Submitted:
17 May 2024
Posted:
17 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
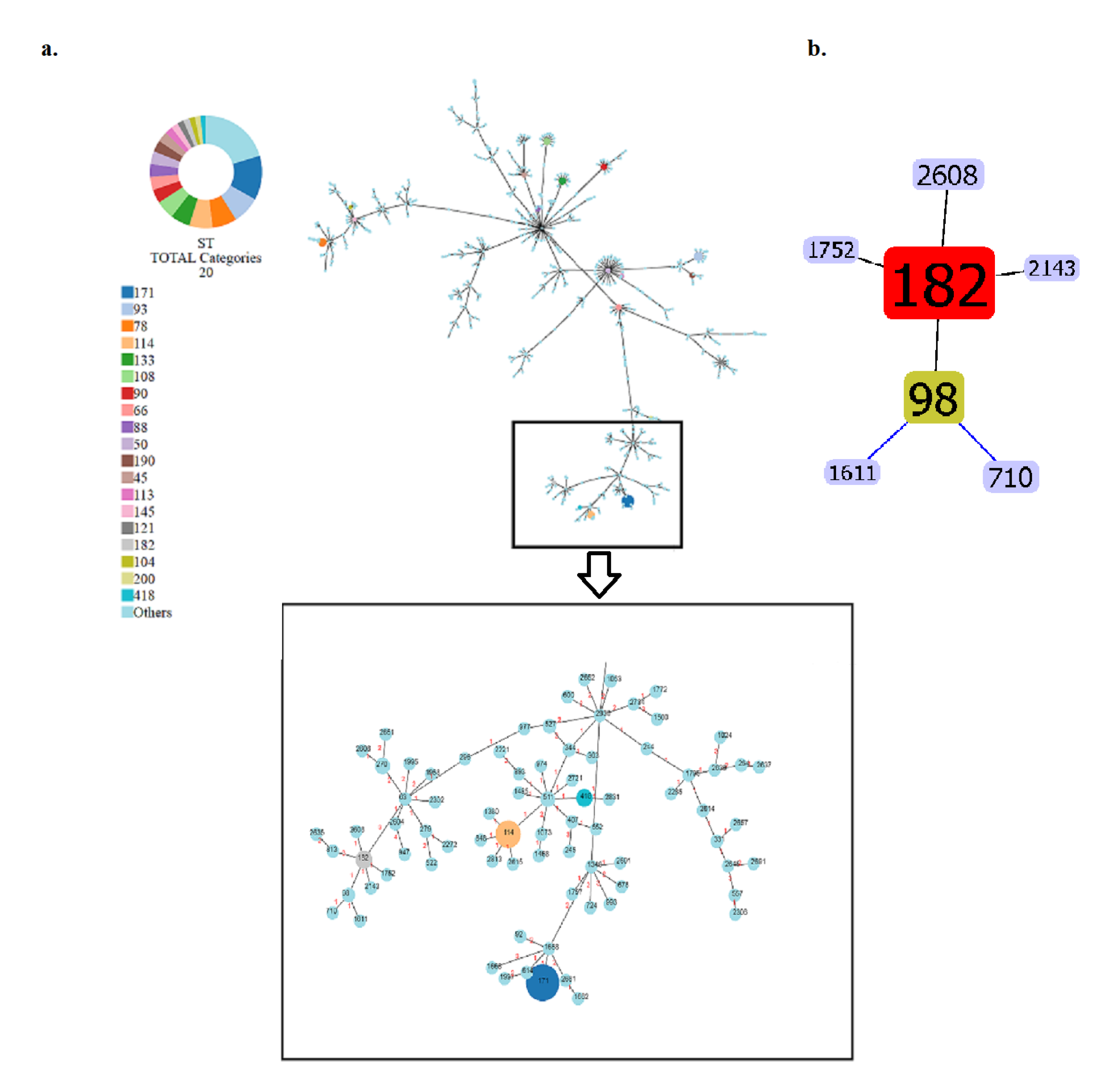
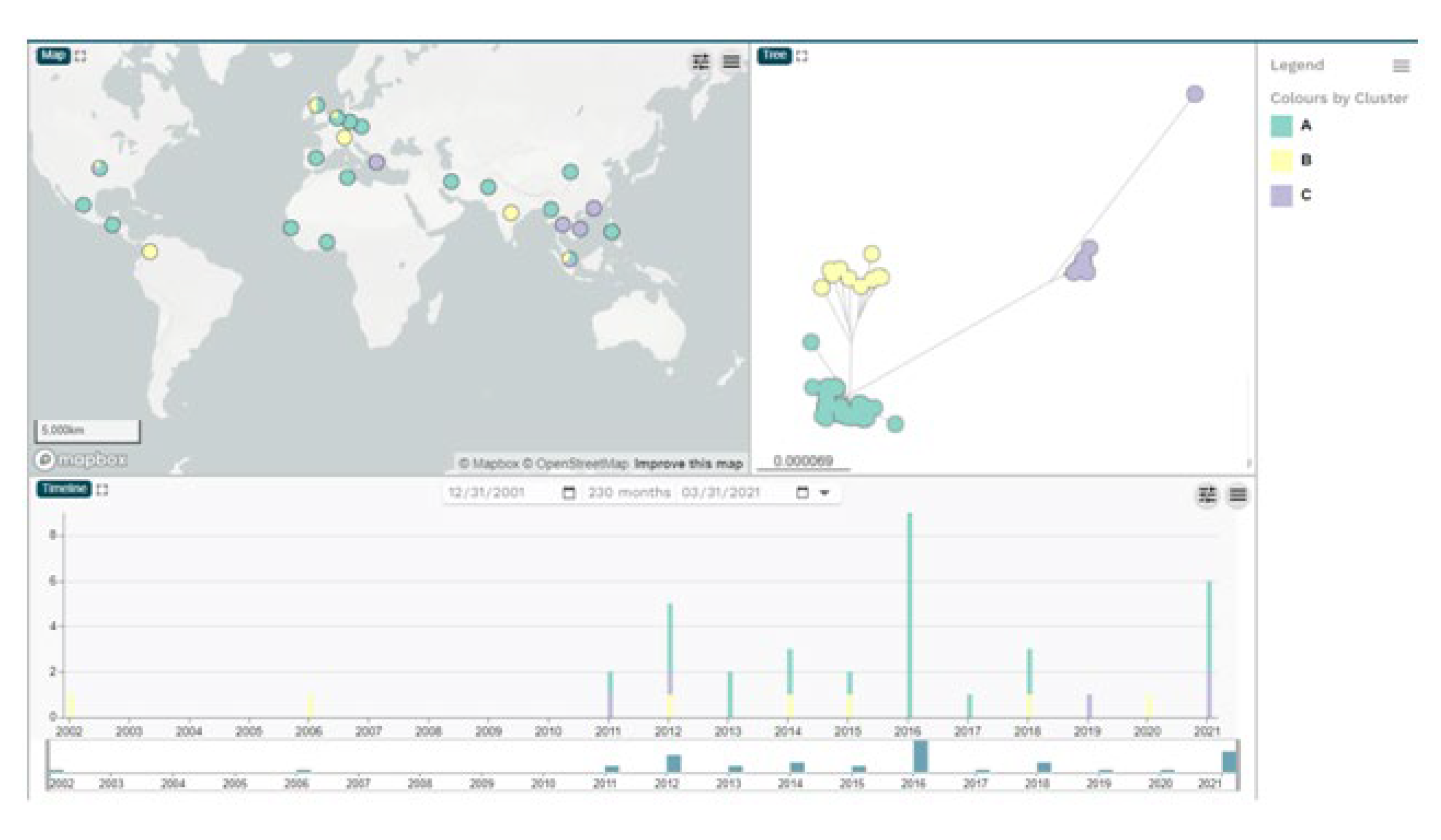
2.1. Bacterial strains, whole genome sequences and phylogenetic analysis of ECC isolates
2.2. In silico identification of plasmids, antimicrobial resistance and virulence genes of the ECC NDM-producing isolates
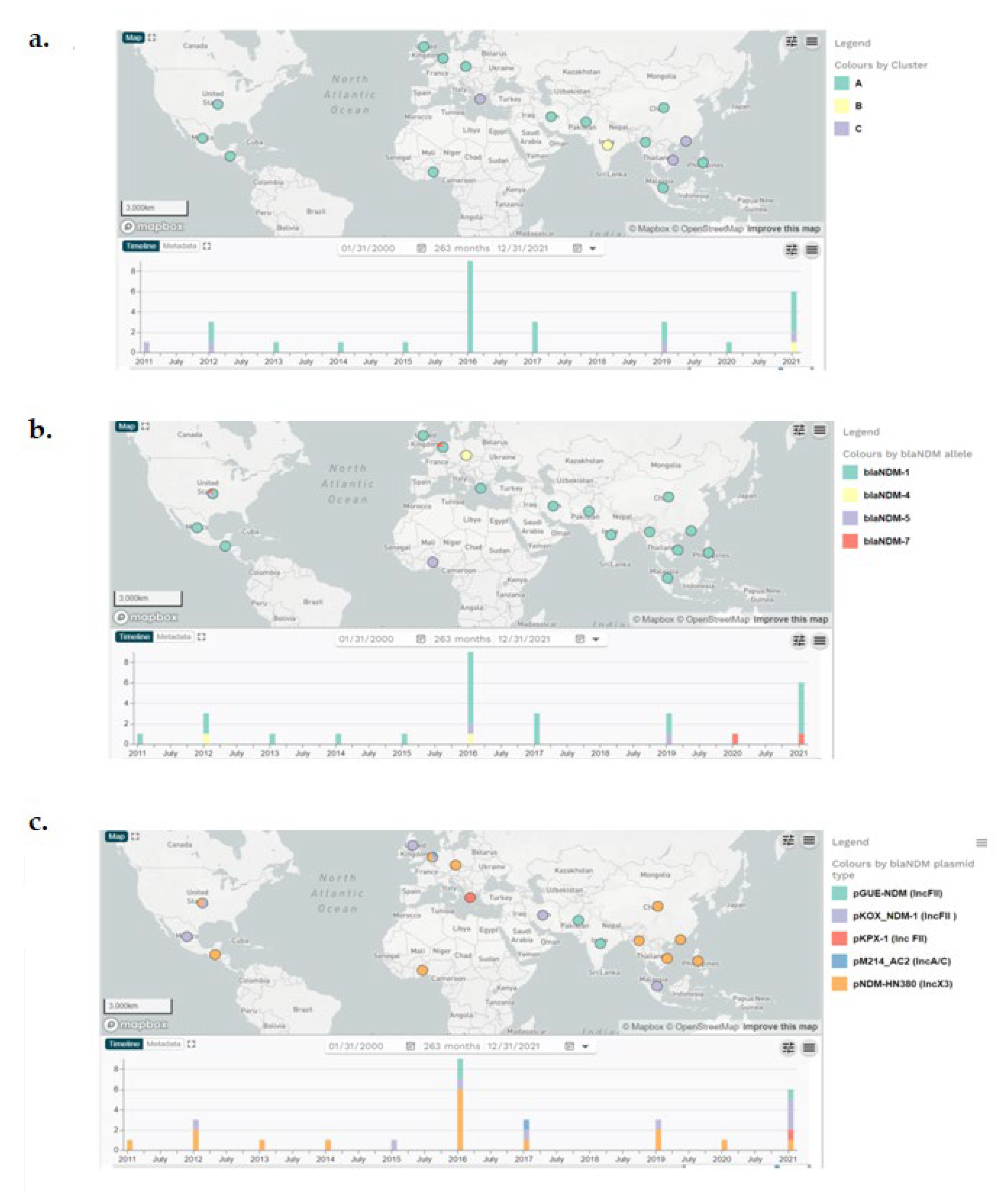
2.3. Genetic background of blaNDM and plasmid analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial isolates, genome sequences and phylogenetic analysis
4.2. Identification of MGEs, antimicrobial resistance genes and virulence factors, and plasmid analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Davin-Regli A, Lavigne JP, Pagès JM. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. ClinMicrobiol Rev. 2019; 32:e00002-19. [CrossRef]
- Paauw A, Caspers MPM, Schuren FHJ, Leverstein-van Hall MA, Delétoile A, Montijn RC, et al. Genomic diversity within the Enterobacter cloacae complex. PLoS One 2008; 3: e3018. [CrossRef]
- De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson, DL, Walker MJ. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 2020; 33: e00181-19. [CrossRef]
- Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant Enterobacter cloacae Complex emerging as a global, diversifying threat. Front Microbiol. 2019; 10: 44. [CrossRef]
- Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. NDM Metallo-β-lactamases and their bacterial producers in health care settings. ClinMicrobiol Rev 2019; 32: e00115-18. [CrossRef]
- Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M; MOSAR WP2, WP3 and WP5 Study Groups. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J AntimicrobChemother 2015; 70:48-56. [CrossRef]
- Peirano G, Matsumura Y, Adams MD, Bradford P, Motyl M, Chen L, et al. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg Infect Dis 2018; 24:1010-19. [CrossRef]
- Bolourchi, N., Giske, C. G., Nematzadeh, S., Mirzaie, A., Abhari, S. S., Solgi, H., &Badmasti, F. Comparative resistome and virulome analysis of clinical NDM-1–producing carbapenem-resistant Enterobacter cloacae complex. J Glob Antimicrob Resist 2022; 28: 254-63. [CrossRef]
- Sugawara Y, Akeda Y, Hagiya H, Sakamoto N, Takeuchi D, Shanmugakani RK, Motooka D, Nishi I, Zin KN, Aye MM, MyintT,Tomono K, Hamada S. Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob Agents Chemother. 2019; 63: e01924-18. [CrossRef]
- Papagiannitsis, CC, Studentova V, Chudackova E. et al. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Enterobacter cloacae from a Czech patient previously hospitalized in Sri Lanka. Folia Microbiol 2013; 58: 547–9. [CrossRef]
- Paskova V, Medvecky M, Skalova A, Chudejova K, Bitar I, Jakubu V, Bergerova T, Zemlickova H, Papagiannitsis CC, Hrabak J. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front Microbiol. 2018; 9:1549. [CrossRef]
- Merhi G, Amayri S, Bitar I, Araj GF, Tokajian S. Whole genome-based characterization of multidrug-resistant Enterobacterand Klebsiellaaerogenes isolates from Lebanon. MicrobiolSpectr 2023; 1: e0291722. [CrossRef]
- Gartzonika K, Politi L, Mavroidi A, Tsantes AG, Spanakis N, Priavali E, Vrioni G, Tsakris A. High prevalence of clonally related ST182 NDM-1-producing Enterobacter cloacae complex clinical isolates in Greece. Int J Antimicrob Agents 2023; 62: 106837. [CrossRef]
- Mavroidi, A., Gartzonika, K., Spanakis, N., Froukala, E., Kittas, C., Vrioni, G., Tsakris, A. Comprehensive analysis of virulence determinants and genomic islands of blaNDM-1-producing Enterobacterhormaechei clinical isolates from Greece. Antibiotics 2023; 12: 1549. [CrossRef]
- Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, Abudahab K, Yeats CA, Goater R, Taylor B, Harste H, Muddyman D, Feil EJ, Brisse S, Holt K, Donado-Godoy P, Ravikumar KL, Okeke IN, Carlos C, Aanensen DM; NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance. 2021. Rapid genomic characterization and global surveillance of Klebsiellausing Pathogenwatch. Clin Infect Dis. 2021; 73 (Suppl_4): S325-S335. [CrossRef] [PubMed]
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. WellcomeOpenRes 2018; 3:124. [CrossRef]
- Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C, PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods.Bioinformatics 2017; 33: 128–9. [CrossRef]
- Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole genome phylogenies from short sequence reads. MolBiol Evol 2014; 31:1077-88.
- Tamura K., Stecher G., and Kumar S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021; 38:3022-27. [CrossRef]
- Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H. Spratt BG, Aanensen DM. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. MicrobGenom 2016; 2: e000093. [CrossRef]
- Clausen PT, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J AntimicrobChemother 2016;71:2484-8. [CrossRef]
- Johansson MHK, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, Petersen TN. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J AntimicrobChemother 2021;76:101-9. [CrossRef]
- Gao F, Zhang CT. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res 2006; 34 (Web Server issue): W686-91. [CrossRef]
- Li X, Xie Y, Liu M, Tai C, Sun J, Zixin Deng, Ou H-Y. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements, Nucleic Acids Research 2018; 46: W229–34. [CrossRef]
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons, BMC Genomics 2011, 12: 402. [CrossRef]
- Liu W-Y, Wong C-F, Chung KM-K, Jiang J-W, Leung FC-C. Comparative genome analysis of Enterobacter cloacae. PLoS One, 2013; 8: e74487. [CrossRef]
- Tao J, Sang Y, Teng Q, Ni J, Yang Y, Tsui SK, Yao YF. Heat shock proteins IbpA and IbpB are required for NlpI-participated cell division in Escherichia coli. Front Microbiol 2015; 6:51. [CrossRef]
- L. Turkovicova, R. Smidak, G. Jung, J. Turna, G. Lubec, J. Aradska, Proteomic analysis of the TerCinteractome: Novel links to tellurite resistance and pathogenicity, J Proteomics 2016; 136:167-73. [CrossRef]
- Sukupolvi S, O′Connor CD. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. MicrobiolRev 1990; 54:331-41. [CrossRef]
- Gual-de-Torrella A, Delgado-Valverde M, Pérez-Palacios P, Oteo-Iglesias J, Rojo-Molinero E, Macià MD, Oliver A, Pascual Á, Fernández-Cuenca F. Prevalence of the fimbrial operon mrkABCD, mrkA expression, biofilm formation and effect of biocides on biofilm formation in carbapenemase-producing Klebsiellapneumoniae isolates belonging or not belonging to high-risk clones. Int J Antimicrob Agents 2022; 60: 106663. [CrossRef]
- Wang J, Huang Y, Guan C, Li J, Yang H, Zhao G, Liu C, Ma J, Tang B. Characterization of an Escherichia coli isolate coharboring the virulence gene astA and tigecycline resistance gene tet(X4) from a dead piglet. Pathogens 2023; 12: 903.
- BingbingZong, Wugang Liu, Yanyan Zhang, Xiangru Wang, Huanchun Chen, Chen Tan, Effect of kpsM on the virulence of porcine extraintestinal pathogenic Escherichia coli, FEMS Microbiology Letters 2016; 363: fnw232.
- Moss JE, Cardozo TJ, Zychlinsky A, Groisman EA. The selC-associated SHI-2 pathogenicity island of Shigellaflexneri. MolMicrobiol 1999; 33; 74-83.
- Pak-Leung Ho, Zhen Li, Wai-U Lo, Yuk-Yam Cheung, Chi-Ho Lin, Pak-Chung Sham, Vincent Chi-Chung Cheng, Tak-Keung Ng, Tak-Lun Que & Kin-Hung Chow (2012). Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 2012; 1: e39. [CrossRef]
- Wailan AM, Paterson DL, Kennedy K, Ingram PR, Bursle E, Sidjabat HE. Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob Agents Chemother. 2015; 60: 136-41. [CrossRef]
- Sugawara Y, Akeda Y, Sakamoto N, Takeuchi D, Motooka D, Nakamura S, et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS ONE 2017; 12: e0184720. [CrossRef]
- Hao Y, Shao C, Geng X, Bai Y, Jin Y, Lu Z. Genotypic and phenotypic characterization of clinical Escherichia coli Sequence Type 405 carrying IncN2 plasmid harboring blaNDM-1. Front Microbiol. 2019;10:788. [CrossRef]
- Krishnaraju M, Kamatchi C, Jha AK, Devasena N, Vennila R, Sumathi G, Vaidyanathan R. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol 2015, 33: 30-8. [CrossRef]
- Huang TW, Wang JT, Lauderdale TL, Liao TL, Lai JF, Tan MC, Lin AC, Chen YT, Tsai SF, Chang SC. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiellaoxytoca isolate from Taiwan. Antimicrob Agents Chemother 2013; 57: 4072-6. [CrossRef]
- Phan HTT, Stoesser N, Maciuca IE, Toma F, Szekely E, Flonta M, Hubbard ATM, Pankhurst L, Do T, Peto TEA, Walker AS, Crook DW, Timofte D. Illumina short-read and MinION long-read WGS to characterize the molecular epidemiology of an NDM-1 Serratiamarcescens outbreak in Romania. J AntimicrobChemother. 2018; 73: 672-679. [CrossRef]
- Bonnin RA, Poirel L, Carattoli A, Nordmann P. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One. 2012; 7: e34752. [CrossRef]
- Shin J, Baek JY, Cho SY, Huh HJ, Lee NY, Song JH, Chung DR, Ko KS. blaNDM-5-bearing IncFII-type plasmids of Klebsiellapneumoniae sequence type 147 transmitted by cross-border transfer of a patient. Antimicrob Agents Chemother. 2016; 60:1932-4. Retraction in: Antimicrob Agents Chemother 2023; 67: e0133022. [CrossRef]
- Huang TW, Chen TL, Chen YT, Lauderdale TL, Liao TL, Lee YT, Chen CP, Liu YM, Lin AC, Chang YH, Wu KM, Kirby R, Lai JF, Tan MC, Siu LK, Chang CM, Fung CP, Tsai SF. Copy number change of the NDM-1 sequence in a multidrug-resistantKlebsiellapneumoniae clinical isolate. PLoS One 2013; 8: e62774.
- Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J SystEvolMicrobiol. 2018; 68:2386-2392. [CrossRef]
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009; 53: 5046-54. [CrossRef]
- Adler A, Ghosh H, Gross A, Rechavi A, Lasnoy M, Assous MV, Geffen Y, Darawshe B, Wiener-Well Y, Grundmann H, Reuter S. Molecular features and transmission of NDM-producing Enterobacterales in Israeli hospitals. J Antimicrob Chemother. 2023; 78: 719-23. [CrossRef]
- Acman M, Wang R, van Dorp L, Shaw LP, Wang Q, Luhmann N, Yin Y, Sun S, Chen H, Wang H, Balloux F. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat Commun 2022; 13:1131. [CrossRef]
- David S, Cohen V, Reuter S, Sheppard AE, Giani T, Parkhill J; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group; ESCMID Study Group for Epidemiological Markers (ESGEM); Rossolini GM, Feil EJ, Grundmann H, Aanensen DM. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl AcadSci U S A. 2020; 117:25043-25054. [CrossRef]
| Genomic clusters |
Continent/country of isolation |
blaNDM variants | Total | ||||
|---|---|---|---|---|---|---|---|
| blaNDM-1 | blaNDM-4 | blaNDM-5 | blaNDM-7 | None | |||
| Cluster A | 19 | 2 | 1 | 2 | 13 | 37 | |
| Africa | 1 | 3 | 4 | ||||
| Senegal | 2 | 2 | |||||
| Togo | 1 | 1 | |||||
| Tunisia | 1 | 1 | |||||
| Asia | 10 | 1 | 11 | ||||
| China | 2 | 1 | 3 | ||||
| Iran | 1 | 1 | |||||
| Myanmar | 3 | 3 | |||||
| Pakistan | 2 | 2 | |||||
| Philippines | 1 | 1 | |||||
| Singapore | 1 | 1 | |||||
| Europe | 4 | 2 | 1 | 5 | 12 | ||
| CzechRepublic | 2 | 2 | |||||
| Germany | 1 | 1 | |||||
| Netherlands | 3 | 1 | 4 | ||||
| Spain | 3 | 3 | |||||
| United Kingdom | 1 | 1 | 2 | ||||
| North America | 4 | 1 | 4 | 9 | |||
| Guatemala | 1 | 1 | |||||
| USA | 3 | 1 | 4 | 8 | |||
| SouthAmerica | 1 | 1 | |||||
| Mexico | 1 | 1 | |||||
| Cluster B | 1 | 9 | 10 | ||||
| Asia | 1 | 1 | 2 | ||||
| India | 1 | 1 | |||||
| Singapore | 1 | 1 | |||||
| Europe | 4 | 4 | |||||
| Netherlands | 1 | 1 | |||||
| Switzerland | 1 | 1 | |||||
| United Kingdom | 2 | 2 | |||||
| North America | 2 | 2 | |||||
| USA | 2 | 2 | |||||
| SouthAmerica | 2 | 2 | |||||
| Colombia | 2 | 2 | |||||
| Cluster C | 4 | 1 | 3 | 8 | |||
| Asia | 3 | 1 | 4 | ||||
| HongKong | 1 | 1 | |||||
| Singapore | 1 | 1 | |||||
| Thailand | 1 | 1 | |||||
| VietNam | 1 | 1 | |||||
| Europe | 1 | 1 | |||||
| Greece | 1 | 1 | |||||
| North America | 1 | 2 | 3 | ||||
| USA | 1 | 2 | 3 | ||||
| Total | 24 | 2 | 2 | 2 | 25 | 55 | |
| Plasmid types, blaNDM variants and continent of isolation |
Genomic clusters (sublineages) |
Number of isolates | ||
|---|---|---|---|---|
| A | B | C | ||
| pNDM-HN380 (IncX3) | 13 | 3 | 16 | |
| blaNDM-1 | 8 | 2 | 10 | |
| Asia | 6 | 2 | 8 | |
| Europe | 1 | 1 | ||
| North America | 1 | 1 | ||
| blaNDM-4 | 2 | 2 | ||
| Europe | 2 | 2 | ||
| blaNDM-5 | 1 | 1 | 2 | |
| Africa | 1 | 1 | ||
| North America | 1 | 1 | ||
| blaNDM-7 | 2 | 2 | ||
| Europe | 1 | 1 | ||
| North America | 1 | 1 | ||
| pKOX_NDM-1 (IncFII ) | 8 | 8 | ||
| blaNDM-1 | 8 | 8 | ||
| Asia | 2 | 2 | ||
| Europe | 2 | 2 | ||
| North America | 3 | 3 | ||
| SouthAmerica | 1 | 1 | ||
| pGUE-NDM (IncFII) | 2 | 1 | 3 | |
| blaNDM-1 | 2 | 1 | 3 | |
| Asia | 2 | 1 | 3 | |
| pKPX-1 (Inc FII) | 1 | 1 | ||
| blaNDM-1 | 1 | 1 | ||
| Europe | 1 | 1 | ||
| pJN24NDM1 (IncN2) | 1 | 1 | ||
| blaNDM-1 | 1 | 1 | ||
| Asia | 1 | 1 | ||
| pM214_AC2 (IncA/C) | 1 | 1 | ||
| blaNDM-1 | 1 | 1 | ||
| Europe | 1 | 1 | ||
| Total | 24 | 1 | 5 | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





