist to Predict SARS-CoV-2 Targets
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCormack, M.E.; Lopez, J.A.; Crocker, T.H.; Mukhtar, M.S. Making the right connections: Network biology and plant immune system dynamics. Current Plant Biology 2016, 5, 2–12. [Google Scholar] [CrossRef]
- Pan, A.; Lahiri, C.; Rajendiran, A.; Shanmugham, B. Computational analysis of protein interaction networks for infectious diseases. Brief Bioinform 2016, 17, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabasi, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Garbutt, C.C.; Bangalore, P.V.; Kannar, P.; Mukhtar, M.S. Getting to the edge: Protein dynamical networks as a new frontier in plant-microbe interactions. Front Plant Sci 2014, 5, 312. [Google Scholar] [CrossRef]
- Majeed, A.; Mukhtar, S. Protein-Protein Interaction Network Exploration Using Cytoscape. Methods Mol Biol 2023, 2690, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Pfefferle, S.; Schopf, J.; Kogl, M.; Friedel, C.C.; Muller, M.A.; Carbajo-Lozoya, J.; Stellberger, T.; von Dall’Armi, E.; Herzog, P.; Kallies, S.; et al. The SARS-coronavirus-host interactome: Identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 2011, 7, e1002331. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012, 487, 491–495. [Google Scholar] [CrossRef]
- Gulbahce, N.; Yan, H.; Dricot, A.; Padi, M.; Byrdsong, D.; Franchi, R.; Lee, D.S.; Rozenblatt-Rosen, O.; Mar, J.C.; Calderwood, M.A.; et al. Viral perturbations of host networks reflect disease etiology. PLoS Comput Biol 2012, 8, e1002531. [Google Scholar] [CrossRef]
- Abreu, A.L.; Souza, R.P.; Gimenes, F.; Consolaro, M.E. A review of methods for detect human Papillomavirus infection. Virol J 2012, 9, 262. [Google Scholar] [CrossRef]
- Calderwood, M.A.; Venkatesan, K.; Xing, L.; Chase, M.R.; Vazquez, A.; Holthaus, A.M.; Ewence, A.E.; Li, N.; Hirozane-Kishikawa, T.; Hill, D.E.; et al. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A 2007, 104, 7606–7611. [Google Scholar] [CrossRef]
- de Chassey, B.; Navratil, V.; Tafforeau, L.; Hiet, M.S.; Aublin-Gex, A.; Agaugue, S.; Meiffren, G.; Pradezynski, F.; Faria, B.F.; Chantier, T.; et al. Hepatitis C virus infection protein network. Mol Syst Biol 2008, 4, 230. [Google Scholar] [CrossRef] [PubMed]
- Roohvand, F.; Maillard, P.; Lavergne, J.P.; Boulant, S.; Walic, M.; Andreo, U.; Goueslain, L.; Helle, F.; Mallet, A.; McLauchlan, J.; et al. Initiation of hepatitis C virus infection requires the dynamic microtubule network: Role of the viral nucleocapsid protein. J Biol Chem 2009, 284, 13778–13791. [Google Scholar] [CrossRef] [PubMed]
- Shapira, S.D.; Gat-Viks, I.; Shum, B.O.; Dricot, A.; de Grace, M.M.; Wu, L.; Gupta, P.B.; Hao, T.; Silver, S.J.; Root, D.E.; et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 2009, 139, 1255–1267. [Google Scholar] [CrossRef]
- Simonis, N.; Rual, J.F.; Lemmens, I.; Boxus, M.; Hirozane-Kishikawa, T.; Gatot, J.S.; Dricot, A.; Hao, T.; Vertommen, D.; Legros, S.; et al. Host-pathogen interactome mapping for HTLV-1 and -2 retroviruses. Retrovirology 2012, 9, 26. [Google Scholar] [CrossRef]
- Arabidopsis Interactome Mapping, C. Evidence for network evolution in an Arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Wessling, R.; Epple, P.; Altmann, S.; He, Y.; Yang, L.; Henz, S.R.; McDonald, N.; Wiley, K.; Bader, K.C.; Glasser, C.; et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 2014, 16, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Tang, K.; Tang, J.; Zeng, J.; Shen, W.; Zou, M.; Zhang, C.; Sun, Q.; Ye, X.; Li, C.; Sun, C.; et al. A network view of human immune system and virus-human interaction. Front Immunol 2022, 13, 997851. [Google Scholar] [CrossRef]
- Bosl, K.; Ianevski, A.; Than, T.T.; Andersen, P.I.; Kuivanen, S.; Teppor, M.; Zusinaite, E.; Dumpis, U.; Vitkauskiene, A.; Cox, R.J.; et al. Common Nodes of Virus-Host Interaction Revealed Through an Integrated Network Analysis. Front Immunol 2019, 10, 2186. [Google Scholar] [CrossRef]
- Kumar, N.; Mukhtar, M.S. Ranking Plant Network Nodes Based on Their Centrality Measures. Entropy 2023, 25, 676. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 2021, 184, 3022–3040. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Borgeson, B.; Phanse, S.; Tu, F.; Drew, K.; Clark, G.; Xiong, X.; Kagan, O.; Kwan, J.; Bezginov, A.; et al. Panorama of ancient metazoan macromolecular complexes. Nature 2015, 525, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Tang, Z.; Fan, W.; Wang, X.; Huang, L.; Jia, Y.; Wang, M.; Hu, Z.; Zhou, Y. Atlas of interactions between SARS-CoV-2 macromolecules and host proteins. Cell Insight 2023, 2, 100068. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Y.; Gupta, S.; Paramo, M.I.; Hou, Y.; Mao, C.; Luo, Y.; Judd, J.; Wierbowski, S.; Bertolotti, M.; et al. A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets. Nat Biotechnol 2023, 41, 128–139. [Google Scholar] [CrossRef]
- Hu, Z.; Snitkin, E.S.; DeLisi, C. VisANT: An integrative framework for networks in systems biology. Brief Bioinform 2008, 9, 317–325. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Sheynkman, G.M.; Zhang, I.; Vidal, M. Proteome-Scale Human Interactomics. Trends Biochem Sci 2017, 42, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.P.; Ding, X.B.; Jin, J.; Zhang, H.B.; Yang, Q.L.; Chen, P.C.; Yao, H.; Ruan, L.I.; Tao, Y.T.; Chen, X. HIR V2: A human interactome resource for the biological interpretation of differentially expressed genes via gene set linkage analysis. Database (Oxford) 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Mishra, B.; Mehmood, A.; Mohammad, A.; Mukhtar, M.S. Integrative Network Biology Framework Elucidates Molecular Mechanisms of SARS-CoV-2 Pathogenesis. iScience 2020, 23, 101526. [Google Scholar] [CrossRef]
- Das, J.K.; Roy, S.; Guzzi, P.H. Analyzing host-viral interactome of SARS-CoV-2 for identifying vulnerable host proteins during COVID-19 pathogenesis. Infect Genet Evol 2021, 93, 104921. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Mukhtar, M.S. Ranking Plant Network Nodes Based on Their Centrality Measures. Entropy 2023, 25, 676. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Kumar, N.; Mukhtar, M.S. Systems Biology and Machine Learning in Plant-Pathogen Interactions. Mol Plant Microbe Interact 2019, 32, 45–55. [Google Scholar] [CrossRef]
- Mishra, B.; Kumar, N.; Mukhtar, M.S. Network biology to uncover functional and structural properties of the plant immune system. Curr Opin Plant Biol 2021, 62, 102057. [Google Scholar] [CrossRef]
- Mishra, B.; Kumar, N.; Shahid Mukhtar, M. A rice protein interaction network reveals high centrality nodes and candidate pathogen effector targets. Comput Struct Biotechnol J 2022, 20, 2001–2012. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. A measure of betweenness centrality based on random walks. arXiv 2003. [Google Scholar] [CrossRef]
- Barman, R.K.; Mukhopadhyay, A.; Maulik, U.; Das, S. A network biology approach to identify crucial host targets for COVID-19. Methods 2022, 203, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Howton, T.C.; Sun, Y.; Weinberger, N.; Belkhadir, Y.; Mukhtar, M.S. Network biology discovers pathogen contact points in host protein-protein interactomes. Nature communications 2018, 9, 2312. [Google Scholar] [CrossRef] [PubMed]
- Smakowska-Luzan, E.; Mott, G.A.; Parys, K.; Stegmann, M.; Howton, T.C.; Layeghifard, M.; Neuhold, J.; Lehner, A.; Kong, J.; Grunwald, K.; et al. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 2018, 553, 342–346. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




