Methods and Materials
Instead of observing the entire space of the image, the human radiologist only carefully observes the area where aneurysm is frequently found on the vessel [
1,
2]. Hence, we mimicked these radiologists' behavior patterns to make deep learning models train the landmarks on the vessels where aneurysm is likely to occur. The easiest way to obtain a deep learning model that predicts landmark is to train the model using human-made landmark ground truth.
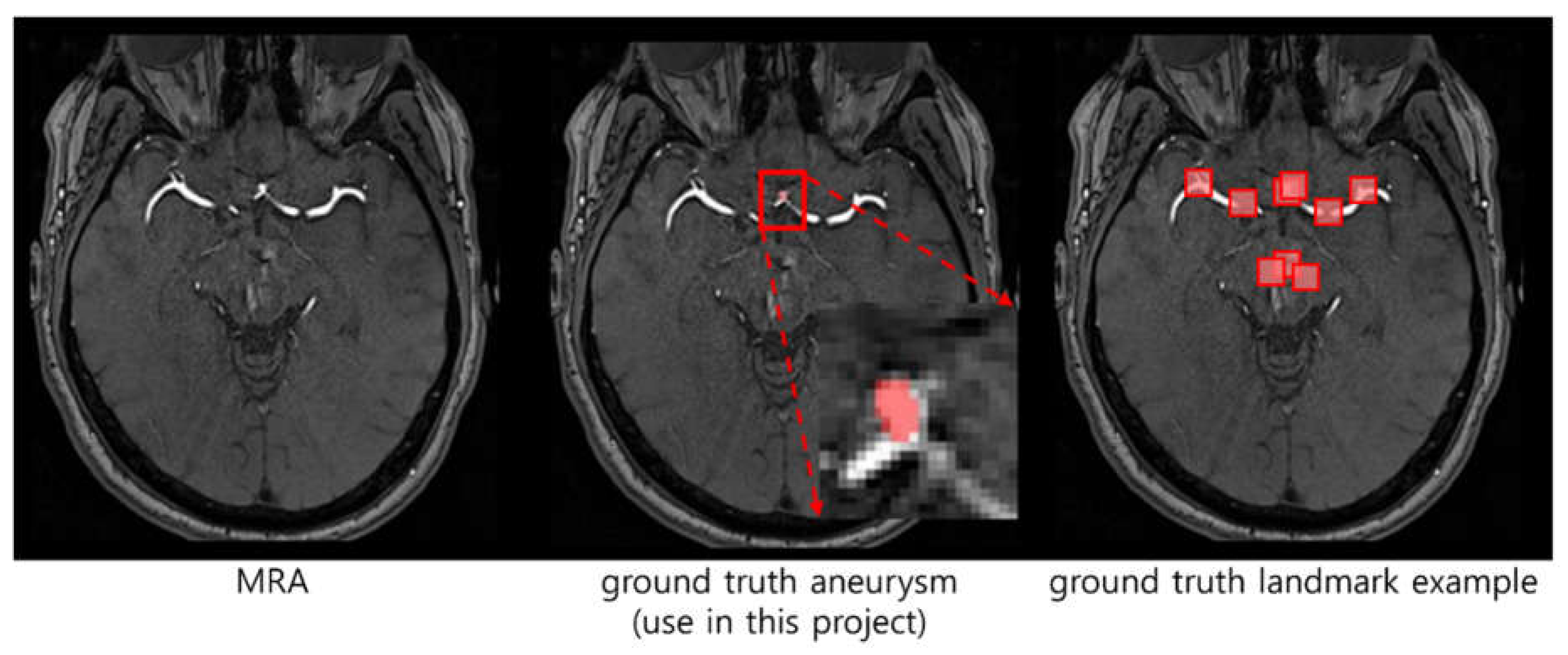
Definition of human-made landmark ground truth: The human radiologist reviewed the patient's MRA image and segmented it by the name of the vessel (ICA, ACA, MCA, etc.) [
3] where aneurysm may occur. As shown in
Figure 1, the ground truth landmark should indicate not only the area with aneurysm but also all areas where aneurysm can occur.
Dataset: 3D TOF-MRA images were retried from one Korean hospital to train and validate a proposed model. Multiple radiologists independently read the presence or absence of cerebral aneurysms (positive and negative), the number of cerebral aneurysms, and location. Only images with consistent opinions of radiologists were selected and used, the final number of selected data is 550. Of the 550 images, 50 images are without aneurysms, 426 images have 1 aneurysm per image, 60 images have 2 aneurysm per image, 12 images have 3 aneurysm per image, 1 image has 4 aneurysm per image, 1 image has 5 aneurysm per image. 55 images, which is 10%, were randomly selected and used as a validation only set, and there are a total of 55 aneurysm included.
However, due to the difficulty of obtaining the human-made landmark ground truth, we propose a semi-supervised learning [
4]-based automatic landmark generation method that uses only aneurysm-specific ground truth.
The proposed method consists of two steps: the first step is clustering the aneurysm-positive position of all MRA images on the 3D space. This cluster center is used as the initial location of the landmark for the deep learning model. The second step is building the deep learning model that outputs predicted landmarks, which do not overlap but simultaneously contain all the aneurysms well. Additionally, we propose two novel loss functions for the landmark-generated model, described in detail below.
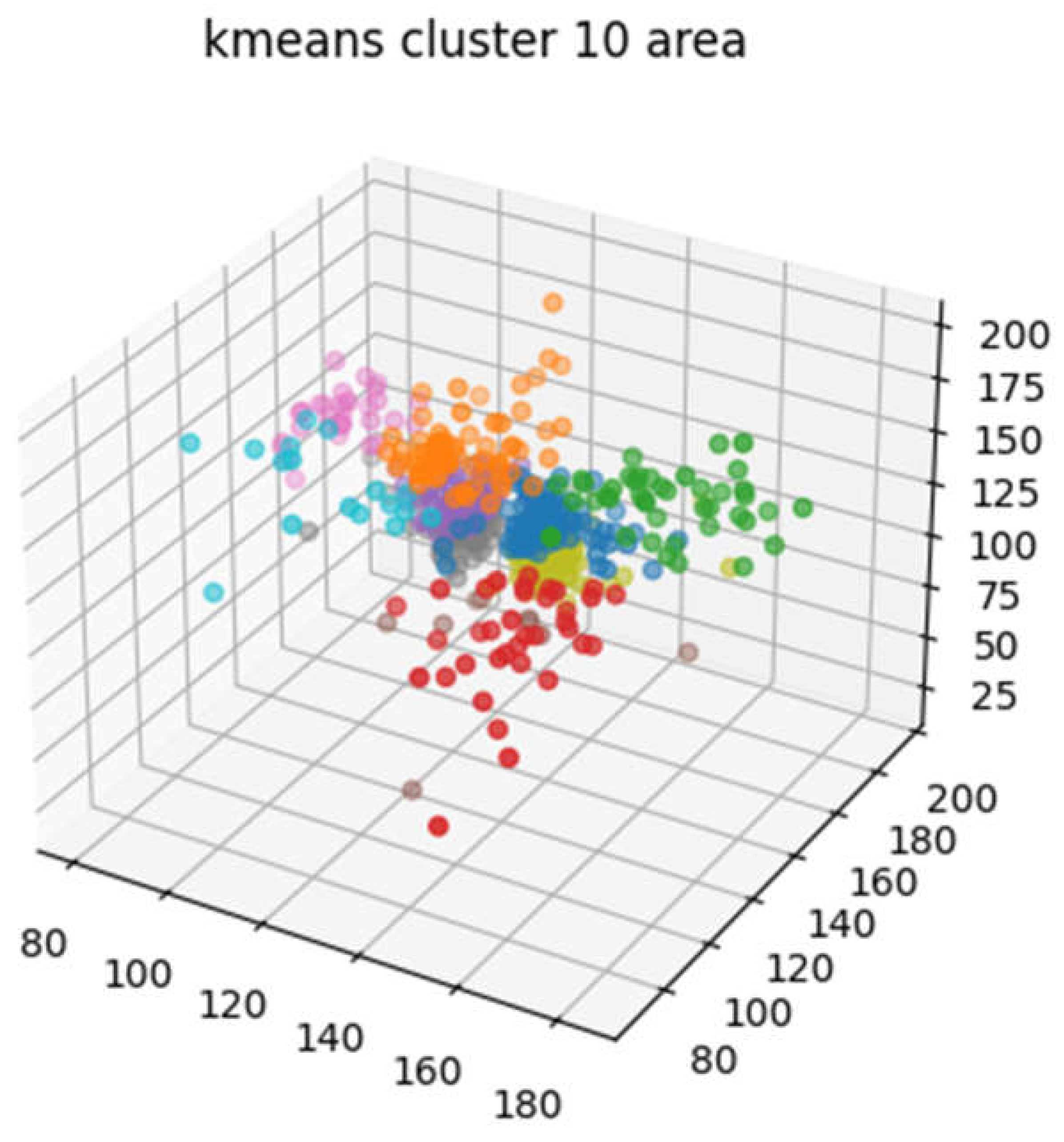
Clustering: Aneurysm-positive position does not occur in all the areas of the MRA images, but in specific vascular areas. The first step is to group these regions into several subgroups that share common properties. We grouped the aneurysm-positive locations close to Euclidean distance using k-means clustering [
5]. As a result, sub-groups are bound with similar blood vessels shape as shown in
Figure 2.
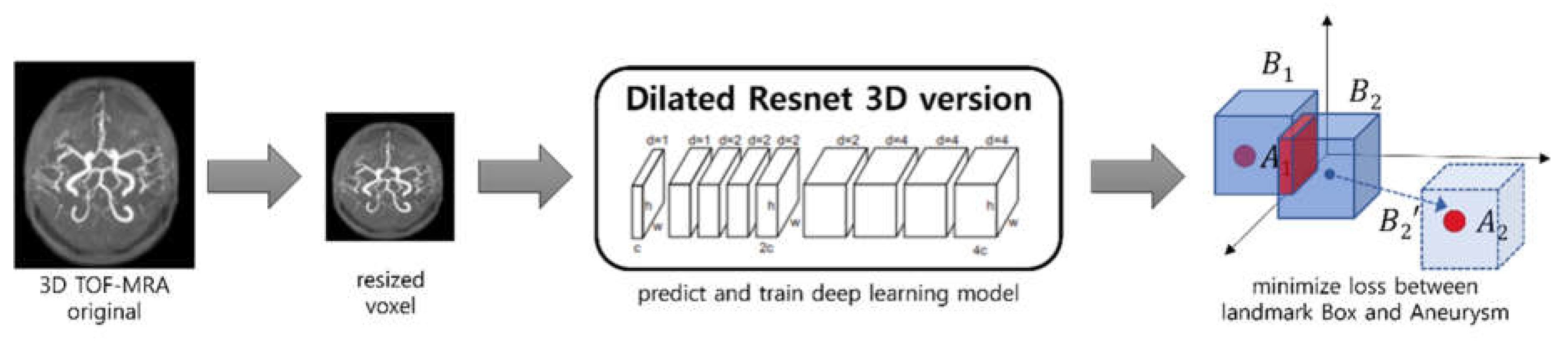
Training a deep Learning model: We create a landmark that contain areas where aneurism is likely to exist using a deep learning model. By considering the shape of blood vessels, the deep learning model can predict more precisely where aneurysms are likely to occur. The deep learning model uses a dilated residual network [
6] as shown in
Figure 3, it receives the MRA image resized to 256
K is the number of landmarks and
R is the size of the landmark. We select experimentally suitable
K and
R, and described the results in the section of Results or Findings.
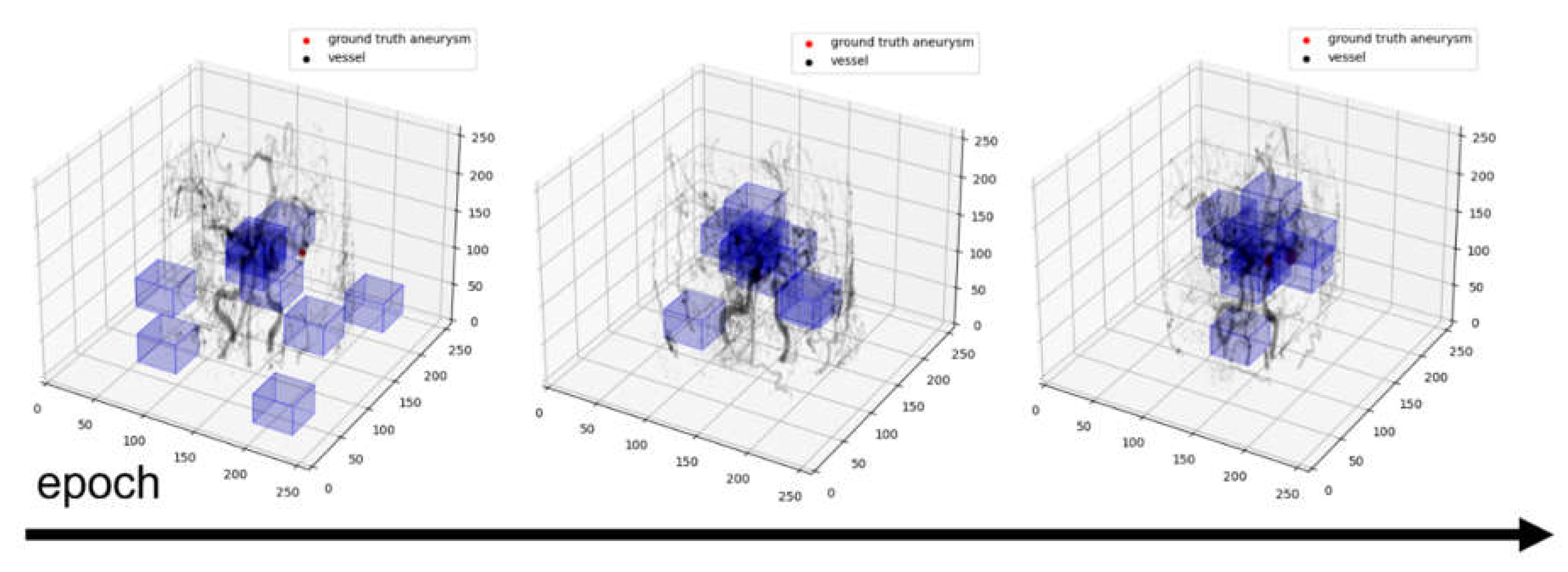
We trained the deep learning model in a semi-supervised manner by proposed the custom loss functions because we did not have the ground truth landmark label. The deep learning model initially mimicked a sub-group simply made of k-means clustering then trains using the proposed custom loss term as shown in
Figure 4.
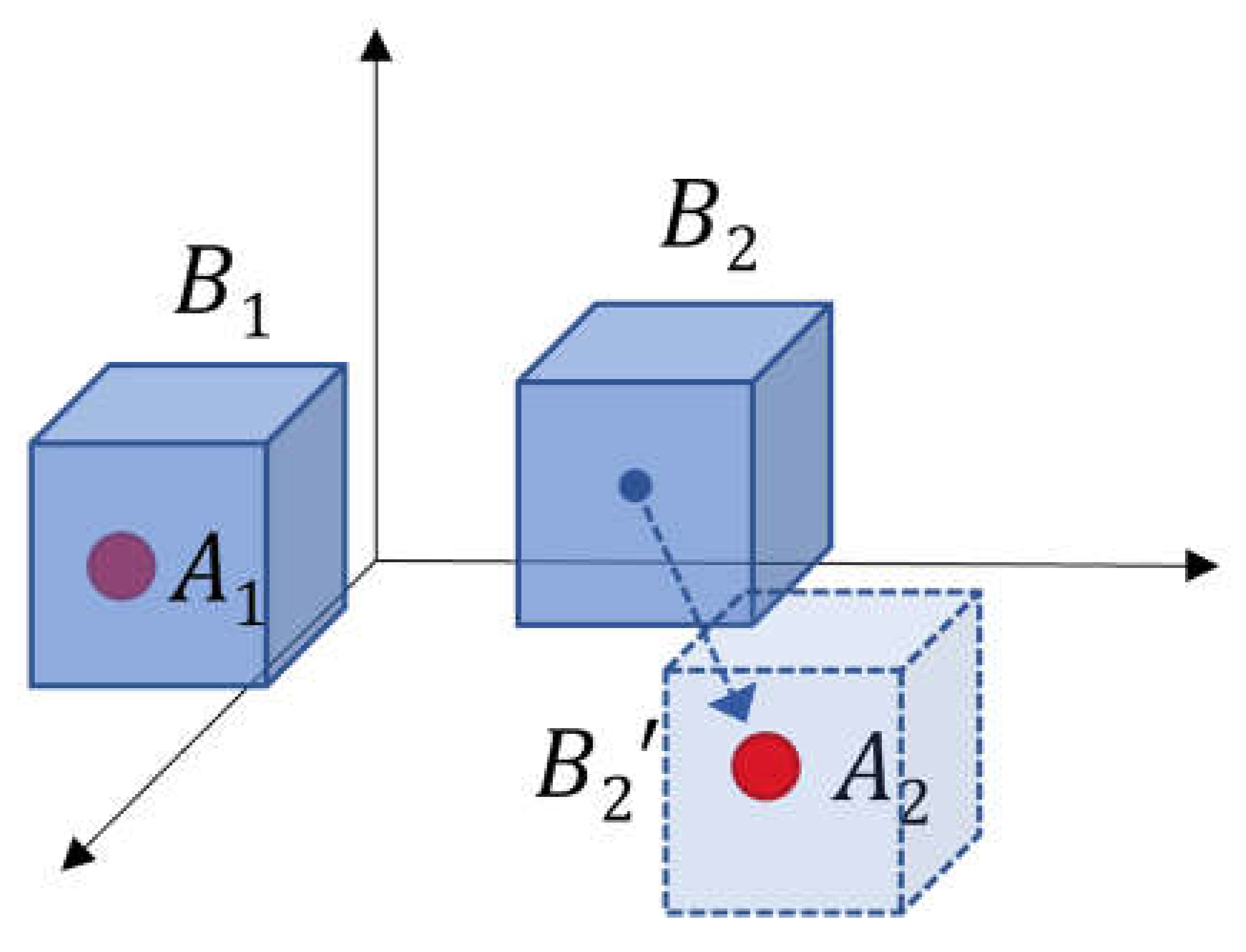
L
1 makes a landmark box
B contain aneurysm-positive location
A, which is not yet included in any landmark. For example, as shown in
Figure 5,
B1 already contains
A1 so it remains in the current state.
B2 is not contained any
A so it moves to a position close to
A2.
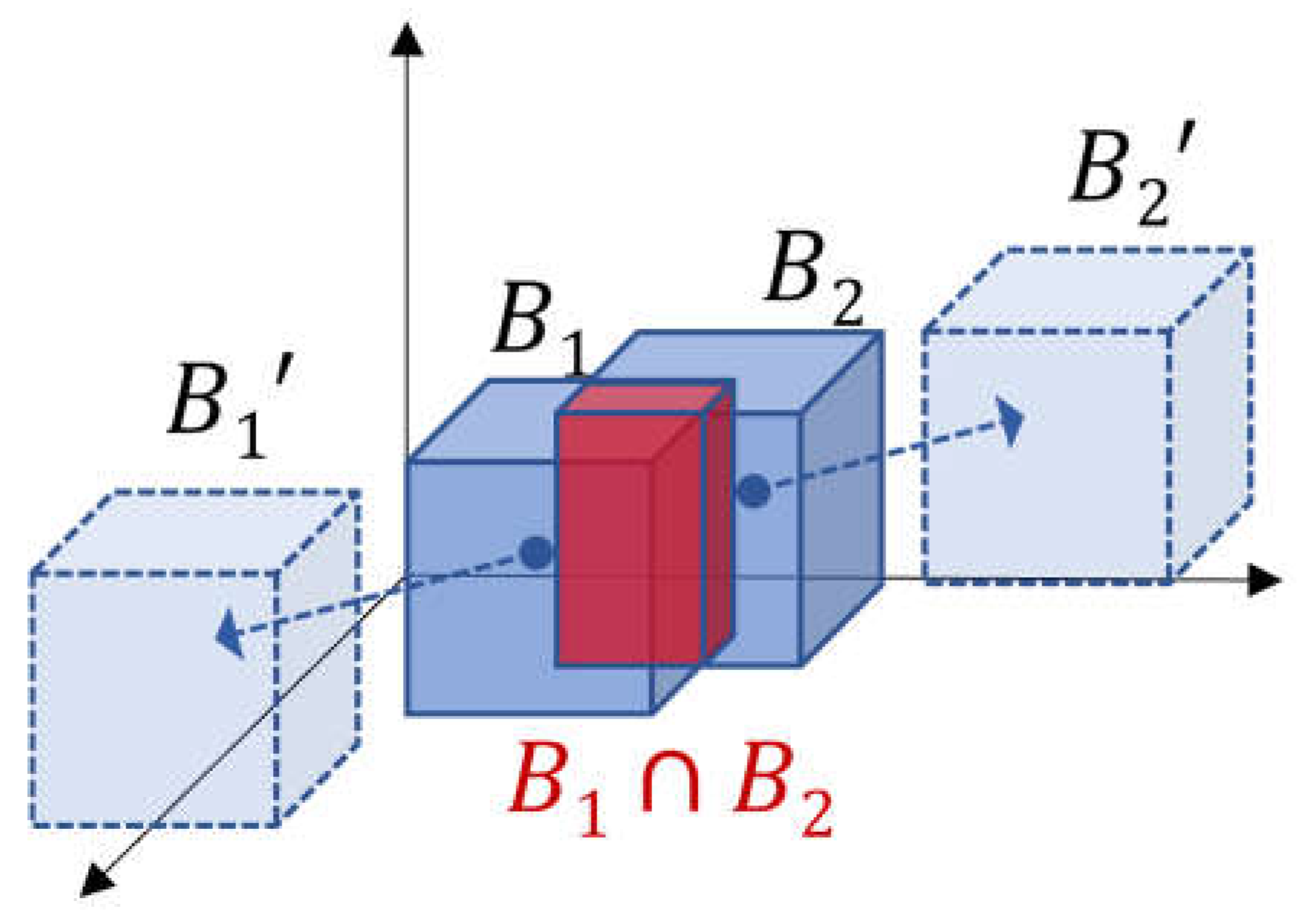
L
2 makes a landmark box B which does not overlap each other. As shown in
Figure 6, boxes with overlapping areas are pushed away from each other.
Boxes where overlapping areas do not exist, are not affected.
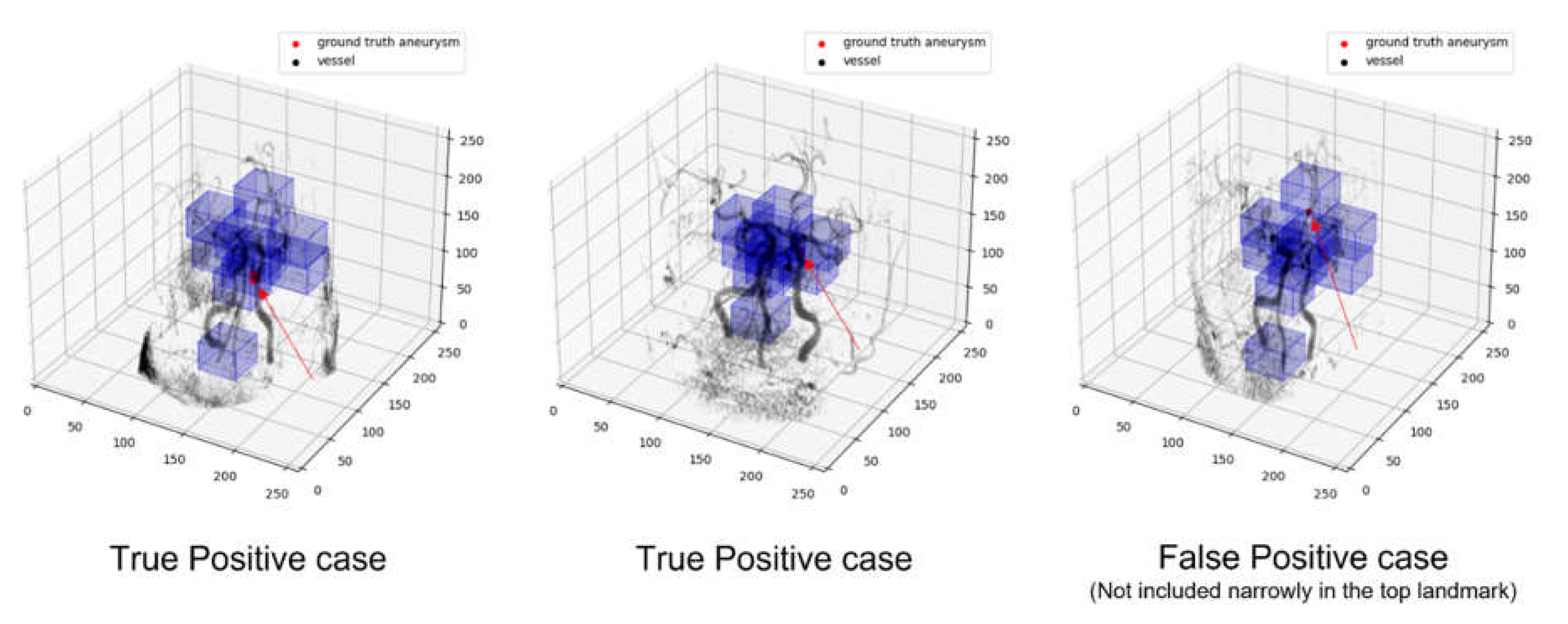
The training process is as shown in
Figure 7, and the landmark is located on a vessel where aneurysm is likely to occur.