Submitted:
06 May 2024
Posted:
06 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Background on Spinal Muscular Atrophy
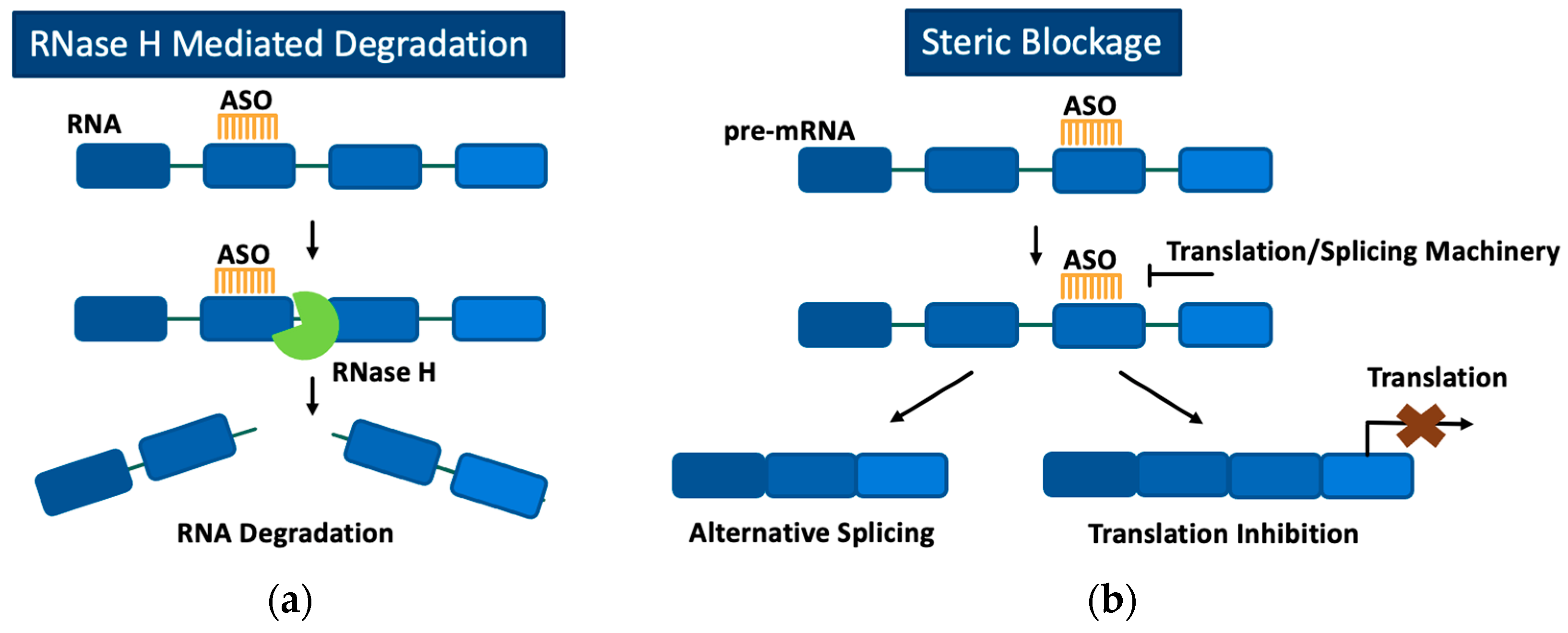
3. Antisense Oligonucleotides
3.1. Chemical Modifications to Improve the Stability, Safety, and Delivery of Antisense Oligonucleotides
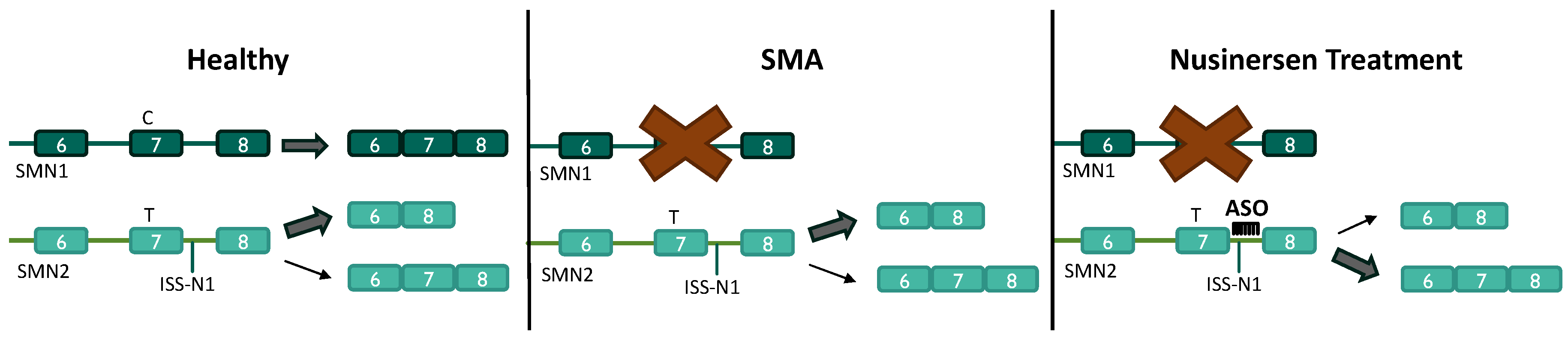
3.2. Antisense Oligonucleotides for the Treatment of SMA
3.3. Limitations of Antisense Oligonucleotides for the Treatment of SMA
4. Cell-Penetrating Peptides
4.1. Types of Cell-Penetrating Peptides
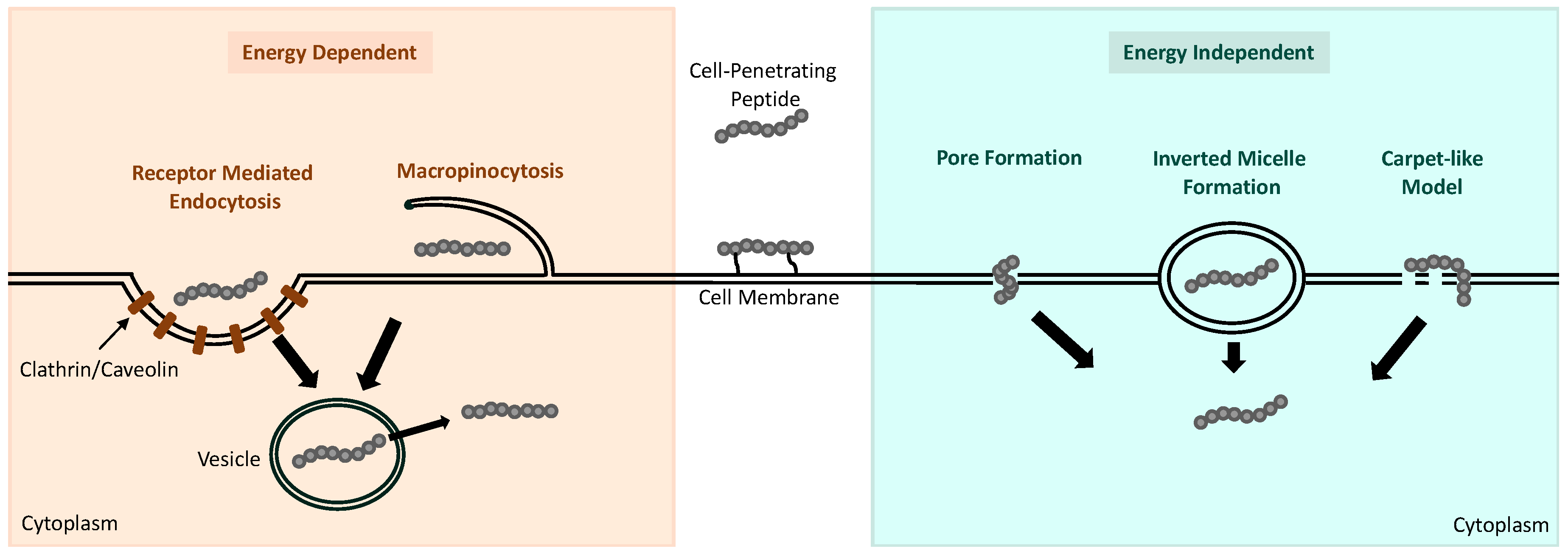
4.2. Internalization Mechanisms of Cell-Penetrating Peptides
4.3. Cell-Penetrating Peptide Conjugation with Antisense Oligonucleotides
5. Cell-Penetrating Peptide Conjugated Antisense Oligonucleotides for the Treatment of Spinal Muscular Atrophy
6. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prior, T.W.; Leach, M.E.; Finanger, E. Spinal Muscular Atrophy. In GeneReviews(®), Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds. University of Washington, Seattle Copyright © 1993-2024, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.: Seattle (WA), 1993.
- Neil, E.E.; Bisaccia, E.K. Nusinersen: A Novel Antisense Oligonucleotide for the Treatment of Spinal Muscular Atrophy. J Pediatr Pharmacol Ther 2019, 24, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. Big win possible for Ionis/Biogen antisense drug in muscular atrophy. Nat Biotechnol 2016, 34, 1002–1003. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, L.S.; Li, P.; Lau, C.H.; Leung, H.M.; Chin, Y.R.; Tin, C.; Lo, P.K. Cellular uptake, tissue penetration, biodistribution, and biosafety of threose nucleic acids: Assessing in vitro and in vivo delivery. Mater Today Bio 2022, 15, 100299. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Hazell, G.; Shabanpoor, F.; Saleh, A.F.; Bowerman, M.; Sleigh, J.N.; Meijboom, K.E.; Zhou, H.; Muntoni, F.; Talbot, K.; et al. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc Natl Acad Sci U S A 2016, 113, 10962–10967. [Google Scholar] [CrossRef] [PubMed]
- Shabanpoor, F.; Hammond, S.M.; Abendroth, F.; Hazell, G.; Wood, M.J.A.; Gait, M.J. Identification of a Peptide for Systemic Brain Delivery of a Morpholino Oligonucleotide in Mouse Models of Spinal Muscular Atrophy. Nucleic Acid Ther 2017, 27, 130–143. [Google Scholar] [CrossRef]
- Dastpeyman, M.; Sharifi, R.; Amin, A.; Karas, J.A.; Cuic, B.; Pan, Y.; Nicolazzo, J.A.; Turner, B.J.; Shabanpoor, F. Endosomal escape cell-penetrating peptides significantly enhance pharmacological effectiveness and CNS activity of systemically administered antisense oligonucleotides. Int J Pharm 2021, 599, 120398. [Google Scholar] [CrossRef] [PubMed]
- Bersani, M.; Rizzuti, M.; Pagliari, E.; Garbellini, M.; Saccomanno, D.; Moulton, H.M.; Bresolin, N.; Comi, G.P.; Corti, S.; Nizzardo, M. Cell-penetrating peptide-conjugated Morpholino rescues SMA in a symptomatic preclinical model. Mol Ther 2022, 30, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Aslesh, T.; Erkut, E.; Ren, J.; Lim, K.R.Q.; Woo, S.; Hatlevig, S.; Moulton, H.M.; Gosgnach, S.; Greer, J.; Maruyama, R.; et al. DG9-conjugated morpholino rescues phenotype in SMA mice by reaching the CNS via a subcutaneous administration. JCI Insight 2023, 8. [Google Scholar] [CrossRef]
- Haque, U.S.; Yokota, T. Enhancing Antisense Oligonucleotide-Based Therapeutic Delivery with DG9, a Versatile Cell-Penetrating Peptide. Cells 2023, 12. [Google Scholar] [CrossRef]
- Oates, E.C.; Reddel, S.; Rodriguez, M.L.; Gandolfo, L.C.; Bahlo, M.; Hawke, S.H.; Lamandé, S.R.; Clarke, N.F.; North, K.N. Autosomal dominant congenital spinal muscular atrophy: a true form of spinal muscular atrophy caused by early loss of anterior horn cells. Brain 2012, 135, 1714–1723. [Google Scholar] [CrossRef]
- Mercuri, E.; Finkel, R.S.; Muntoni, F.; Wirth, B.; Montes, J.; Main, M.; Mazzone, E.S.; Vitale, M.; Snyder, B.; Quijano-Roy, S.; et al. Diagnosis and management of spinal muscular atrophy: Part 1: Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscul Disord 2018, 28, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Wilson, R.B. Genetic testing and risk assessment for spinal muscular atrophy (SMA). Hum Genet 2002, 111, 477–500. [Google Scholar] [CrossRef] [PubMed]
- Sugarman, E.A.; Nagan, N.; Zhu, H.; Akmaev, V.R.; Zhou, Z.; Rohlfs, E.M.; Flynn, K.; Hendrickson, B.C.; Scholl, T.; Sirko-Osadsa, D.A.; et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet 2012, 20, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Mailman, M.D.; Heinz, J.W.; Papp, A.C.; Snyder, P.J.; Sedra, M.S.; Wirth, B.; Burghes, A.H.; Prior, T.W. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med 2002, 4, 20–26. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Osman, E.Y.; Bolding, M.R.; Villalón, E.; Kaifer, K.A.; Lorson, Z.C.; Tisdale, S.; Hao, Y.; Conant, G.C.; Pires, J.C.; Pellizzoni, L.; et al. Functional characterization of SMN evolution in mouse models of SMA. Sci Rep 2019, 9, 9472. [Google Scholar] [CrossRef]
- Howell, M.D.; Singh, N.N.; Singh, R.N. Advances in therapeutic development for spinal muscular atrophy. Future Med Chem 2014, 6, 1081–1099. [Google Scholar] [CrossRef]
- Singh, R.N.; Howell, M.D.; Ottesen, E.W.; Singh, N.N. Diverse role of survival motor neuron protein. Biochim Biophys Acta Gene Regul Mech 2017, 1860, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Pellizzoni, L.; Kataoka, N.; Charroux, B.; Dreyfuss, G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 1998, 95, 615–624. [Google Scholar] [CrossRef]
- Zhang, R.; So, B.R.; Li, P.; Yong, J.; Glisovic, T.; Wan, L.; Dreyfuss, G. Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell 2011, 146, 384–395. [Google Scholar] [CrossRef]
- Tisdale, S.; Lotti, F.; Saieva, L.; Van Meerbeke, J.P.; Crawford, T.O.; Sumner, C.J.; Mentis, G.Z.; Pellizzoni, L. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3'-end formation of histone mRNAs. Cell Rep 2013, 5, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Fayzullina, S.; Martin, L.J. Skeletal muscle DNA damage precedes spinal motor neuron DNA damage in a mouse model of Spinal Muscular Atrophy (SMA). PLoS One 2014, 9, e93329. [Google Scholar] [CrossRef]
- Takizawa, Y.; Qing, Y.; Takaku, M.; Ishida, T.; Morozumi, Y.; Tsujita, T.; Kogame, T.; Hirota, K.; Takahashi, M.; Shibata, T.; et al. GEMIN2 promotes accumulation of RAD51 at double-strand breaks in homologous recombination. Nucleic Acids Res 2010, 38, 5059–5074. [Google Scholar] [CrossRef]
- Genabai, N.K.; Ahmad, S.; Zhang, Z.; Jiang, X.; Gabaldon, C.A.; Gangwani, L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum Mol Genet 2015, 24, 6986–7004. [Google Scholar] [CrossRef]
- Sen, A.; Yokokura, T.; Kankel, M.W.; Dimlich, D.N.; Manent, J.; Sanyal, S.; Artavanis-Tsakonas, S. Modeling spinal muscular atrophy in Drosophila links Smn to FGF signaling. J Cell Biol 2011, 192, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Rossoll, W.; Jablonka, S.; Andreassi, C.; Kröning, A.K.; Karle, K.; Monani, U.R.; Sendtner, M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol 2003, 163, 801–812. [Google Scholar] [CrossRef]
- Gabanella, F.; Pisani, C.; Borreca, A.; Farioli-Vecchioli, S.; Ciotti, M.T.; Ingegnere, T.; Onori, A.; Ammassari-Teule, M.; Corbi, N.; Canu, N.; et al. SMN affects membrane remodelling and anchoring of the protein synthesis machinery. J Cell Sci 2016, 129, 804–816. [Google Scholar] [CrossRef]
- Monani, U.R.; Coovert, D.D.; Burghes, A.H. Animal models of spinal muscular atrophy. Hum Mol Genet 2000, 9, 2451–2457. [Google Scholar] [CrossRef] [PubMed]
- Lorson, C.L.; Hahnen, E.; Androphy, E.J.; Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A 1999, 96, 6307–6311. [Google Scholar] [CrossRef]
- Burnett, B.G.; Muñoz, E.; Tandon, A.; Kwon, D.Y.; Sumner, C.J.; Fischbeck, K.H. Regulation of SMN protein stability. Mol Cell Biol 2009, 29, 1107–1115. [Google Scholar] [CrossRef]
- Calucho, M.; Bernal, S.; Alías, L.; March, F.; Venceslá, A.; Rodríguez-Álvarez, F.J.; Aller, E.; Fernández, R.M.; Borrego, S.; Millán, J.M.; et al. Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord 2018, 28, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Butchbach, M.E. Copy Number Variations in the Survival Motor Neuron Genes: Implications for Spinal Muscular Atrophy and Other Neurodegenerative Diseases. Front Mol Biosci 2016, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Prior, T.W.; Krainer, A.R.; Hua, Y.; Swoboda, K.J.; Snyder, P.C.; Bridgeman, S.J.; Burghes, A.H.; Kissel, J.T. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet 2009, 85, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Darras, B.T. Spinal muscular atrophies. Pediatr Clin North Am 2015, 62, 743–766. [Google Scholar] [CrossRef] [PubMed]
- D'Amico, A.; Mercuri, E.; Tiziano, F.D.; Bertini, E. Spinal muscular atrophy. Orphanet J Rare Dis 2011, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Kolb, S.J.; Kissel, J.T. Spinal Muscular Atrophy. Neurol Clin 2015, 33, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Dubowitz, V. Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype. Eur J Paediatr Neurol 1999, 3, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Grotto, S.; Cuisset, J.M.; Marret, S.; Drunat, S.; Faure, P.; Audebert-Bellanger, S.; Desguerre, I.; Flurin, V.; Grebille, A.G.; Guerrot, A.M.; et al. Type 0 Spinal Muscular Atrophy: Further Delineation of Prenatal and Postnatal Features in 16 Patients. J Neuromuscul Dis 2016, 3, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Stein, H.; Hausen, P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science 1969, 166, 393–395. [Google Scholar] [CrossRef]
- Wu, H.; Lima, W.F.; Zhang, H.; Fan, A.; Sun, H.; Crooke, S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J Biol Chem 2004, 279, 17181–17189. [Google Scholar] [CrossRef]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A 1978, 75, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Rigo, F.; Hua, Y.; Chun, S.J.; Prakash, T.P.; Krainer, A.R.; Bennett, C.F. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat Chem Biol 2012, 8, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Nichols, J.G.; Hsu, C.W.; Vickers, T.A.; Crooke, S.T. mRNA levels can be reduced by antisense oligonucleotides via no-go decay pathway. Nucleic Acids Res 2019, 47, 6900–6916. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Wang, J.; Lynch, D.R.; Shen, X.; Corey, D.R.; Parekh, D.; Bhat, B.; Woo, C.; Cherry, J.J.; et al. Targeting 3' and 5' untranslated regions with antisense oligonucleotides to stabilize frataxin mRNA and increase protein expression. Nucleic Acids Res 2021, 49, 11560–11574. [Google Scholar] [CrossRef]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 2012, 11, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M. Antisense Oligonucleotide-Mediated Exon-skipping Therapies: Precision Medicine Spreading from Duchenne Muscular Dystrophy. Jma j 2021, 4, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, O.; Thieme, A.; Claeys, K.G.; Wenninger, S.; Kley, R.A.; Kuhn, M.; Lukacs, Z.; Deschauer, M.; Gaeta, M.; Toscano, A.; et al. Homozygosity for the common GAA gene splice site mutation c.-32-13T>G in Pompe disease is associated with the classical adult phenotypical spectrum. Neuromuscul Disord 2015, 25, 719–724. [Google Scholar] [CrossRef]
- van der Wal, E.; Bergsma, A.J.; Pijnenburg, J.M.; van der Ploeg, A.T.; Pijnappel, W. Antisense Oligonucleotides Promote Exon Inclusion and Correct the Common c.-32-13T>G GAA Splicing Variant in Pompe Disease. Mol Ther Nucleic Acids 2017, 7, 90–100. [Google Scholar] [CrossRef]
- Lauffer, M.C.; van Roon-Mom, W.; Aartsma-Rus, A. Possibilities and limitations of antisense oligonucleotide therapies for the treatment of monogenic disorders. Commun Med (Lond) 2024, 4, 6. [Google Scholar] [CrossRef]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res 2016, 44, 6518–6548. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Wang, S.; Vickers, T.A.; Shen, W.; Liang, X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol 2017, 35, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev 2000, 10, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther 2014, 24, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Bennett, C.F. Progress in antisense oligonucleotide therapeutics. Annu Rev Pharmacol Toxicol 1996, 36, 107–129. [Google Scholar] [CrossRef] [PubMed]
- Prakash, T.P. An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem Biodivers 2011, 8, 1616–1641. [Google Scholar] [CrossRef] [PubMed]
- Owczarzy, R.; You, Y.; Groth, C.L.; Tataurov, A.V. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry 2011, 50, 9352–9367. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J Clin Med 2020, 9. [Google Scholar] [CrossRef]
- Hebb, M.O.; Robertson, H.A. End-capped antisense oligodeoxynucleotides effectively inhibit gene expression in vivo and offer a low-toxicity alternative to fully modified phosphorothioate oligodeoxynucleotides. Brain Res Mol Brain Res 1997, 47, 223–228. [Google Scholar] [CrossRef]
- Moulton, J.D.; Jiang, S. Gene knockdowns in adult animals: PPMOs and vivo-morpholinos. Molecules 2009, 14, 1304–1323. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Vickers, T.A.; Okunola, H.L.; Bennett, C.F.; Krainer, A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 2008, 82, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Vickers, T.A.; Baker, B.F.; Bennett, C.F.; Krainer, A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 2007, 5, e73. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther 2017, 24, 520–526. [Google Scholar] [CrossRef]
- Sintusek, P.; Catapano, F.; Angkathunkayul, N.; Marrosu, E.; Parson, S.H.; Morgan, J.E.; Muntoni, F.; Zhou, H. Histopathological Defects in Intestine in Severe Spinal Muscular Atrophy Mice Are Improved by Systemic Antisense Oligonucleotide Treatment. PLoS One 2016, 11, e0155032. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Hung, G.; Rigo, F.; Passini, M.A.; Bennett, C.F.; Krainer, A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 2010, 24, 1634–1644. [Google Scholar] [CrossRef]
- Passini, M.A.; Bu, J.; Richards, A.M.; Kinnecom, C.; Sardi, S.P.; Stanek, L.M.; Hua, Y.; Rigo, F.; Matson, J.; Hung, G.; et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 2011, 3, 72ra18. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Rigo, F.; Hung, G.; Horev, G.; Bennett, C.F.; Krainer, A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 2011, 478, 123–126. [Google Scholar] [CrossRef]
- Porensky, P.N.; Mitrpant, C.; McGovern, V.L.; Bevan, A.K.; Foust, K.D.; Kaspar, B.K.; Wilton, S.D.; Burghes, A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet 2012, 21, 1625–1638. [Google Scholar] [CrossRef]
- Zhou, H.; Janghra, N.; Mitrpant, C.; Dickinson, R.L.; Anthony, K.; Price, L.; Eperon, I.C.; Wilton, S.D.; Morgan, J.; Muntoni, F. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum Gene Ther 2013, 24, 331–342. [Google Scholar] [CrossRef]
- Chiriboga, C.A.; Swoboda, K.J.; Darras, B.T.; Iannaccone, S.T.; Montes, J.; De Vivo, D.C.; Norris, D.A.; Bennett, C.F.; Bishop, K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology 2016, 86, 890–897. [Google Scholar] [CrossRef]
- Finkel, R.S.; Chiriboga, C.A.; Vajsar, J.; Day, J.W.; Montes, J.; De Vivo, D.C.; Yamashita, M.; Rigo, F.; Hung, G.; Schneider, E.; et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 2016, 388, 3017–3026. [Google Scholar] [CrossRef] [PubMed]
- Luu, K.T.; Norris, D.A.; Gunawan, R.; Henry, S.; Geary, R.; Wang, Y. Population Pharmacokinetics of Nusinersen in the Cerebral Spinal Fluid and Plasma of Pediatric Patients With Spinal Muscular Atrophy Following Intrathecal Administrations. J Clin Pharmacol 2017, 57, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Nusinersen: First Global Approval. Drugs 2017, 77, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Goyal, N.; Narayanaswami, P. Making sense of antisense oligonucleotides: A narrative review. Muscle Nerve 2018, 57, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wahane, A.; Alhamadani, F.; Zhang, K.; Parikh, R.; Lee, S.; McCabe, E.M.; Rasmussen, T.P.; Bahal, R.; Zhong, X.B.; et al. Nephrotoxicity of marketed antisense oligonucleotide drugs. Curr Opin Toxicol 2022, 32. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Carver, K.; Fisher, M.; Noel, R.; Cintrat, J.C.; Gillet, D.; Barbier, J.; Cao, C.; Bauman, J.; Juliano, R.L. The small molecule Retro-1 enhances the pharmacological actions of antisense and splice switching oligonucleotides. Nucleic Acids Res 2013, 41, 3673–3687. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, C.; Wood, M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol 2018, 14, 9–21. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Jimenez-Mallebrera, C.; van Roon, W.; Sewing, S.; Krieg, A.M.; Arechavala-Gomeza, V.; Andersson, P. Considerations in the Preclinical Assessment of the Safety of Antisense Oligonucleotides. Nucleic Acid Ther 2023, 33, 1–16. [Google Scholar] [CrossRef]
- Fox, D.; To, T.M.; Seetasith, A.; Patel, A.M.; Iannaccone, S.T. Adherence and Persistence to Nusinersen for Spinal Muscular Atrophy: A US Claims-Based Analysis. Adv Ther 2023, 40, 903–919. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Ishino, M.; Loewenstein, P.M. Mutational analysis of HIV-1 Tat minimal domain peptides: identification of trans-dominant mutants that suppress HIV-LTR-driven gene expression. Cell 1989, 58, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Lodigiani, G.; Colombarolli, S.G.; Bergamaschi, G.; Vitali, A. Cell Penetrating Peptides: Classification, Mechanisms, Methods of Study, and Applications. ChemMedChem 2023, 18, e202300236. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, M.; Rosenthal-Aizman, K.; Saar, K.; Eiríksdóttir, E.; Jiang, Y.; Sassian, M.; Ostlund, P.; Hällbrink, M.; Langel, U. Overcoming methotrexate resistance in breast cancer tumour cells by the use of a new cell-penetrating peptide. Biochem Pharmacol 2006, 71, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Depollier, J.; Mery, J.; Heitz, F.; Divita, G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol 2001, 19, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, J.; Fu, L.; Ran, R.; Liu, Y.; Yuan, M.; He, Q. A pH-responsive α-helical cell penetrating peptide-mediated liposomal delivery system. Biomaterials 2013, 34, 7980–7993. [Google Scholar] [CrossRef] [PubMed]
- Margus, H.; Padari, K.; Pooga, M. Cell-penetrating peptides as versatile vehicles for oligonucleotide delivery. Mol Ther 2012, 20, 525–533. [Google Scholar] [CrossRef]
- Chavda, V.P.; Solanki, H.K.; Davidson, M.; Apostolopoulos, V.; Bojarska, J. Peptide-Drug Conjugates: A New Hope for Cancer Management. Molecules 2022, 27. [Google Scholar] [CrossRef]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem Biol Drug Des 2013, 81, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front Pharmacol 2020, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, W.B.; Fuselier, T.; He, J.; Wimley, W.C. Mechanism Matters: A Taxonomy of Cell Penetrating Peptides. Trends Biochem Sci 2015, 40, 749–764. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.; Gautam, A. CPPsite 2.0: a repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res 2016, 44, D1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, A.; Tornesello, A.L.; Tornesello, M.L.; Buonaguro, F.M. Cell Penetrating Peptides as Molecular Carriers for Anti-Cancer Agents. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Verdurmen, W.P.; Brock, R. Biological responses towards cationic peptides and drug carriers. Trends Pharmacol Sci 2011, 32, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Tammam, S.N.; Azzazy, H.M.E.; Lamprecht, A. The effect of nanoparticle size and NLS density on nuclear targeting in cancer and normal cells; impaired nuclear import and aberrant nanoparticle intracellular trafficking in glioma. J Control Release 2017, 253, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Rhee, M.; Davis, P. Mechanism of uptake of C105Y, a novel cell-penetrating peptide. J Biol Chem 2006, 281, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Mao, S.; Ditzel, H.J.; Farnaes, L.; Wirsching, P.; Lerner, R.A.; Janda, K.D. A cell-penetrating peptide from a novel pVII-pIX phage-displayed random peptide library. Bioorg Med Chem 2002, 10, 4057–4065. [Google Scholar] [CrossRef]
- Gao, S.; Simon, M.J.; Hue, C.D.; Morrison, B., 3rd; Banta, S. An unusual cell penetrating peptide identified using a plasmid display-based functional selection platform. ACS Chem Biol 2011, 6, 484–491. [Google Scholar] [CrossRef]
- Guo, Z.; Peng, H.; Kang, J.; Sun, D. Cell-penetrating peptides: Possible transduction mechanisms and therapeutic applications. Biomed Rep 2016, 4, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.H.; Park, I.S.; Hahm, K.S.; Shin, S.Y. Antimicrobial activity, bactericidal mechanism and LPS-neutralizing activity of the cell-penetrating peptide pVEC and its analogs. J Pept Sci 2011, 17, 812–817. [Google Scholar] [CrossRef]
- Johansson, H.J.; El-Andaloussi, S.; Holm, T.; Mäe, M.; Jänes, J.; Maimets, T.; Langel, U. Characterization of a novel cytotoxic cell-penetrating peptide derived from p14ARF protein. Mol Ther 2008, 16, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Magzoub, M.; Sandgren, S.; Lundberg, P.; Oglecka, K.; Lilja, J.; Wittrup, A.; Göran Eriksson, L.E.; Langel, U.; Belting, M.; Gräslund, A. N-terminal peptides from unprocessed prion proteins enter cells by macropinocytosis. Biochem Biophys Res Commun 2006, 348, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Milletti, F. Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov Today 2012, 17, 850–860. [Google Scholar] [CrossRef]
- Oehlke, J.; Krause, E.; Wiesner, B.; Beyermann, M.; Bienert, M. Extensive cellular uptake into endothelial cells of an amphipathic beta-sheet forming peptide. FEBS Lett 1997, 415, 196–199. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J Control Release 2014, 174, 126–136. [Google Scholar] [CrossRef]
- Ruseska, I.; Zimmer, A. Internalization mechanisms of cell-penetrating peptides. Beilstein J Nanotechnol 2020, 11, 101–123. [Google Scholar] [CrossRef]
- Richard, J.P.; Melikov, K.; Brooks, H.; Prevot, P.; Lebleu, B.; Chernomordik, L.V. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem 2005, 280, 15300–15306. [Google Scholar] [CrossRef]
- Zhao, X.L.; Chen, B.C.; Han, J.C.; Wei, L.; Pan, X.B. Delivery of cell-penetrating peptide-peptide nucleic acid conjugates by assembly on an oligonucleotide scaffold. Sci Rep 2015, 5, 17640. [Google Scholar] [CrossRef]
- McClorey, G.; Banerjee, S. Cell-Penetrating Peptides to Enhance Delivery of Oligonucleotide-Based Therapeutics. Biomedicines 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Yesylevskyy, S.; Marrink, S.J.; Mark, A.E. Alternative mechanisms for the interaction of the cell-penetrating peptides penetratin and the TAT peptide with lipid bilayers. Biophys J 2009, 97, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Sharmin, S.; Moniruzzaman, M.; Yamazaki, M. Elementary processes for the entry of cell-penetrating peptides into lipid bilayer vesicles and bacterial cells. Appl Microbiol Biotechnol 2018, 102, 3879–3892. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, S.; Morris, M.C.; Divita, G.; Heitz, F. Interactions of amphipathic CPPs with model membranes. Biochim Biophys Acta 2006, 1758, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.C.; Zhang, J.; Min, K.A.; Lee, K.; Byun, Y.; David, A.E.; He, H.; Yang, V.C. Cell-penetrating peptides: achievements and challenges in application for cancer treatment. J Biomed Mater Res A 2014, 102, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, G.D.; Arzumanov, A.; Abes, R.; Yin, H.; Wood, M.J.; Lebleu, B.; Gait, M.J. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res 2008, 36, 6418–6428. [Google Scholar] [CrossRef]
- Leger, A.J.; Mosquea, L.M.; Clayton, N.P.; Wu, I.H.; Weeden, T.; Nelson, C.A.; Phillips, L.; Roberts, E.; Piepenhagen, P.A.; Cheng, S.H.; et al. Systemic delivery of a Peptide-linked morpholino oligonucleotide neutralizes mutant RNA toxicity in a mouse model of myotonic dystrophy. Nucleic Acid Ther 2013, 23, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Tsoumpra, M.K.; Fukumoto, S.; Matsumoto, T.; Takeda, S.; Wood, M.J.A.; Aoki, Y. Peptide-conjugate antisense based splice-correction for Duchenne muscular dystrophy and other neuromuscular diseases. EBioMedicine 2019, 45, 630–645. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Krieg, A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther 2017, 27, 1–3. [Google Scholar] [CrossRef]
- Lim, K.R.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther 2017, 11, 533–545. [Google Scholar] [CrossRef]
- Yin, H.; Saleh, A.F.; Betts, C.; Camelliti, P.; Seow, Y.; Ashraf, S.; Arzumanov, A.; Hammond, S.; Merritt, T.; Gait, M.J.; et al. Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice. Mol Ther 2011, 19, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Betts, C.; Saleh, A.F.; Arzumanov, A.A.; Hammond, S.M.; Godfrey, C.; Coursindel, T.; Gait, M.J.; Wood, M.J. Pip6-PMO, A New Generation of Peptide-oligonucleotide Conjugates With Improved Cardiac Exon Skipping Activity for DMD Treatment. Mol Ther Nucleic Acids 2012, 1, e38. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.R.Q.; Woo, S.; Melo, D.; Huang, Y.; Dzierlega, K.; Shah, M.N.A.; Aslesh, T.; Roshmi, R.R.; Echigoya, Y.; Maruyama, R.; et al. Development of DG9 peptide-conjugated single- and multi-exon skipping therapies for the treatment of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2022, 119. [Google Scholar] [CrossRef] [PubMed]
- Sazani, P.; Kang, S.H.; Maier, M.A.; Wei, C.; Dillman, J.; Summerton, J.; Manoharan, M.; Kole, R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res 2001, 29, 3965–3974. [Google Scholar] [CrossRef]
- Hsieh-Li, H.M.; Chang, J.G.; Jong, Y.J.; Wu, M.H.; Wang, N.M.; Tsai, C.H.; Li, H. A mouse model for spinal muscular atrophy. Nat Genet 2000, 24, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Mitrpant, C.; Porensky, P.; Zhou, H.; Price, L.; Muntoni, F.; Fletcher, S.; Wilton, S.D.; Burghes, A.H. Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLoS One 2013, 8, e62114. [Google Scholar] [CrossRef] [PubMed]
- Villaseñor, R.; Ozmen, L.; Messaddeq, N.; Grüninger, F.; Loetscher, H.; Keller, A.; Betsholtz, C.; Freskgård, P.O.; Collin, L. Trafficking of Endogenous Immunoglobulins by Endothelial Cells at the Blood-Brain Barrier. Sci Rep 2016, 6, 25658. [Google Scholar] [CrossRef]
- Nizzardo, M.; Simone, C.; Salani, S.; Ruepp, M.D.; Rizzo, F.; Ruggieri, M.; Zanetta, C.; Brajkovic, S.; Moulton, H.M.; Müehlemann, O.; et al. Effect of combined systemic and local morpholino treatment on the spinal muscular atrophy Δ7 mouse model phenotype. Clin Ther 2014, 36, 340–356. [Google Scholar] [CrossRef]
| Cell-Penetrating Peptide (CPP) | CPP Sequence | Route of Administration | Reference |
|---|---|---|---|
| Pip6a | RXRRBRRXRYQFLIRXRBRXRB | IV Injection | [5] |
| Br-ApoE (K->A) | LRALRARLLR-G*GKX-Bpg-G (LRALRARLLR-G*G) |
IV Injection | [6] |
| HA2-ApoE (133-150) | GLFHAIAHFIHGGWH-X-LRVRLASHLRKLRKRLLR | IV Injection | [7] |
| RXR | [RXRRBR]2XB | IP and ICV Injection | [8] |
| r6 | RRRRRR | IP and ICV Injection | [8] |
| DG9 | YArVRRrGPRGYArVRRrGPRr | SQ Injection | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




