1. Introduction
Healthcare Associated Infections (HAIs) are a concern for patients treated in medical facilities, including veterinary physical rehabilitation facilities. Pathogens can be especially concerning for post-operative patients or those that are hospitalized for prolonged time periods. These concerns are important in facilities for human and animal patients with potential transmission from equipment or other external surfaces to the patients as possible causes for HAIs [
1,
2]. In human medicine, certain pathogens tend to be of most concern for serious medical problems, including:
Clostridium difficile (Cdiff),
Staphylococcus aureus (SA, especially methicillin resistant variants [e.g., MRSA]),
Staphylococcus pseudintermedius (SIM)
, and multidrug resistant Gram-negative rods (e.g. Enterics that may include
Enterobacter sp. and
Klebsiella sp.). While not necessarily of equal concern in human and veterinary clinics, the presence of these bacterial species on surfaces in any clinic represents contamination that could lead to HAIs [
3].
In veterinary clinics bacterial contamination of surfaces can be problematic to animals being treated, but also to the human care givers. For example, Cdiff is a pathogen that can cause severe disease for humans. Infection by Cdiff can cause severe diarrhea and pseudomembranous colitis [
4,
5]. Although Cdiff has been isolated from dogs, a link between human infection with Cdiff and companion animals is not well documented and should be further explored [
6,
7,
8].
Staphylococci, including SA and SIM are opportunistic pathogens in both humans and animals [
9,
10,
11,
12]. Typically SA infections have been successfully treated with topical or systemic courses of antibiotics [
10]. However, antibiotic resistant Staphylococcal strains, including MRSA represent an increasing problem throughout all of healthcare. These pathogens may also be isolated from companion animal veterinary patients, in addition to other pathogens that affect animals, and can also be transmitted from humans to animals.
Pathogens with known transmission between humans and animals include: SA, MRSA, and
S. pseudintermedius. Orden et al. [
6] isolated Cdiff from 12% of the dogs they studied. In a similar study, Álvarez-Perez et al. [
3] found Cdiff infections in 5% of dogs they sampled. Cdiff can cause enteritis, diarrhea, and hemorrhagic diarrhea in both humans and dogs [
6,
13]. Since Cdiff can be cultured from dogs, a major concern is the possible transfer of this pathogen to human care givers. Since Cdiff spores may be found on environmental surfaces of both human and animal clinics, knowledge of this contamination could be important to prevent HAIs of both humans and animals.
Staphylococcal infections in canine patients are often the result of surgical procedures, that may cause moderate to severe morbidity. With the continued impact of MRSA in human medicine and the close contact between humans and household pets, there has been an increase in MRSA in household pets [
14]. Most infections associated with MRSA in veterinary patients are community acquired often transmitted from pet owners to their pets [
15]. The prevalence of MRSA infections in veterinary patients ranges from 0-9% and may result in different symptoms related to skin and soft tissue infections, especially surgical site wounds, otitis, and pyoderma [
10,
15,
16].
Staphylococcus pseudintermedius is a more common form of Staphylococcus isolated from dogs and is responsible for infections such as pyoderma and otitis [
17,
18]. Beginning in 2005
S. pseudintermedius became a new species within the
S. intermedius group of Staphylococci [
19]. It was formerly believed that dogs were colonized by
S. intermedius, but in fact the most common Staphylococcal opportunistic pathogen associated with dogs is
S. pseudintermedius [
20,
21,
22]. Similar to other Staphylococcal isolates,
S. pseudintermedius causes urinary tract infections, otitis, wound infection, soft tissue infections, and surgical site infections, and is the leading cause of pyoderma in dogs [
23,
24]. Similar to strains of SA,
S. pseudintermedius has developed resistance to methicillin resulting in more complex and costly treatment options for veterinary patients [
1,
16,
25]. It is known that the spread of
S. pseudintermedius usually involves contact between two hosts. With increased exposure between humans and veterinary patients, the risk of transmission of pathogens will likely rise, especially with MRSA, antibiotic resistant
S. pseudintermedius, and
C. difficile [
7,
15,
26]. Zoonotic pathogens are a huge public health concern and interspecies transfer of these pathogens between animals and humans could enhance horizontal exchange of resistance factors between these pathogens.
With the close contact between pets and their owners, other animal patients, and veterinary medical personnel in veterinary rehabilitation facilities, it is important to know if surfaces within the facility are contaminated by potential pathogens. Knowledge of problematic surface spots in these clinics should encourage managers of these clinics to proactively clean and disinfect sites known to be contaminated by these pathogens. The aim of this study was to determine the prevalence of contamination by potential pathogens of both humans and animals from environmental surfaces and equipment commonly found in veterinary physical rehabilitation clinics. Overall, we found bacterial contamination by potential pathogens to be commonplace throughout the clinics sampled. Future studies should seek any links between this background contamination and the incidence of HAIs in clinic patients.
2. Materials and Methods
Healthcare Associated Infections (HAIs) are a concern for patients treated in medical facilities, including veterinary physical rehabilitation facilities. Pathogens can be especially concerning for post-operative patients or those that are hospitalized for prolonged time periods. These concerns are important in facilities for human and animal patients with potential transmission from equipment or other external surfaces to the patients as possible causes for HAIs [
1,
2]. In human medicine, certain pathogens tend to be of most concern for serious medical problems, including:
Clostridium difficile (Cdiff),
Staphylococcus aureus (SA, especially methicillin resistant variants [e.g., MRSA]),
Staphylococcus pseudintermedius (SIM)
, and multidrug resistant Gram-negative rods (e.g. Enterics that may include
Enterobacter sp. and
Klebsiella sp.). While not necessarily of equal concern in human and veterinary clinics, the presence of these bacterial species on surfaces in any clinic represents contamination that could lead to HAIs [
3].
In veterinary clinics bacterial contamination of surfaces can be problematic to animals being treated, but also to the human care givers. For example, Cdiff is a pathogen that can cause severe disease for humans. Infection by Cdiff can cause severe diarrhea and pseudomembranous colitis [
4,
5]. Although Cdiff has been isolated from dogs, a link between human infection with Cdiff and companion animals is not well documented and should be further explored [
6,
7,
8].
Staphylococci, including SA and SIM are opportunistic pathogens in both humans and animals [
9,
10,
11,
12]. Typically SA infections have been successfully treated with topical or systemic courses of antibiotics [
10]. However, antibiotic resistant Staphylococcal strains, including MRSA represent an increasing problem throughout all of healthcare. These pathogens may also be isolated from companion animal veterinary patients, in addition to other pathogens that affect animals, and can also be transmitted from humans to animals.
Pathogens with known transmission between humans and animals include: SA, MRSA, and
S. pseudintermedius. Orden et al. [
6] isolated Cdiff from 12% of the dogs they studied. In a similar study, Álvarez-Perez et al. [
3] observed that 5% of dogs were infected with Cdiff. This Clostridial species can cause enteritis, diarrhea, and hemorrhagic diarrhea in both humans and dogs [
6,
13]. Since Cdiff can be cultured from dogs, a major concern is the possible transfer of this pathogen to human care givers. Since Cdiff spores may be found on environmental surfaces of both human and animal clinics, knowledge of this contamination could be important to prevent HAIs of both humans and animals.
Staphylococcal infections in canine patients are often the result of surgical procedures, that may cause moderate to severe morbidity. With the continued impact of MRSA in human medicine and the close contact between humans and household pets, there has been an increase in MRSA in household pets [
14]. Most infections associated with MRSA in veterinary patients are community acquired often transmitted from pet owners to their pets [
15]. The prevalence of MRSA infections in veterinary patients ranges from 0-9% and may result in different symptoms related to skin and soft tissue infections, especially surgical site wounds, otitis, and pyoderma [
10,
15,
16].
Staphylococcus pseudintermedius is a more common form of Staphylococcus isolated from dogs and is responsible for infections such as pyoderma and otitis [
17,
18]. Beginning in 2005
S. pseudintermedius became a new species within the
S. intermedius group of Staphylococci [
19]. It was formerly believed that dogs were colonized by
S. intermedius, but in fact the most common Staphylococcal opportunistic pathogen associated with dogs is
S. pseudintermedius [
20,
21,
22]. Similar to other Staphylococcal isolates,
S. pseudintermedius causes urinary tract infections, otitis, wound infection, soft tissue infections, and surgical site infections, and is the leading cause of pyoderma in dogs [
23,
24]. Similar to strains of SA,
S. pseudintermedius has developed resistance to methicillin resulting in more complex and costly treatment options for veterinary patients [
1,
16,
25]. It is known that the spread of
S. pseudintermedius usually involves contact between two hosts. With increased exposure between humans and veterinary patients, the risk of transmission of pathogens will likely rise, especially with MRSA, antibiotic resistant
S. pseudintermedius, and
C. difficile [
7,
15,
26]. Zoonotic pathogens are a huge public health concern and interspecies transfer of these pathogens between animals and humans could enhance horizontal exchange of resistance factors between these pathogens.
With the close contact between pets and their owners, other animal patients, and veterinary medical personnel in veterinary rehabilitation facilities, it is important to know if surfaces within the facility are contaminated by potential pathogens. Knowledge of problematic surface spots in these clinics should encourage managers of these clinics to proactively clean and disinfect sites known to be contaminated by these pathogens. The aim of this study was to determine the prevalence of contamination by potential pathogens of both humans and animals from environmental surfaces and equipment commonly found in veterinary physical rehabilitation clinics.
3. Results
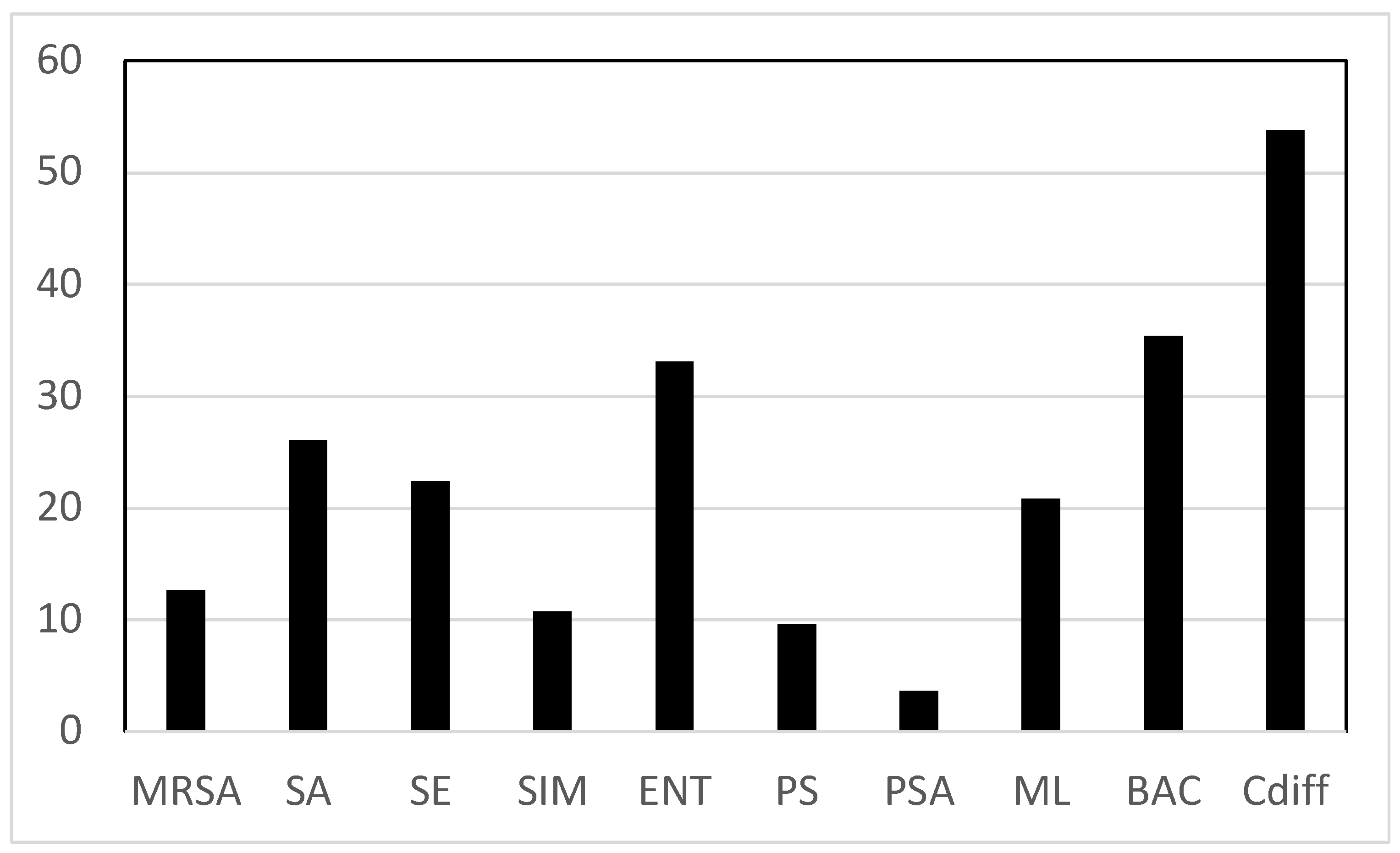
The most common contaminating species based on an average of percent positive swabs for all sites was Cdiff, with BAC and ENT the next two most common contaminating bacteria observed. Contamination by Cdiff was found at 58.3% of all sites sampled, while BAC and ENT were found on 35.4% and 33.1% of sites (
Figure 1). At the lower end of the contamination range we found PSA contaminating only 3.6% of sites.
When looking at clinic contamination by site and species, Cdiff contaminated 94.7% of the floors and 83.3% of the HVAC return air ducts (
Table 1). Enteric bacteria and Bacillus sp. were the next most encountered contaminants, found on 100% of swabs from the return air ducts. The floors and return air ducts were consistently contaminated by other species, including Cdiff, SA, MRSA, and SIM. The highest levels of contamination by SA was found on the HVAC return air duct (83.3%) and the scales (83.3%).
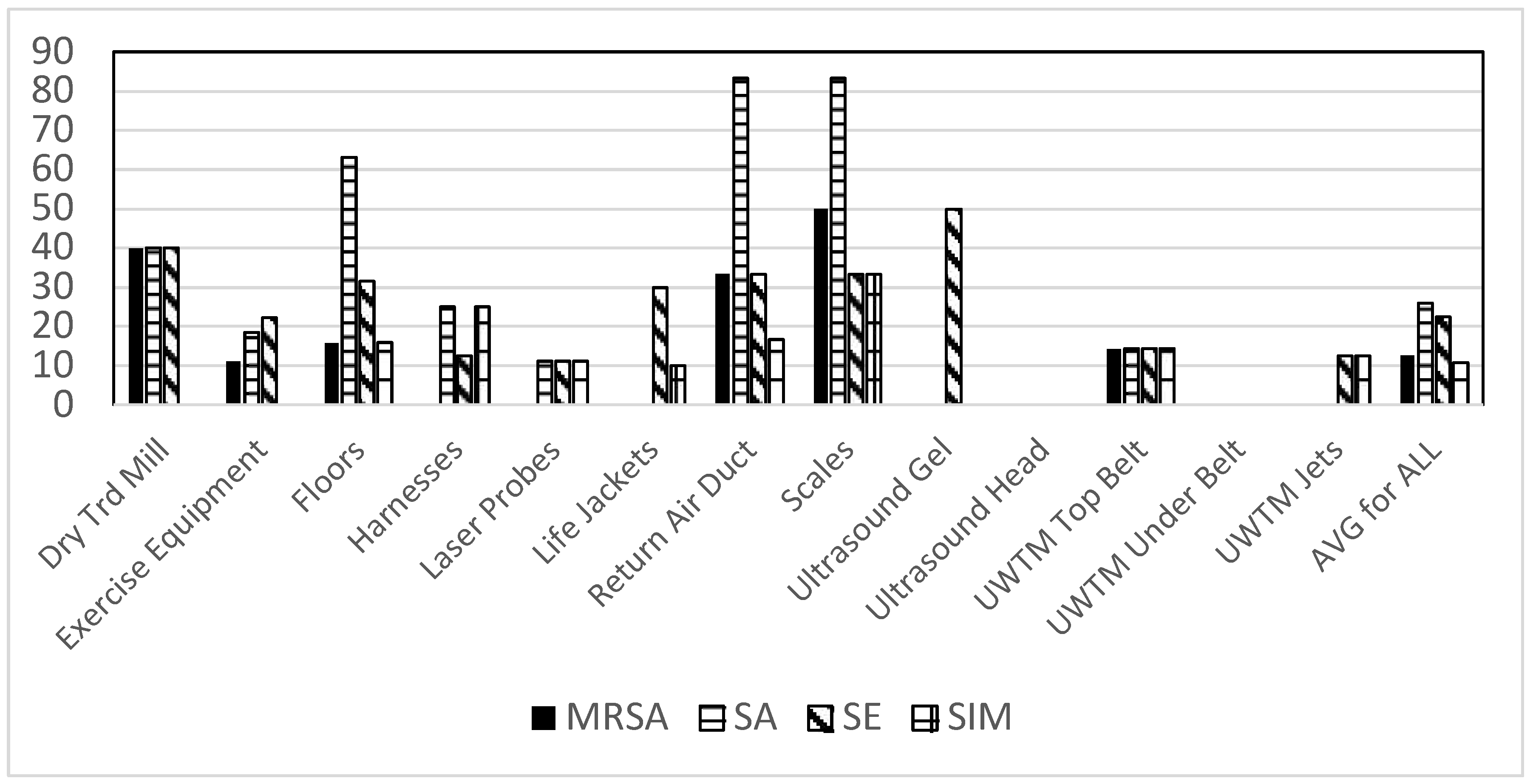
When looking at bacterial contamination of different clinic sites a large range of contamination was observed. Staphylococci were found most prominently on the floors, with SA found on the largest number of swabs (
Figure 2). The only sites where Staphylococci were not found were the ultrasound gel bottles and heads, and the bottom surface of the belt on underwater treadmills.
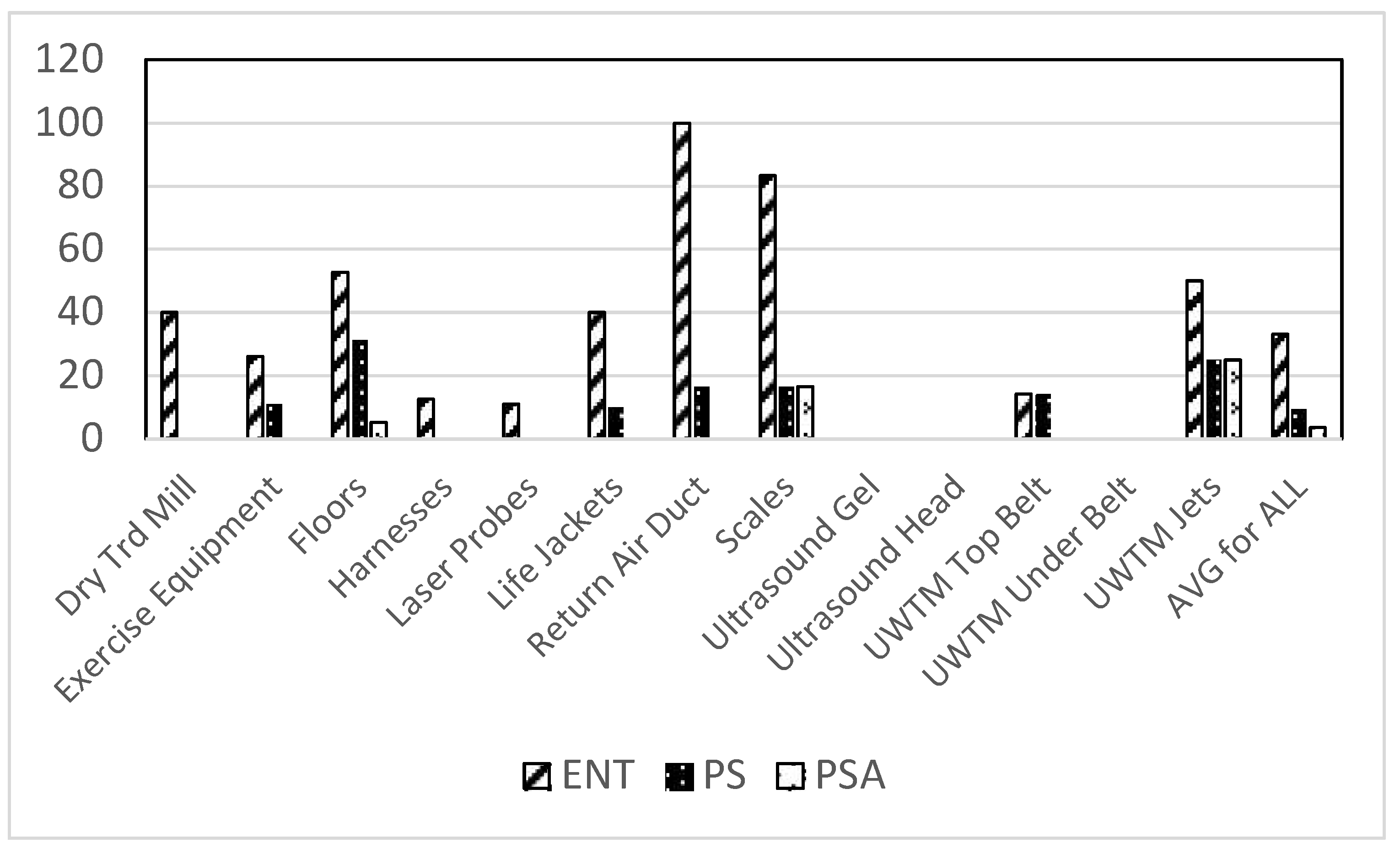
Bacterial contamination due to select Gram-negative rods was also found throughout the veterinary clinics. Enteric contamination (lactose positive cells, e.g.,
Escherichia coli) was found on the greatest number of sampled sites of the Gram-negative bacteria studied (
Figure 3). Again, the floors were the most contaminated sites in these clinics.
Pseudomonas aeruginosa was also found in the clinics, but in relatively low numbers. Other species of
Pseudomonas sp. were generally widespread in the clinics at slightly higher numbers.
Bacterial contamination by other select Gram positive species was also found throughout the clinics. Most notable, Cdiff was found in very high numbers of sites throughout the clinics (
Figure 4). Another spore-forming genera of Gram-positive bacteria,
Bacillus sp. was also found contaminating many of the same sites as Cdiff.
Micrococcus luteus, a Gram-positive coccus often associated with human skin was also found on many sites throughout the clinic.
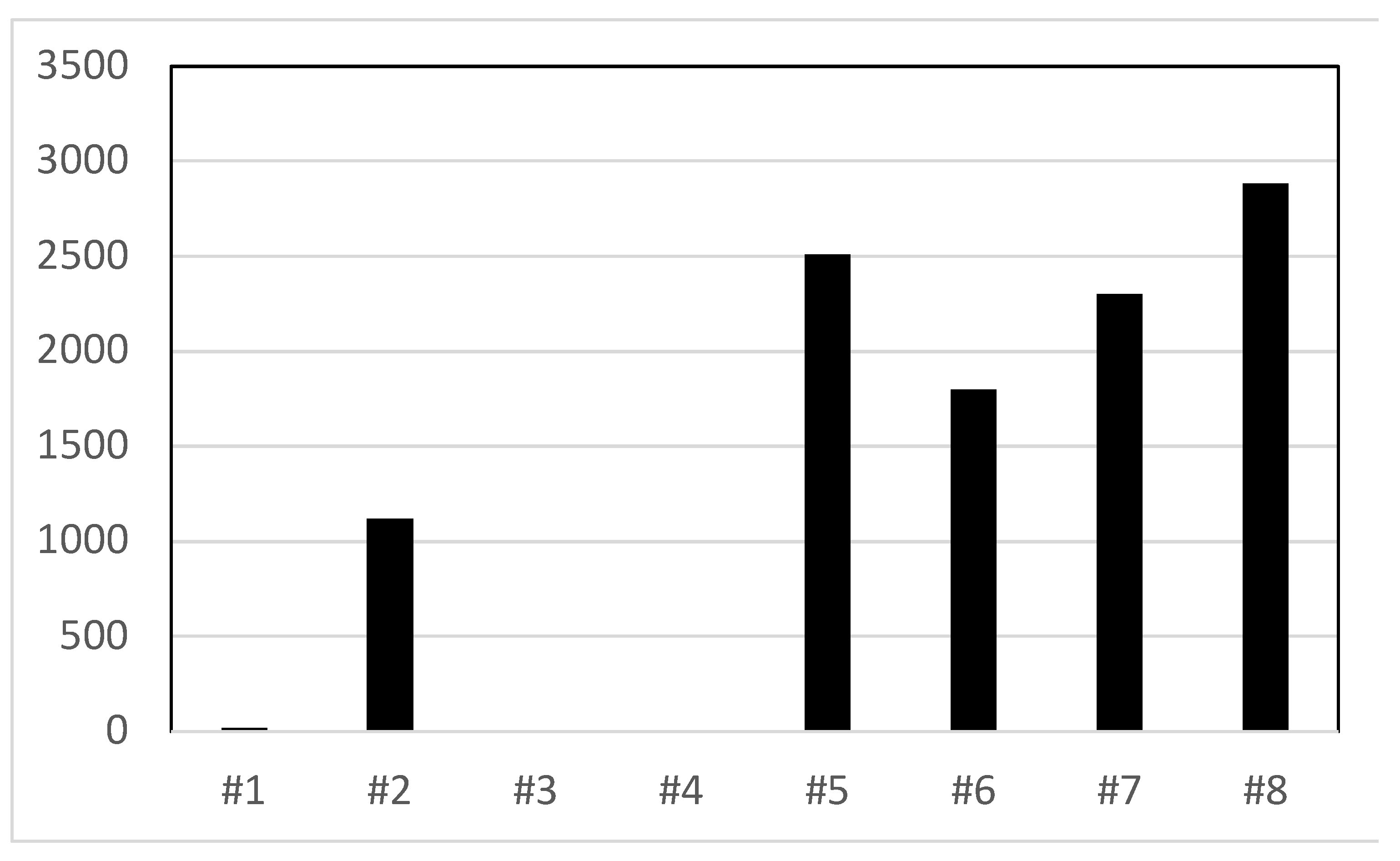
When water from underwater treatmill tanks was streaked onto TSA and EMB agar plates colony counts as high as 2800 cfu/ml were detected (
Figure 5). The largest number of bacterial colonies observed were for enteric bacteria.
4. Discussion
This study assessed patterns of bacterial contamination medical equipment and environmental sites in veterinary physical rehabilitation clinics. Bacterial species that were isolated and grown included both Gram-negative and Gram-positive bacteria. Of the Gram-positive cocci, several species of Staphylococci were observed, including SA, MRSA, SE, and SIM. Of the Gram-negative bacteria detected enterics (e.g.,
E. coli) and several species of
Pseudomonas were represented. In general, contamination of the veterinary clinics studied was mostly by Gram-positive bacteria. Staphylococcal (a Gram-positive coccus) contamination was prevalent throughout the sites sampled. Notably,
S. pseudintermedius can be problematic for both dogs and humans, possibly causing disease in both species [
29]. Two of the Gram-positive rods detected, Cdiff and BAC are spore-forming species. This is important since bacterial spores offer the species a higher degree of resistance to many abiotic factors that may be employed to control bacterial contamination. In addition, many Bacillus sp. are often associated with soil, and since dogs are known to dig in soil quite often, veterinary patients may carry these bacteria into the clinics on their skin or feet. The other spore-forming bacteria, Cdiff, is a notable human pathogen, and could pose a threat to care-giving humans in these clinics [
30].
Within the clinics several “hot spot areas” such as HVAC return air ducts, floors, and scales are areas that are common to veterinary patients and represent sites where human caregivers also interact. As the results demonstrate, these areas had 50-94.7% positive swabs for multiple potential pathogenic bacteria. Of the bacteria cultured from HAVC return air ducts and from the floors, spore-forming bacteria (e.g., Cdiff and BAC) were very prevalent. When bacterial species that may be from soil are found on both floors and HAVC return air ducts, this suggests that levels of dust in the facilities might be high. When dust is suspended in air it can help transmit airborne pathogens. Air borne pathogens in human hospitals has been found to contribute to infections in both patients and their caregivers [
31]. Targeted cleaning and disinfection of veterinary clinics to reduce dust may be a good strategy to reduce the potential for contamination of animals and their caregivers.
Water in the underwater treadmill tanks was contaminated by both enteric and non-specific bacteria. If the enterics observed were of fecal origin, dogs using the treadmills may be contaminated by a wide range of fecally-transmitted dog pathogens [
32]. One measure that can be used to reduce waterborne contamination in underwater treadmills is to use shock treatments with either chlorine, bromine or hydrogen peroxide based chemical treatments. Periodic emptying of the tanks and refilling with treated water (saltwater system or low levels of bromine or chlorine) may help reduce the potential for contamination with enteric pathogens.
A recent study performed by Lord et al. [
1] found increased numbers of antimicrobial resistant bacteria in hospitals, particularly Staphylococcal species. In some cases, resistance against last tier antimicrobial therapies (e.g., Fluoroquinolones and Phenicols) was observed in strains of Staphylococci [
1]. Although methicillin resistant
S. pseudintermedius was not a focus of this study, it is highly probable that horizontal exchange of resistant factors between strains of MRSA and this dog pathogen could occur [
33]. Antimicrobial and multidrug resistant pathogens are of a huge public health concern to both veterinary patients and their caregivers and need to be addressed in all clinics.
This current study was based on the use of viable bacterial culture techniques to monitor contamination in the veterinary clinics sampled. Many contemporary studies of the presence of bacteria (and other pathogens) in human health care facilities use some form of molecular approach to extract DNA from samples [
34]. Culture techniques provide useful data with regard to the presence of viable cells on surfaces from the sampled sites. Using molecular data to describe bacterial diversity of a site provides little or no evidence of whether the cells are viable. There is no evidence that DNA from Staphylococci alone causes HAIs, however, viable Staphylococci could cause HAIs. Thus, the data presented here represent living contaminants present on clinical surfaces that could easily cause infections in open wounds or other sites on animal patients. Measures to address contamination in these clinics should be a priority.
Areas of the veterinary clinics studied here that need to have focused cleaning and disinfection include the HVAC return air vents, scales, exercise equipment, and floors. Although ultrasound gel can be obtained in sterile packets, the clinics studied use gel in reusable bottles. A previous study of human physical therapy clinic ultrasound devices found that the tips of coupling gel bottles were often contaminated with MRSA [
35]. Scales are frequently used by all patients entering the clinic and increase the risk of acquiring pathogens that may result in an infection. All these factors increase risk to veterinary patients and their human caregivers and warrant further investigation and care with disinfection.
5. Conclusions
These findings suggest that veterinary clinical environmental surfaces and water are generally contaminated by both Gram-positive and Gram-negative bacterial species. Some of the contaminating species of bacteria are known pathogens of both dogs and humans (e.g., MRSA, SA, PSA), or specifically pathogenic to humans (e.g., Cdiff). This contamination has the potential to contribute to HAIs that may occur in veterinary patients or in their human caregivers. Further research is warranted to investigate the extent of bacterial contamination in veterinary clinics, and any potential links to HAIs occurring in animals being treated in those clinics.
Author Contributions
Conceptualization, H. Spratt and D. Levine; methodology, H. Spratt; software, D. Levine; validation, N. Millis and D. Millis; formal analysis, H. Spratt and D. Levine; investigation, H. Spratt, D. Levine, and J. Brackett.; resources, D. Levine; data curation, H. Spratt and D. Levine.; writing—original draft preparation, H. Spratt, D. Levine, and J. Brackett.; writing—review and editing, N. Millis and D. Millis.; visualization, H. Spratt and D. Levine; supervision, H. Spratt and D. Levine; project administration, H. Spratt; funding acquisition, D. Levine. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Although this study did not involve any human or animal subjects, we were concerned that our close association with clinic environments where animals were present in veterinary rehabilitation clinics might constitute a need to obtain approval from the University of Tennessee at Chattanooga’s Institutional Animal Care and Use Committee (IACUC). We submitted a request for a review of our proposed work to UTC’s IACUC on February 7, 2020. After reviewing our request, we received a letter of exemption from the IACUC on February 11, 2020, where the committee “determined that it [this study] does not meet the threshold of research with animal subjects as defined by the United States Department of Agriculture and the Public Health Service. Therefore, your proposed activity will not require review and monitoring by the UTC IACUC.”
Data Availability Statement
We intend to make data generated in this study available via a cloud-based system provided by the University of Tennessee.
Acknowledgments
This project was funded and conducted by UTC’s Clinical Infectious Disease Control research group.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lord, J.; Millis, N.; Jones, R.; Johnson, B.; Kania, S.; Odoi, A. Patterns of antimicrobial, multidrug and methicillin resistance among Staphylococcus spp. isolated from canine specimens submitted to a diagnostic laboratory in Tennessee, USA: a descriptive study. BMC Vet Res. 2022, 18, 91. [Google Scholar] [CrossRef] [PubMed]
- Spratt, Jr., H.; Levine, D.; McDonald, S.; Drake, S.; Duke, K.; Kluttz, C.; Noonan, K. Survival of Staphylococcus aureus on therapeutic ultrasound heads. Am J Infect Control. 2019, 47, 1157–1159. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Pérez, S.; Blanco, J.; Harmanus, C.; Kuijper, E.; García, M. Data from a survey of Clostridium perfringens and Clostridium difficile shedding by dogs and cats in the Madrid region (Spain), including phenotypic and genetic characteristics of recovered.isolates. Data Brief. 2017, 14, 88–100. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lim, S.; Knight, D.; Riley, T. Clostridium difficile and One Health. Clin Microbiol Infect. 2019, 26, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Alalawi, M.; Aljahdali, S.; Alharbi, B.; Fagih, L.; Fatani, R.; Aljuhani, O. Clostridium difficile infection in an academic medical center in Saudi Arabia: Prevalence and risk factors. Ann Saudi Med. 2020, 40, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Orden, C.; Blanco, J.; Álvarez-Pérez, S.; Garcia, M.; Blanco, J.; Garcia-Sancho, M.; Rodriguez-Franco, F.; Sainz, A.; Villaescusa, A.; Harmanus, C.; Kuijper, E.; Garcia, M. Isolation of Clostridium difficile from dogs with digestive disorders, including stable metronidazole-resistant strains. Anaerobe. 2017, 43, 78–81. [Google Scholar] [CrossRef]
- Rabold, D.; Espelage, W.; Sin, M.; Eckmanns, T.; Schneeberg, A.; Neubauer, H.; Mobius, N.; Hille, K.; Wieler, L.; Seyboldt, C.; Lubke-Becker, A. The zoonotic potential of Clostridium difficile from small companion animals and their owners. PLoS One. 2018, 13. [Google Scholar] [CrossRef]
- Weese, J. Methicllin-Resistant Staphylococcus Aureus in Animals. ILAR J. 2010, 51, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.; Lautenbach, E.; Zaoutis, T.; Leckerman, K.; Edelstein, P.; Rankin, S. Potential for pet animals to harbour methicillin-resistant Staphylococcus aureus when residing with human MRSA patients. Zoonoses Public Health. 2012, 59, 286–293. [Google Scholar] [CrossRef]
- Worthing, K.; Brown, J.; Gerber, L.; Trott, D.; Abraham, S.; Norris, J. Methicillin-resistant staphylococci amongst veterinary personnel, personnel-owned pets, patients and the hospital environment of two small animal veterinary hospitals. Vet Microbiol. 2018, 223, 79–85. [Google Scholar] [CrossRef]
- Tong, S.; Davis, J.; Eichenberger, E.; Holland, T.; Fowler, V. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sanz, E.; Torres, C.; Lozano, C.; Zarazaga, M. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp Immunol Microbiol Infect Dis. 2013, 36, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; de Oliveira Júnior, C.; Blanc, D.; Pereira, S.; de Araujo, M.; Vasconcelos, A.; Lobato, F. Clostridioides difficile infection in dogs with chronic-recurring diarrhea responsive to dietary changes. Anaerobe. 2018, 51, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Mork, R.; Hogan, P.; Muenks, C.; Boyle, M.; Thompson, R.; Sullivan, M.; Morelli, J.; Seigel, J.; Orschein, R.; Wardenburg, J.; Gehlert, S.; Burnham, C-A. ; Rzhetsky, A.; Fritz, S. Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S. aureus skin and soft tissue infection: a prospective cohort study. Lancet Infect Dis. 2020, 20, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Weese, J. Bacterial enteritis in dogs and cats: Diagnosis, therapy, and zoonotic potential. Vet Clin North Am - Small Anim Pract. 2011, 41, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Gingrich, E.; Kurt, T.; Hyatt, D.; Lappin, M.; Ruch-Gallie, R. Prevalence of methicillin-resistant staphylococci in northern Colorado shelter animals. J Vet Diagnostic Investig. 2011, 23, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Nienhoff, U.; Kadlec, K.; Chaberny, I.; Verspohl, J.; Gerlach, G-F. ; Kreienbrock, L.; Schwarz, S.; Simon, D.; Nolte, I. Methicillin-resistant Staphylococcus pseudintermedius among dogs admitted to a small animal hospital. Vet Microbiol. 2011, 150, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Zur, G.; Gurevich, B.; Elad, D. Prior antimicrobial use as a risk factor for resistance in selected Staphylococcus pseudintermedius isolates from the skin and ears of dogs. Vet Dermatol. 2016, 27, 468–e125. [Google Scholar] [CrossRef]
- Somayaji, R.; Rubin, J.; Priyantha, M.; Church, D. Exploring Staphylococcus pseudintermedius: an emerging zoonotic pathogen? Fut. Microbiol. 2016, 11, 1371–1374. [Google Scholar] [CrossRef]
- Bond, R.; Loeffler, A. What’s happened to Staphylococcus intermedius? Taxonomic revision and emergence of multi-drug resistance. J Small Anim Pract. 2012, 53, 147–154. [Google Scholar] [CrossRef]
- Sasaki, T.; Kikuchi, K.; Tanaka, Y.; Takahashi, N.; Kamata, S.; Hiramatsu, K. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J Clin Microbiol. 2007, 45, 1118–1125. [Google Scholar] [CrossRef]
- Bannoehr, J.; Franco, A.; Iurescia, M.; Battisti, A.; Fitzgerald, J. Molecular diagnostic identification of Staphylococcus pseudintermedius. J Clin Microbiol. 2009, 47, 469–471. [Google Scholar] [CrossRef]
- Bannoehr, J.; Guardabassi, L. Staphylococcus pseudintermedius in the dog: Taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2021, 2012, 23. [Google Scholar] [CrossRef]
- Abouelkhair, M.; Frank, L.; Bemis, D.; Giannone, R.; Kania, S. Staphylococcus pseudintermedius 5′-nucleotidase suppresses canine phagocytic activity. Vet Microbiol. 2020, 246. [Google Scholar] [CrossRef] [PubMed]
- Iverson, S.; Brazil, A.; Ferguson, J.; Nelson, K.; Lautenbach, E.; Rankin, S.; Morris, D.; Davis, M. Anatomical patterns of colonization of pets with staphylococcal species in homes of people with methicillin-resistant Staphylococcus aureus (MRSA) skin or soft tissue infection (SSTI). Vet Microbiol. 2015, 176, 202–208. [Google Scholar] [CrossRef]
- Kmieciak, W.; Szewczyk, E. Are zoonotic Staphylococcus pseudintermedius strains a growing threat for humans? Folia Microbiol (Praha). 2018, 63, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Keilman, R.; Harding, S.; Rowin, M.; Reade, E.; Klingborg, P.; Levine, D.; Spratt, Jr., H. Investigations of Staphylococcal contamination on environmental surfaces of a neonatal intensive care unit of a children’s hospital. Am J Infect Cont. 2021, 49, 1450–1453. [Google Scholar] [CrossRef]
- Flayhart, D.; Hindler, J.; Bruckner, D.; Hall, G.; Shrestha, R.; Vogel, S.; Richter, S.; Howard, W.; Walther, R.; Carroll, K. Multicenter evaluation of BBL CHROMagar MRSA medium for direct detection of methicillin-resistant Staphylococcus aureus from surveillance cultures of the anterior nares. J Clin Microbiol. 2005, 43, 5536–5540. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kelesidis, T.; Tsiodras, S. Staphylococcus intermedius is not only a zoonotic pathogen, but may also cause skin abscesses in humans after exposure to saliva. Int J Infect Dis. 2010, 14, e838–e841. [Google Scholar] [CrossRef]
- Álvarez-Pérez, S.; Blanco, J.; Harmanus, C.; Kuijper, E.; García, M. Prevalence and characteristics of Clostridium perfringens and Clostridium difficile in dogs and cats attended in diverse veterinary clinics from the Madrid region. Anaerobe. 2017, 48, 47–55. [Google Scholar] [CrossRef]
- Bonadonna, L.; Briancesco, R.; Coccia, A.; Meloni, P.; Rosa, G.; Moscato, U. Microbial Air Quality in Healthcare Facilities. Int. J. Environ. Res. Public Health 2021, 18, 6226. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.; Reid-Smith, R.; Boerlin, P.; Weese, J.; Prescott, J.; Janecko, N.; Hassard, L.; McEwen, S. Escherichia coli and selected veterinary and zoonotic pathogens isolated from environmental sites in companion animal veterinary hospitals in southern Ontario. Can Vet J. 2010, 51, 963–972. [Google Scholar] [PubMed] [PubMed Central]
- Souza-Silva, T.; Rossi, C.; Andrade-Oliveira, A.; Vilar, L.; Pereira, M.; de Araujo Penna, B.; Giambiagi-deMarval, M. Interspecies transfer of plasmid-borne gentamicin resistance between Staphylococcus isolated from Staphylococcus aureus. Infection, Genetics and Evolution 2022, 98, 105230. [Google Scholar] [CrossRef] [PubMed]
- Lax, S.; Sangwan, N.; Smith, D.; Larsen, P.; Handley, K.; Richardson, M.; Guyton, K.; Krezalek, M.; Shogan, B.; Defazio, J.; Flemming, I.; Shakhsheer, B.; Weber, S.; Landon, E.; Garcia-Houchins, S.; Siegel, J.; Alverdy, J.; Knight, R.; Stephens, B.; Gilbert, J. Bacterial colonization and succession in a newly opened hospital. Science translational medicine 2017, 9. [Google Scholar] [CrossRef]
- Spratt, Jr., H.; Levine, D.; Tillman, L. Physical therapy clinic therapeutic ultrasound equipment as a source for bacterial contamination. Physiotherapy Theory and Practice. 2014, 30, 507–511. [Google Scholar] [CrossRef]
Figure 1.
Average of the percent positive swabs for all sites by bacterial species. Key: MRSA = Methicillin resistant S. aureus, SA = S. aureus, SE = S. epidermidis (mannitol negative),. SIM = S. pseudintermedius, ENT = Enteric bacteria (lactose positive Gram-negative rods), PS = Pseudomonas sp., PSA = P. aeruginosa, ML = Micrococcus luteus, BAC = Bacillus sp., Cdiff = Clostridium difficile.
Figure 1.
Average of the percent positive swabs for all sites by bacterial species. Key: MRSA = Methicillin resistant S. aureus, SA = S. aureus, SE = S. epidermidis (mannitol negative),. SIM = S. pseudintermedius, ENT = Enteric bacteria (lactose positive Gram-negative rods), PS = Pseudomonas sp., PSA = P. aeruginosa, ML = Micrococcus luteus, BAC = Bacillus sp., Cdiff = Clostridium difficile.
Figure 2.
Total positive swabs by site having Staphylococci. Presumptive species identification: MRSA = methicillin resistant S. aureus, SA = S. aureus, SE = S. epidermidis, SIM = S. pseudintermedius.
Figure 2.
Total positive swabs by site having Staphylococci. Presumptive species identification: MRSA = methicillin resistant S. aureus, SA = S. aureus, SE = S. epidermidis, SIM = S. pseudintermedius.
Figure 3.
Total positive swabs by site having select Gram negative bacteria. Presumptive species identification: ENT = Lactose positive (e.g., E. coli), PS = Pseudomonas sp., PSA = Pseudomonas aeruginosa.
Figure 3.
Total positive swabs by site having select Gram negative bacteria. Presumptive species identification: ENT = Lactose positive (e.g., E. coli), PS = Pseudomonas sp., PSA = Pseudomonas aeruginosa.
Figure 4.
Total positive swabs by site having select Gram Positive bacteria. Presumptive species identification: ML = Micrococcus luteus, BAC = Bacillus sp., Cdiff = Clostridium difficile.
Figure 4.
Total positive swabs by site having select Gram Positive bacteria. Presumptive species identification: ML = Micrococcus luteus, BAC = Bacillus sp., Cdiff = Clostridium difficile.
Figure 5.
Water sample contamination for underwater treadmills (UWTM) at five veterinary clinic sites. Individual UWTMs indicated by number (#1 through #4 were at one site, while #5 to #8 were each a different sites). Total colony counts (per ml) for TSA and EMB media streaked with 0.1 ml samples from the treadmill tanks.
Figure 5.
Water sample contamination for underwater treadmills (UWTM) at five veterinary clinic sites. Individual UWTMs indicated by number (#1 through #4 were at one site, while #5 to #8 were each a different sites). Total colony counts (per ml) for TSA and EMB media streaked with 0.1 ml samples from the treadmill tanks.
Table 1.
Percentage of positive swabs having viable bacteria by site and bacterial type or species. Legend: UWTM = Underwater Treadmill; Exercise Equipment = Peanuts, balance boards, Physiorolls, Donuts, etc.; MRSA = Methicillin-resistant S. aureus, SA = S. aureus, SE = S. epidermidis (mannitol negative),.
Table 1.
Percentage of positive swabs having viable bacteria by site and bacterial type or species. Legend: UWTM = Underwater Treadmill; Exercise Equipment = Peanuts, balance boards, Physiorolls, Donuts, etc.; MRSA = Methicillin-resistant S. aureus, SA = S. aureus, SE = S. epidermidis (mannitol negative),.
| |
MRSA |
SA |
SE |
SIM |
ENT |
PS |
PSA |
ML |
BAC |
Cdiff |
| Dry Treadmill (belt, n = 5) |
40% |
40% |
40% |
0% |
40% |
0% |
0% |
100% |
60% |
80% |
| Exercise Equipment (n = 27) |
11.1% |
18.5% |
22.2% |
0% |
25.9% |
11.1% |
0% |
22.2% |
44.4% |
74.1% |
| Floors (n = 19) |
15.8% |
52.6% |
31.6% |
15.8% |
52.6% |
31.6% |
5.3% |
21.1% |
63.2% |
94.7% |
| Harnesses (n = 8) |
0% |
25% |
12.5% |
25% |
12.5% |
0% |
0% |
12.5% |
12.5% |
75% |
| Laser Probes (tip of probe, n = 9) |
0% |
11.1% |
11.1% |
11.1% |
11.1% |
0% |
0% |
0% |
0% |
33.3% |
| Life Jackets (n=10) |
0% |
0% |
30% |
10% |
40% |
10% |
0% |
40% |
10% |
60% |
| Return Air Ducts (n = 6) |
15.8% |
83.3% |
33.3% |
16.7% |
100% |
16.7% |
0% |
16.7% |
100% |
16.7% |
| Scales (n = 6) |
50% |
83.3% |
33.3% |
33.3% |
83.3% |
16.7% |
16.7% |
16.7% |
100% |
66.7% |
| Ultrasound Gel (bottle tip, n=4) |
0% |
0% |
50% |
0% |
0% |
0% |
0% |
0% |
0% |
25% |
| Ultrasound Heads (n = 6) |
0% |
0% |
0% |
0% |
0% |
0% |
0% |
0% |
16.7% |
16.7% |
| UWTM Top Belt (n = 7) |
14.3% |
14.3% |
14.3% |
14.3% |
14.3% |
14.3% |
0% |
28.6% |
28.6% |
28.6% |
| UWTM Bottom Surface of Belt (n = 7) |
0% |
0% |
0% |
0% |
0% |
0% |
0% |
0% |
0% |
28.6% |
| UWTM Jets (inside surface) n = 8) |
0% |
0% |
12.5% |
12.5% |
50% |
25% |
25% |
12.5% |
25% |
12.5% |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).