Submitted:
21 May 2024
Posted:
23 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
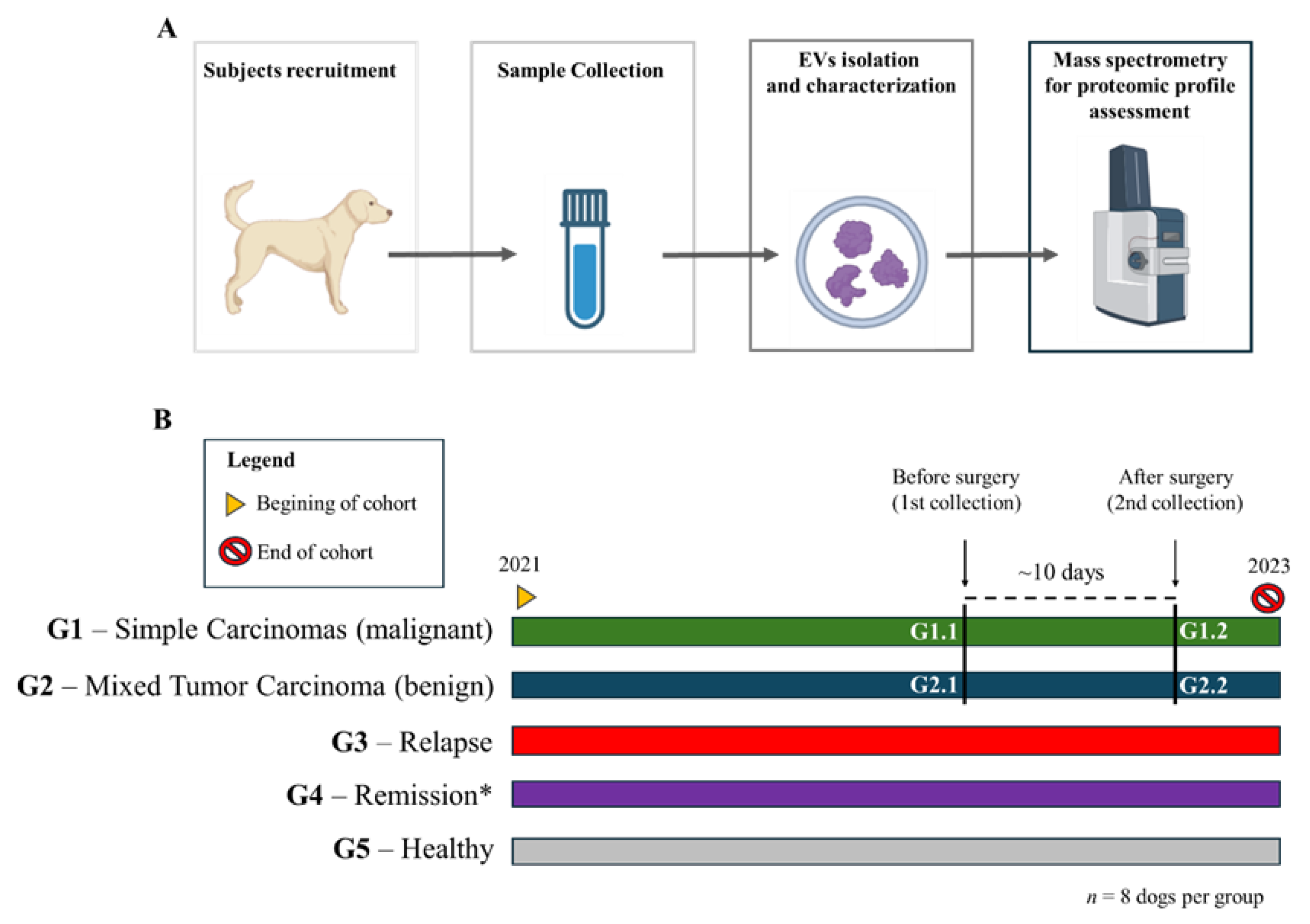
2. Materials and Methods
Experimental Groups
3. Results
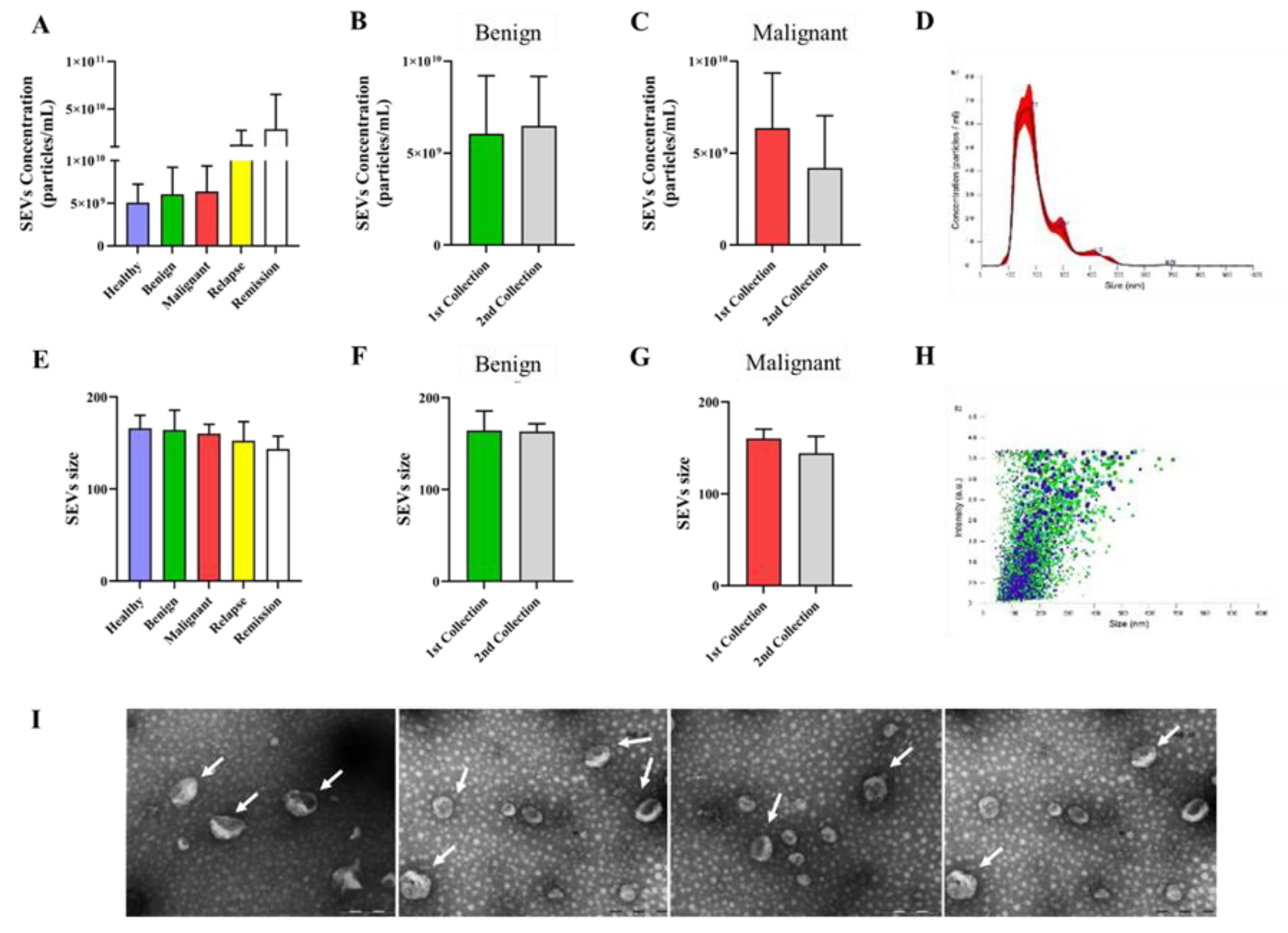
3.1. Extracellular Vesicles Isolated from Canine Plasma Exhibited Characteristics Typical of Small Extracellular Vesicles (SEVs), with Consistent Concentration and Size across Different Groups
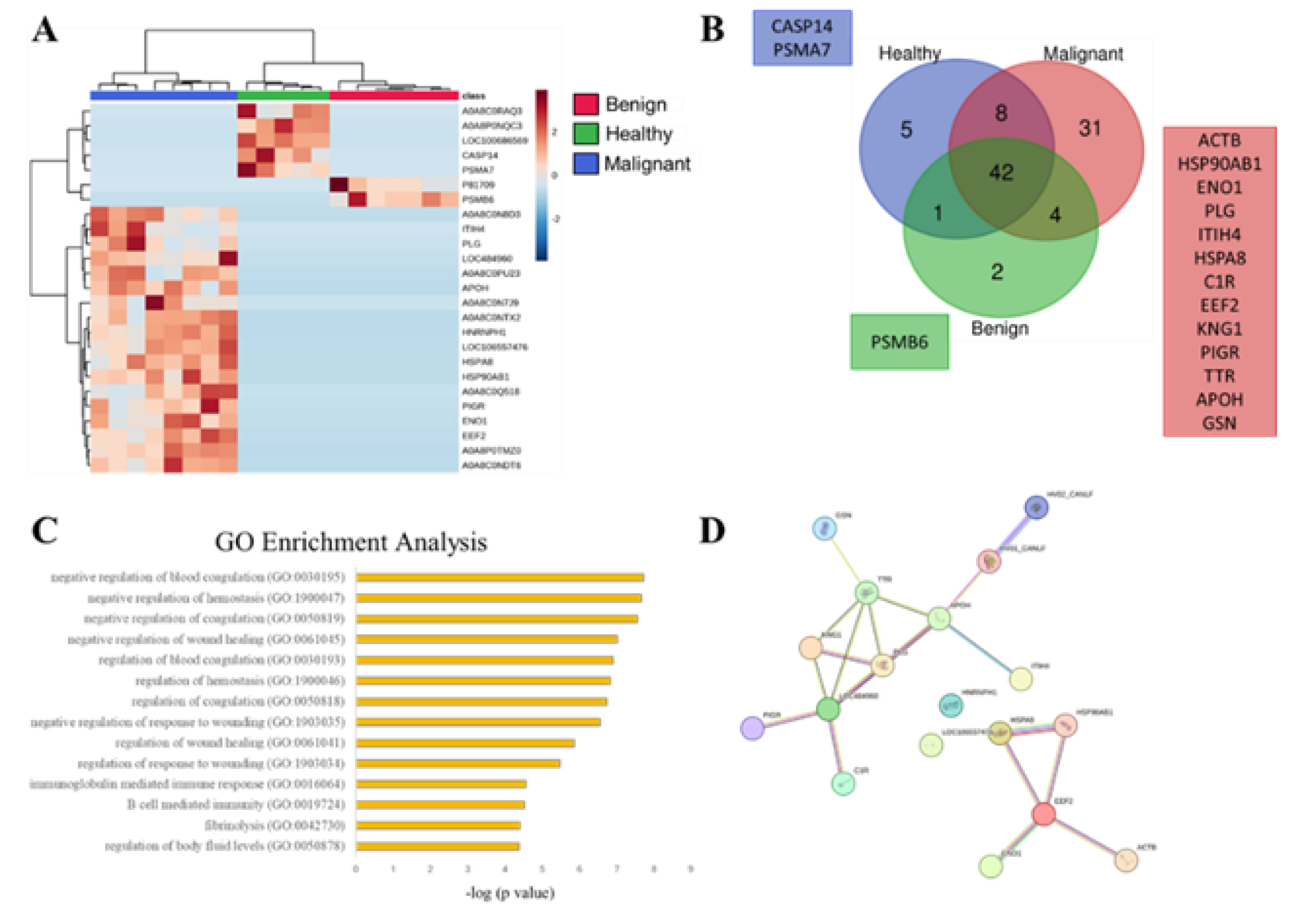
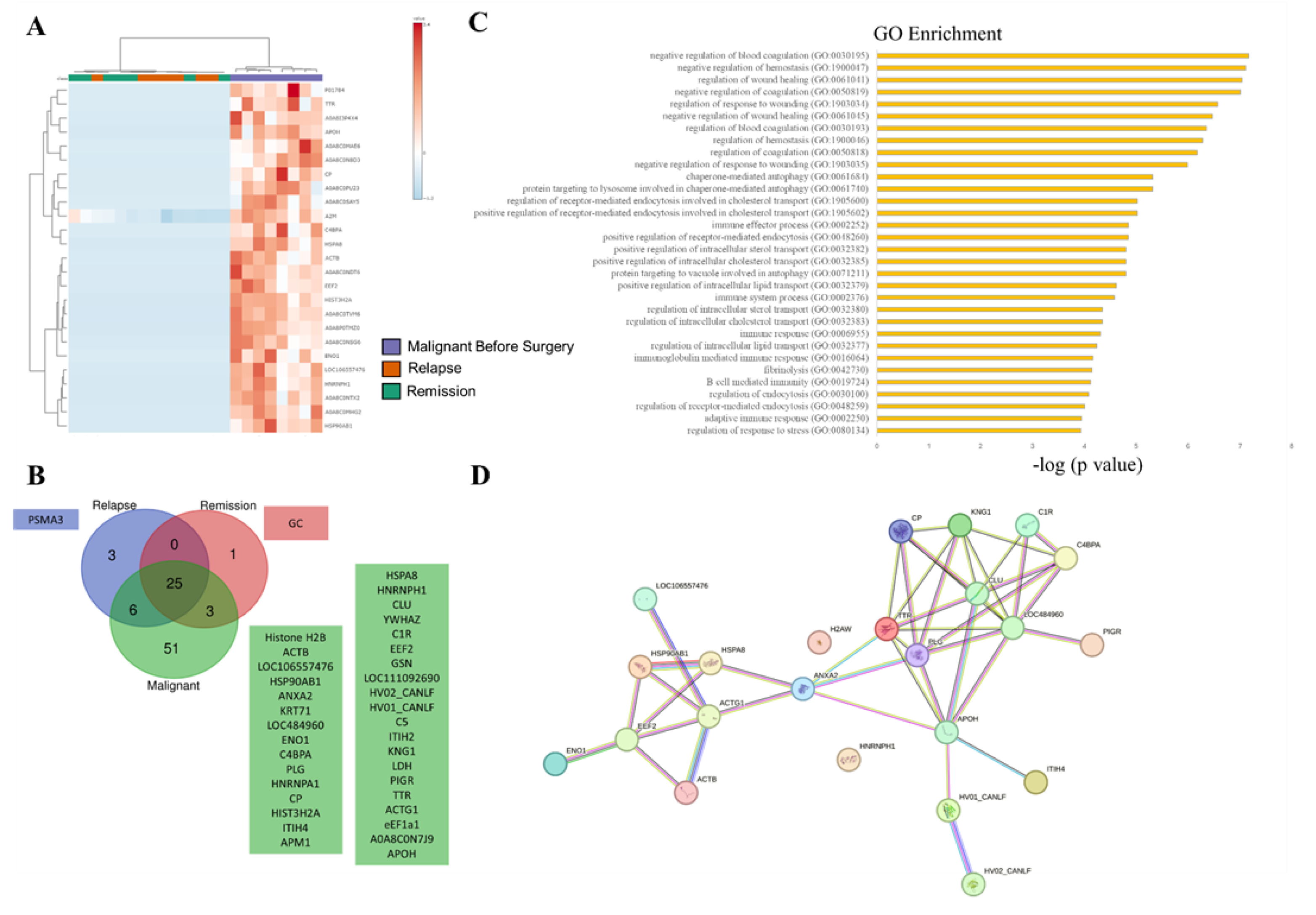
3.2. SEVs` Proteomic Profile from Dogs with Malignant Tumors Exhibit a Larger Number of Unique Proteins, Most of Them Associated with Immune and Wound Healing Processes
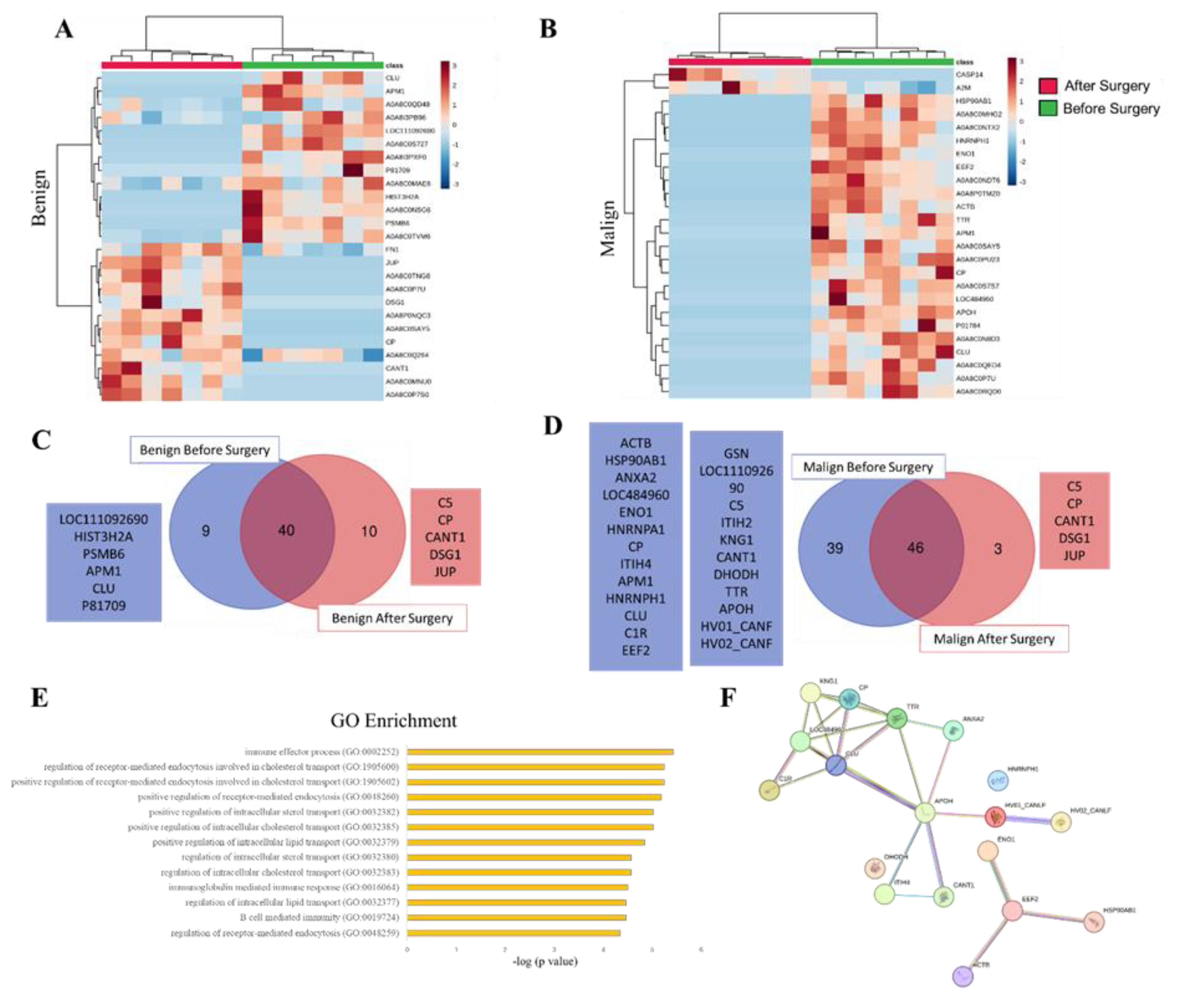
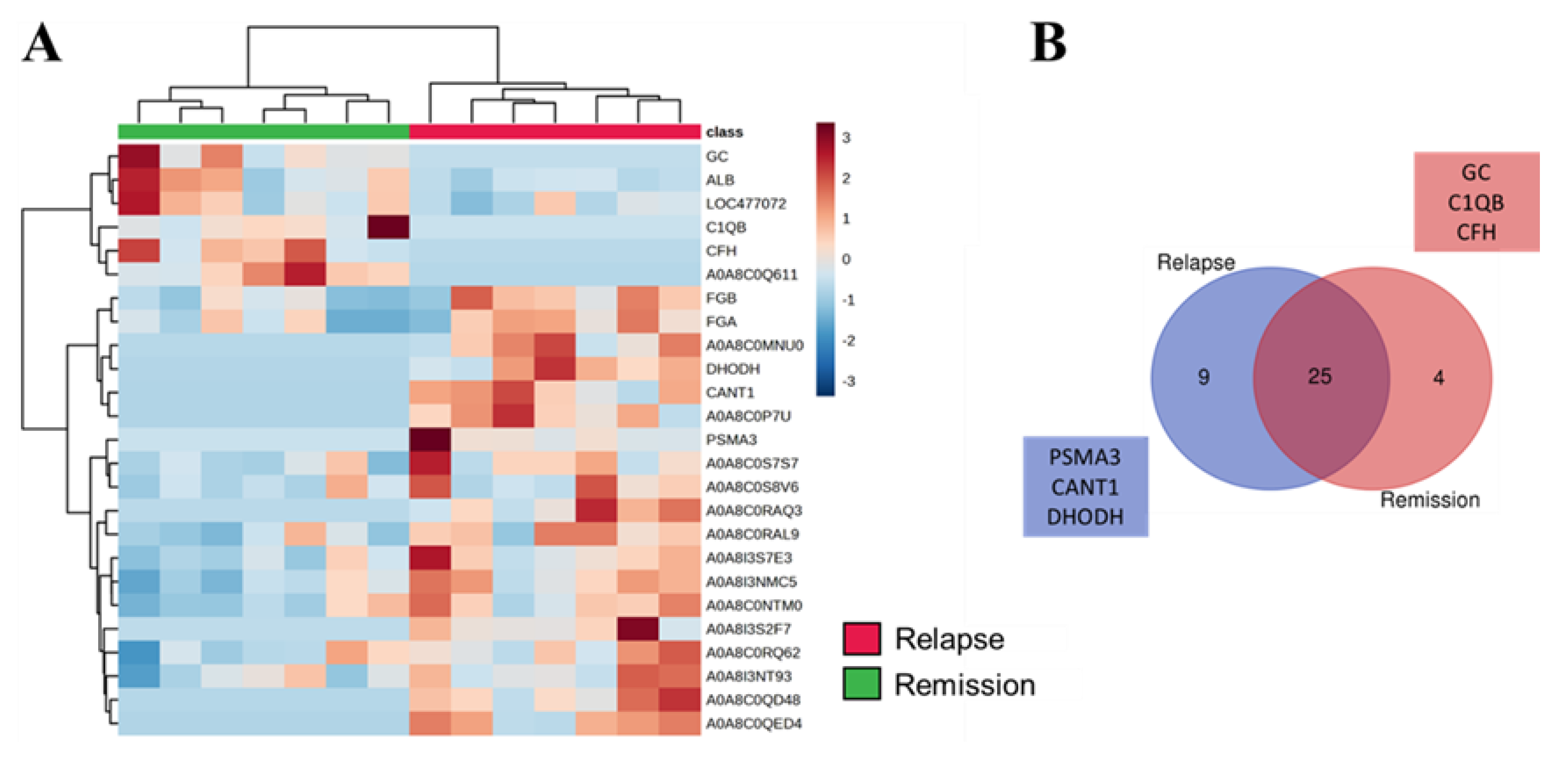
3.3. SEV`s Cargo from Dogs with Malignant Tumor Decreases after Mastectomy, and This Trend Persists Even after Surgery in Cases of Remission or Relapse
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- L. Zeng, W. Li, C.-S. Chen, Breast cancer animal models and applications., Zool. Res. 41 (2020) 477–494. [CrossRef]
- J.Y. Kwon, N. Moskwa, W. Kang, T.M. Fan, C. Lee, Canine as a comparative and translational model for human mammary tumor., J. Breast Cancer. 26 (2023) 1–13. [CrossRef]
- L. Neves Rebello Alves, D. Dummer Meira, L. Poppe Merigueti, M. Correia Casotti, D. do Prado Ventorim, J. Ferreira Figueiredo Almeida, V. Pereira de Sousa, M. Cindra Sant’Ana, R. Gonçalves Coutinho da Cruz, L. Santos Louro, G. Mendonça Santana, T. Erik Santos Louro, R. Evangelista Salazar, D. Ribeiro Campos da Silva, A. Stefani Siqueira Zetum, R. Silva Dos Reis Trabach, F. Imbroisi Valle Errera, F. de Paula, E. de Vargas Wolfgramm Dos Santos, E. Fagundes de Carvalho, I. Drumond Louro, Biomarkers in Breast Cancer: An Old Story with a New End., Genes. 14 (2023). [CrossRef]
- S. Afzal, M. Hassan, S. Ullah, H. Abbas, F. Tawakkal, M.A. Khan, Breast cancer; discovery of novel diagnostic biomarkers, drug resistance, and therapeutic implications., Front. Mol. Biosci. 9 (2022) 783450. [CrossRef]
- T.K.Y. Tay, P.H. Tan, Liquid biopsy in breast cancer: A focused review., Arch. Pathol. Lab. Med. 145 (2021) 678–686. [CrossRef]
- D. Yu, Y. Li, M. Wang, J. Gu, W. Xu, H. Cai, X. Fang, X. Zhang, Exosomes as a new frontier of cancer liquid biopsy., Mol. Cancer. 21 (2022) 56. [CrossRef]
- E. Diomaiuto, V. Principe, A. De Luca, F. Laperuta, C. Alterisio, A. Di Loria, Exosomes in Dogs and Cats: An Innovative Approach to Neoplastic and Non-Neoplastic Diseases., Pharmaceuticals (Basel). 14 (2021). [CrossRef]
- R.J. Simpson, S.S. Jensen, J.W.E. Lim, Proteomic profiling of exosomes: current perspectives., Proteomics. 8 (2008) 4083–4099. [CrossRef]
- A.A. Novais, G.H. Tamarindo, L.G. de A. Chuffa, D.A.P. de C. Zuccari, Decoding hidden messengers: proteomic profiling of exosomes in mammary cancer research., Biomedicines. 11 (2023). [CrossRef]
- S. Rontogianni, E. Synadaki, B. Li, M.C. Liefaard, E.H. Lips, J. Wesseling, W. Wu, M. Altelaar, Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping., Commun. Biol. 2 (2019) 325. [CrossRef]
- M. Goldschmidt, L. Peña, R. Rasotto, V. Zappulli, Classification and grading of canine mammary tumors., Vet. Pathol. 48 (2011) 117–131. [CrossRef]
- J. Xia, N. Psychogios, N. Young, D.S. Wishart, MetaboAnalyst: a web server for metabolomic data analysis and interpretation., Nucleic Acids Res. 37 (2009) W652-60. [CrossRef]
- P.D. Thomas, D. Ebert, A. Muruganujan, T. Mushayahama, L.-P. Albou, H. Mi, PANTHER: Making genome-scale phylogenetics accessible to all., Protein Sci. 31 (2022) 8–22. [CrossRef]
- D. Szklarczyk, R. Kirsch, M. Koutrouli, K. Nastou, F. Mehryary, R. Hachilif, A.L. Gable, T. Fang, N.T. Doncheva, S. Pyysalo, P. Bork, L.J. Jensen, C. von Mering, The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest., Nucleic Acids Res. 51 (2023) D638–D646. [CrossRef]
- C. Théry, K.W. C. Théry, K.W. Witwer, E. Aikawa, M.J. Alcaraz, J.D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G.K. Atkin-Smith, D.C. Ayre, J.-M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N.N. Bauer, A.A. Baxter, M. Bebawy, C. Beckham, A. Bedina Zavec, A. Benmoussa, A.C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F.E. Borràs, S. Bosch, C.M. Boulanger, X. Breakefield, A.M. Breglio, M.Á. Brennan, D.R. Brigstock, A. Brisson, M.L. Broekman, J.F. Bromberg, P. Bryl-Górecka, S. Buch, A.H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E.I. Buzás, J.B. Byrd, G. Camussi, D.R. Carter, S. Caruso, L.W. Chamley, Y.-T. Chang, C. Chen, S. Chen, L. Cheng, A.R. Chin, A. Clayton, S.P. Clerici, A. Cocks, E. Cocucci, R.J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F.A. Coumans, B. Coyle, R. Crescitelli, M.F. Criado, C. D’Souza-Schorey, S. Das, A. Datta Chaudhuri, P. de Candia, E.F. De Santana, O. De Wever, H.A. Del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L.C. Dieterich, V. Dolo, A.P. Dominguez Rubio, M. Dominici, M.R. Dourado, T.A. Driedonks, F.V. Duarte, H.M. Duncan, R.M. Eichenberger, K. Ekström, S. El Andaloussi, C. Elie-Caille, U. Erdbrügger, J.M. Falcón-Pérez, F. Fatima, J.E. Fish, M. Flores-Bellver, et al., Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines., J. Extracell. Vesicles. 7 (2018) 1535750. [CrossRef]
- M. Szajnik, M. Derbis, M. Lach, P. Patalas, M. Michalak, H. Drzewiecka, D. Szpurek, A. Nowakowski, M. Spaczynski, W. Baranowski, T.L. Whiteside, Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy., Gynecol Obstet (Sunnyvale). Suppl 4 (2013) 3. [CrossRef]
- M. Aguilera-Rojas, B. Badewien-Rentzsch, J. Plendl, B. Kohn, R. Einspanier, Exploration of serum- and cell culture-derived exosomes from dogs., BMC Vet. Res. 14 (2018) 179. [CrossRef]
- J. Maia, S. Caja, M.C. Strano Moraes, N. Couto, B. Costa-Silva, Exosome-Based Cell-Cell Communication in the Tumor Microenvironment., Front. Cell Dev. Biol. 6 (2018) 18. [CrossRef]
- M. Żmigrodzka, O. Witkowska-Piłaszewicz, A. Rzepecka, A. Cywińska, D. Jagielski, A. Winnicka, Extracellular Vesicles in the Blood of Dogs with Cancer-A Preliminary Study., Animals (Basel). 9 (2019). [CrossRef]
- A. Hamm, J. Veeck, N. Bektas, P.J. Wild, A. Hartmann, U. Heindrichs, G. Kristiansen, T. Werbowetski-Ogilvie, R. Del Maestro, R. Knuechel, E. Dahl, Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis., BMC Cancer. 8 (2008) 25. [CrossRef]
- X. Jiang, X.-Y. Bai, B. Li, Y. Li, K. Xia, M. Wang, S. Li, H. Wu, Plasma Inter-Alpha-Trypsin Inhibitor Heavy Chains H3 and H4 Serve as Novel Diagnostic Biomarkers in Human Colorectal Cancer., Dis. Markers. 2019 (2019) 5069614. [CrossRef]
- A.T. Kopylov, A.A. Stepanov, K.A. Malsagova, D. Soni, N.E. Kushlinsky, D.V. Enikeev, N.V. Potoldykova, A.V. Lisitsa, A.L. Kaysheva, Revelation of proteomic indicators for colorectal cancer in initial stages of development., Molecules. 25 (2020). [CrossRef]
- I.van den Broek, R.W. Sparidans, A.W.J. van Winden, M.-C.W. Gast, E.J. van Dulken, J.H.M. Schellens, J.H. Beijnen, The absolute quantification of eight inter-α-trypsin inhibitor heavy chain 4 (ITIH4)-derived peptides in serum from breast cancer patients., Proteomics Clin. Appl. 4 (2010) 931–939. [CrossRef]
- A.W.J. Opstal-van Winden, E.J.M. Krop, M.H. Kåredal, M.-C.W. Gast, C.H. Lindh, M.C. Jeppsson, B.A.G. Jönsson, D.E. Grobbee, P.H.M. Peeters, J.H. Beijnen, C.H. van Gils, R.C.H. Vermeulen, Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study., BMC Cancer. 11 (2011) 381. [CrossRef]
- J. Yang, X. Xiong, S. Liu, J. Zhu, M. Luo, L. Liu, L. Zhao, Y. Qin, T. Song, C. Huang, Identification of novel serum peptides biomarkers for female breast cancer patients in Western China., Proteomics. 16 (2016) 925–934. [CrossRef]
- A.T. Hoang, B. Vizio, L. Chiusa, A. Cimino, D. Solerio, N.H. Do, S. Pileci, M. Camandona, G. Bellone, Impact of tissue enolase 1 protein overexpression in esophageal cancer progression., Int. J. Med. Sci. 18 (2021) 1406–1414. [CrossRef]
- Y. Song, Q. Luo, H. Long, Z. Hu, T. Que, X. Zhang, Z. Li, G. Wang, L. Yi, Z. Liu, W. Fang, S. Qi, Correction: Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma., Mol. Cancer. 13 (2015) 235. [CrossRef]
- Y. Zhang, Q. Li, Z. Huang, B. Li, E.C. Nice, C. Huang, L. Wei, B. Zou, Targeting glucose metabolism enzymes in cancer treatment: current and emerging strategies., Cancers (Basel). 14 (2022). [CrossRef]
- G. Qiao, A. Wu, X. Chen, Y. Tian, X. Lin, Enolase 1, a moonlighting protein, as a potential target for cancer treatment., Int. J. Biol. Sci. 17 (2021) 3981–3992. [CrossRef]
- G.H. Tamarindo, A.A. Novais, L.G.A. Chuffa, D.A.P.C. Zuccari, Metabolic alterations in canine mammary tumors., Animals (Basel). 13 (2023). [CrossRef]
- P.-Y. Chu, N.C. Hsu, A.T. Liao, N.-Y. Shih, M.-F. Hou, C.-H. Liu, Overexpression of α-enolase correlates with poor survival in canine mammary carcinoma., BMC Vet. Res. 7 (2011) 62. [CrossRef]
- M. Zamani-Ahmadmahmudi, S.M. Nassiri, R. Rahbarghazi, Serological proteome analysis of dogs with breast cancer unveils common serum biomarkers with human counterparts., Electrophoresis. 35 (2014) 901–910. [CrossRef]
- H. Zhu, X. Yang, J. Liu, L. Zhou, C. Zhang, L. Xu, Q. Qin, L. Zhan, J. Lu, H. Cheng, X. Sun, Eukaryotic elongation factor 2 kinase confers tolerance to stress conditions in cancer cells., Cell Stress Chaperones. 20 (2015) 217–220. [CrossRef]
- B. Zhang, J. Zou, Q. Zhang, Z. Wang, N. Wang, S. He, Y. Zhao, C.B. Naman, Progress in the development of eukaryotic elongation factor 2 kinase (eef2k) natural product and synthetic small molecule inhibitors for cancer chemotherapy., Int. J. Mol. Sci. 22 (2021). [CrossRef]
- R.-X. Wang, X.-E. Xu, L. Huang, S. Chen, Z.-M. Shao, eEF2 kinase mediated autophagy as a potential therapeutic target for paclitaxel-resistant triple-negative breast cancer., Ann. Transl. Med. 7 (2019) 783. [CrossRef]
- C. Guo, S. Liu, J. Wang, M.-Z. Sun, F.T. Greenaway, ACTB in cancer., Clin. Chim. Acta. 417 (2013) 39–44. [CrossRef]
- C. Fang, J.-J. Li, T. Deng, B.-H. Li, P.-L. Geng, X.-T. Zeng, Actinin-4 as a Diagnostic Biomarker in Serum of Breast Cancer Patients., Med. Sci. Monit. 25 (2019) 3298–3302. [CrossRef]
- Lenčo, A. Fučíková, J. Dresler, L. Čápková, R. Hrstka, R. Nenutil, P. Bouchal, Targeted proteomics driven verification of biomarker candidates associated with breast cancer aggressiveness., Biochim. Biophys. Acta Proteins Proteom. 1865 (2017) 488–498. [CrossRef]
- D. Tentler, E. Lomert, K. Novitskaya, N.A. Barlev, Role of ACTN4 in tumorigenesis, metastasis, and EMT., Cells. 8 (2019). [CrossRef]
- N. Wang, Q. Wang, H. Tang, F. Zhang, Y. Zheng, S. Wang, J. Zhang, Z. Wang, X. Xie, Direct inhibition of ACTN4 by ellagic acid limits breast cancer metastasis via regulation of β-catenin stabilization in cancer stem cells., J. Exp. Clin. Cancer Res. 36 (2017) 172. [CrossRef]
- L. Chung, K. Moore, L. Phillips, F.M. Boyle, D.J. Marsh, R.C. Baxter, Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer., Breast Cancer Res. 16 (2014) R63. [CrossRef]
- F.-U.-H. Nasim, S. Ejaz, M. Ashraf, A.R. Asif, M. Oellerich, G. Ahmad, G.A. Malik, Attiq-Ur-Rehman, Potential biomarkers in the sera of breast cancer patients from bahawalpur, pakistan., Biomark. Cancer. 4 (2012) 19–34. [CrossRef]
- S. Sharma, L. Malhotra, P. Mukherjee, N. Kaur, T. Krishanlata, C.V. Srikanth, V. Mishra, B.D. Banerjee, A.S. Ethayathulla, R.S. Sharma, Putative interactions between transthyretin and endosulfan II and its relevance in breast cancer., Int. J. Biol. Macromol. 235 (2023) 123670. [CrossRef]
- L. Ren, J. Yi, W. Li, X. Zheng, J. Liu, J. Wang, G. Du, Apolipoproteins and cancer., Cancer Med. 8 (2019) 7032–7043. [CrossRef]
- J.J.P. Kastelein, W.A. van der Steeg, I. Holme, M. Gaffney, N.B. Cater, P. Barter, P. Deedwania, A.G. Olsson, S.M. Boekholdt, D.A. Demicco, M. Szarek, J.C. LaRosa, T.R. Pedersen, S.M. Grundy, TNT Study Group, IDEAL Study Group, Lipids, apolipoproteins, and their ratios in relation to cardiovascular events with statin treatment., Circulation. 117 (2008) 3002–3009. [CrossRef]
- Y. He, J. Chen, Y. Ma, H. Chen, Apolipoproteins: New players in cancers., Front. Pharmacol. 13 (2022) 1051280. [CrossRef]
- Y.-C. Lee, C.-T. Tang, J.-Y. Kan, C.-P. Chiang, H.-T. Li, J.-N. Sung, W.-C. Chiu, Correlation of Beta2-Glycoprotein I With Tumor Prognosis in Breast Cancer Patients., Anticancer Res. 43 (2023) 3455–3462. [CrossRef]
- X. Lin, S. Hong, J. Huang, Y. Chen, Y. Chen, Z. Wu, Plasma apolipoprotein A1 levels at diagnosis are independent prognostic factors in invasive ductal breast cancer., Discov. Med. 23 (2017) 247–258.
- R.M. Baig, I. Mahjabeen, M. Sabir, N. Masood, K. Ali, F.A. Malik, M.A. Kayani, Mutational spectrum of Gelsolin and its down regulation is associated with breast cancer., Dis. Markers. 34 (2013) 71–80. [CrossRef]
- L.M. Mielnicki, A.M. Ying, K.L. Head, H.L. Asch, B.B. Asch, Epigenetic regulation of gelsolin expression in human breast cancer cells., Exp. Cell Res. 249 (1999) 161–176. [CrossRef]
- J.S. Winston, H.L. Asch, P.J. Zhang, S.B. Edge, A. Hyland, B.B. Asch, Downregulation of gelsolin correlates with the progression to breast carcinoma., Breast Cancer Res. Treat. 65 (2001) 11–21. [CrossRef]
- A.-M. Stock, F. Klee, K. Edlund, M. Grinberg, S. Hammad, R. Marchan, C. Cadenas, B. Niggemann, K.S. Zänker, J. Rahnenführer, M. Schmidt, J.G. Hengstler, F. Entschladen, Gelsolin Is Associated with Longer Metastasis-free Survival and Reduced Cell Migration in Estrogen Receptor-positive Breast Cancer., Anticancer Res. 35 (2015) 5277–5285.
- J. Rao, D. Seligson, H. Visapaa, S. Horvath, M. Eeva, K. Michel, A. Pantuck, A. Belldegrun, A. Palotie, Tissue microarray analysis of cytoskeletal actin-associated biomarkers gelsolin and E-cadherin in urothelial carcinoma., Cancer. 95 (2002) 1247–1257. [CrossRef]
- A. Van den Abbeele, V. De Corte, K. Van Impe, E. Bruyneel, C. Boucherie, M. Bracke, J. Vandekerckhove, J. Gettemans, Downregulation of gelsolin family proteins counteracts cancer cell invasion in vitro., Cancer Lett. 255 (2007) 57–70. [CrossRef]
- Z.-Y. Chen, P.-W. Wang, D.-B. Shieh, K.-Y. Chiu, Y.-M. Liou, Involvement of gelsolin in TGF-beta 1 induced epithelial to mesenchymal transition in breast cancer cells., J. Biomed. Sci. 22 (2015) 90. [CrossRef]
- Y. Zhang, X. Luo, J. Lin, S. Fu, P. Feng, H. Su, X. He, X. Liang, K. Liu, W. Deng, Gelsolin Promotes Cancer Progression by Regulating Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma and Correlates with a Poor Prognosis., J. Oncol. 2020 (2020) 1980368. [CrossRef]
- E. Tagliabue, S. Raimondi, S. Gandini, Meta-analysis of vitamin D-binding protein and cancer risk., Cancer Epidemiol. Biomarkers Prev. 24 (2015) 1758–1765. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




