Introduction
An Introduction to the Problem
Before a new medicine can be used, it needs to be tested in the context it has been developed for, very often a whole human. In principle this is to examine two things: 1/ that it is effective against the problem, for example a virus or cancer, and 2/ that it does not appear to cause harm. We might imagine two cell types: those afflicted or infected and those not. However, while this often appears to be an oversimplification, any medicine should function with specificity to a degree able to neutralize diseased cells or make them better while not affecting vital cells or tissue to any extent that could compromise viability of the organism. Thus, it can be seen that however which way we look at this type of problem when it comes to the consideration of the context and searching for medicines there is a mutually exclusive conditional requirement for specificity. And interestingly a specific we are informed is an outdated term for a medicine (OED, 1989).
This requirement is not a problem per se but hitherto the way to achieve this I believe does present a problem and helps to explain not only the lengthy and costly process of research and development (R&D) but also the fact that many potential medicines fail. Thus any method that can speed up this process and importantly enable a better determination of potential therapeutic targets and medicines and their potential for mutually exclusive specificity with respect to the context at an early stage in R&D does appear sought after and should be of high interest.
Some Reasons for the Problem: The Consideration of the Context, Relationship and the Use of Scientific Induction
Context can be everything including the cells and things (e.g. components such as nucleotides, proteins, lipids, sugars etc. or any other functional entity) they consist of and their relationships with each other and the sum of this in space-time. Thus, context can be described by the sum of relationships or simply: context = relationship (
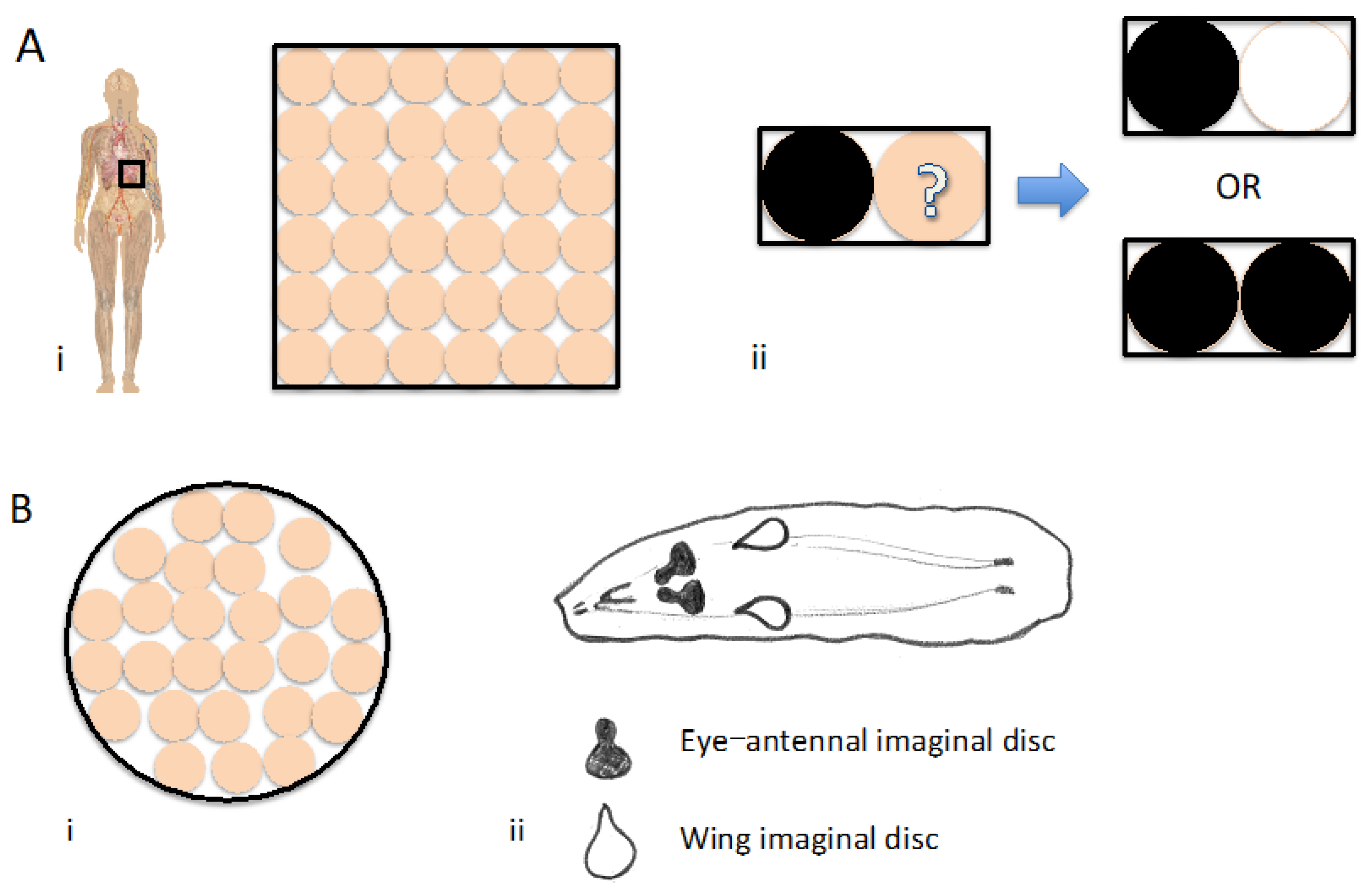
Figure 1a). Therefore it seems apparent that any consideration of a context should include a corresponding relationship.
Regardless of the number of units that make-up a system and the complex relationships that describe them, these can still be considered as few if considered together, even as few as one context of the whole organism. In terms of a relationship to describe any part of this space-time it can be summed up most simply as between two parts: the part in question and the rest. Therefore, if a difference between two parts of the system is the minimum required for a description (
Figure 1a) or with the desired aim in mind to elucidate specificity, then it should become apparent that a mutually exclusive relationship is not only sufficient but required to meet our aim.
Thus, when it comes to studies in the life sciences that can be categorized as in the earlier stages of R&D, a reasonable question might be: what can we say with respect to the context studied in terms of the relationships they include (
Figure 1b)? The Scientific method used in Life science relies heavily on a process of logic involving inductive reasoning something that Alan Turing explains and terms Scientific induction (Turing, 1950). Essentially, any attempt at a conclusion is limited and can only be said to appear to be. In many instances, additional validation studies are essentially only a repeat of the first type of one-dimensional type of study i.e. examination in another part of the organism or in a part or parts of different model systems generally accepted to resemble more closely humans. A mutually exclusive relationship in the context is often not considered in the study, until a clinical trial. Importantly, when it comes to consideration of the context, Scientific induction for the most part does not consider the whole and does not include a mutually exclusive relationship required to examine where a phenomenon does not occur – this I believe is the important limitation, not necessarily an oversight since previously technology has been lacking. Taken together, this I believe helps explain why on a higher proportion of occasions many potential therapeutics appearing to hold great promise, proceed to the later stages of R&D but fail.
Figure 1.
The consideration of context in the life sciences A) i: A schematic of a human body with a representation of a field of cells. ii: A system can be described by considering the relationship between two parts. B) i: A schematic of human cells in a cell culture dish. ii: a drawing of a Drosophila wandering 3rd instar larva with depictions showing the eye-antennae and the wing imaginal discs and their approximate positions. C) Fly mutually exclusive computing. i: Cross scheme. ii: A representation of the four possible results or outputs. D) A schematic representation of cells showing how directed expression (green) and the use of a repressor system (red) can be used to generate a complementary pattern.
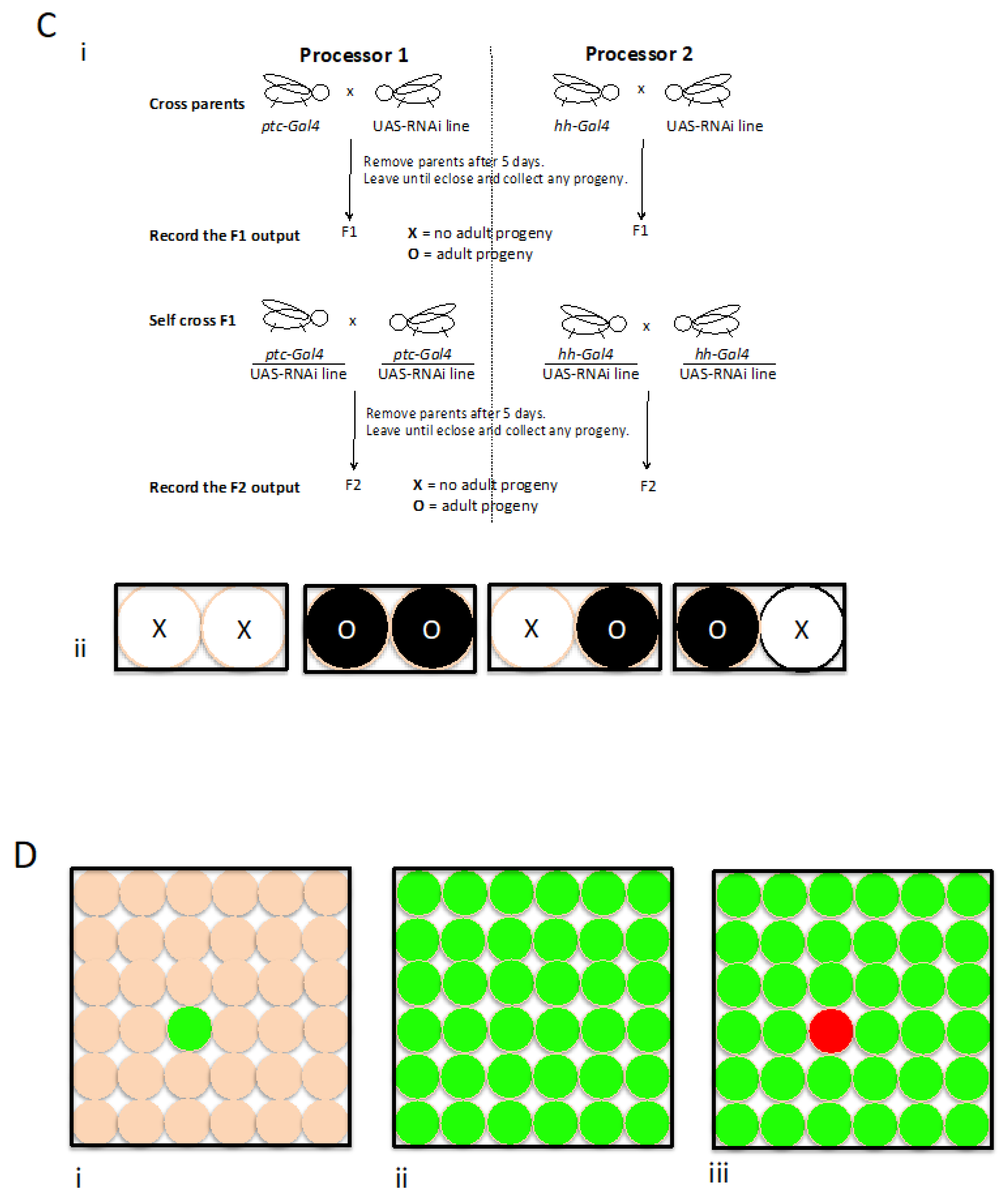
Figure 1.
The consideration of context in the life sciences A) i: A schematic of a human body with a representation of a field of cells. ii: A system can be described by considering the relationship between two parts. B) i: A schematic of human cells in a cell culture dish. ii: a drawing of a Drosophila wandering 3rd instar larva with depictions showing the eye-antennae and the wing imaginal discs and their approximate positions. C) Fly mutually exclusive computing. i: Cross scheme. ii: A representation of the four possible results or outputs. D) A schematic representation of cells showing how directed expression (green) and the use of a repressor system (red) can be used to generate a complementary pattern.
A Solution to the Problem: Drosophila and Mutually Exclusive Computation
The Fruit fly Drosophila Melanogaster is a powerful cost effective in vivo research tool with a quick generation time and technologies for controlling directed gene expression both endogenous and of exogenously introduced nucleotides (Spratt, 2013), not to mention a possibility to easily inject things (Garen & Gehring, 1972; Zalokar, 1971). Thus, the use of Drosophila as a general tool to assess things in real life is probably unsurpassed compared to the possibilities and practical limitations of other systems. In addition, the biology important for life is vital and therefore by definition is highly conserved meaning that most, if not all things or at least parts of the things involved in viability of the system will share a common history with all other life systems i.e. there is commonality. Hence, the inexhaustible effort to record every nucleotide sequence possible and thus generating the consensus genome sequence for comparative genomics and traceability of things vital. This reasoning, allows us to use Drosophila as a model to study human disease, identifying interacting genes for things or components that translate directly to the physiological requirements for putative orthologous components in mammalian cells: for example for parts of a virus (Hao et al., 2008; Merkling et al., 2019) or genes implicated in cancer (Ingham, 2018).
To clarify what I am trying to ask here: if the context and relationship we have interest in, is the whole organism and a mutually exclusive conditional requirement for specificity, this often being the ultimate aim of many studies, then is there an ethical way to include this in our study at an early stage before clinical trials? Thus, with this question in mind I would like to highlight in this report an example of technology able to achieve this (Spratt, 2013) (
Figure 1c). Here I also report the identification, of the putative protein encoding gene CG43658. I believe this emphasizes further the power of this, for the most part overlooked, Scientific method and it`s suitability for generating sufficient data to support the prioritization of things identified.
Materials and Methods
RNAi line with TRiP ID HMS00332 (Drosophila RNAi Screening Center). All other materials and methods are as described in Spratt, 2013.
Results and Discussion
A Computer Able to Identify Components with Mutually Exclusive Specificity
In this instance, with respect to testing components in the context including a mutually exclusive relationship this can be modeled by the use of two directed expression drivers describing the pattern of expression of the receptor Patched (Ptc) and the signal Hedgehog (Hh) for the highly conserved and medically relevant Hedgehog signaling -
ptc-Gal4 and
hh-Gal4, respectively (
Figure 1c; Spratt, 2013). Briefly, this method is able to process nucleotides and does so in this instance using RNA interference (RNAi) and describes the context in terms of viability once processed through both parts, i.e it can be said to consider the whole of the system giving an output or result to describe the sum of everything in it absolutely recorded using unambiguous terms: using X for non-viable and O for viable. The two parts share a relationship, enabling us to interpret something meaningful from the results. This can return four possible results XX, OO, XO or OX with the later two being of interest with respect to specificity. Furthermore, I would like to reiterate because I find it striking that in this Hh signalling context that we know is used again and again throughout development and in the adult, sometimes the information or thing being processed causes the organism to be non-viable appearing to change the cell fate or is toxic when processed through one part of the system while in the complementary part, through two generations, the flies are viable or the information or thing being processed does not appear to change the cell fate or is not toxic to at least enough cells for the organism to be viable. I am always somehow humbled at the thought of this and it may be worth considering whether in all instances this can be explained sufficiently by a component sharing a relationship with Hh signaling or not.
Identification of CG43658
Previously this method identified
CG31522, a putative Elongation of Very Long Chain fatty acids (ELOVL), whose protein is shown to physical interact with ATP synthase and protein encoded by it`s adjacent and highly similar sequence
CG31523 (see flybase.org). Interestingly, CG31523 has since been found to co-immunoprecipitate the Hh signalling component Rab23 (Cicek, et al., 2016), a GTPase. Furthermore, ELOVL6 has been shown to regulate mitochondrial function and heat generation (Tan et al., 2015) and Hh pathway activation can increases mitochondrial capacity (Yao et al., 2017). Here, with the notion that I am describing to you a Computer able to identify things, beyond a reasonable doubt, relevant to the context and showing specificity, I would like to report the identification of
CG43658 a putative Rho guanine nucleotide exchange factor, not reported in the initial study because it was deemed to be insufficiently characterized, with an XO output (
Table 1). Interestingly,
CG43658`s orthologue
ARHGEF10 is an inherited peripheral neuropathy gene associated with Charcot-Marie-Tooth disease and Drosophila
CG43658 appears to offer an appropriate model (Yamaguchi and Takashima, 2018). Furthermore, Schwann cell derived Desert Hedgehog (DHh) controls development of the peripheral nerve sheath (Parmantier et al., 1999) and
DHh is associated with peripheral neuropathy (Umehara et al., 2000).
A Turing Machine Able to Consider the Whole and Describe it Absolutely in Unambiguous Terms
This method can use other mutually exclusive relationships, for example using drivers representing other cell to cell communication mechanisms. Furthermore, by use of a repressor technology such as GAL80, a GAL4 repressor (Ma and Ptashne, 1987; Lee and Luo, 1999) in combination with a global driver e.g.
tubulin-GAL4 the method might be able be used to study the context while including potentially all possible mutually exclusive relationships, at least as determined by the site of a driver`s insertion into the genome and it`s particular expression pattern e.g. if a gene of interest, let`s call it
x can drive the GAL80 repressor in a global GAL4 background then you can have the tools to direct the expression of things in those cells expressing
x i.e. using
x-GAL4 and also now have the ability to direct expression everywhere else or NOT
x by using
x-GAL80;
tubulin-GAL4 (
Figure 1d; Spratt, 2013). This is how we might envisage that this target and potential therapeutic testing device will evolve, adjustable or tuned both on the level of the inputs and on the level of control according to the directed expression and with a possibility to adjust the background: an imaginable Computer able to search the context for mutually exclusive relationships of interest and use them to identify things with specificity.
Interestingly, with respect to finding new things and then feeding the information back to carry out new computations what we can see is that this description is a Turing machine (Turing, 1936). I think also it is worth mention considering Alan`s comments when it comes to Scientific induction (Turing, 1950): this is not Scientific induction. This is a computer or more precisely, as Alan cleverly explains when it comes to definitions – this concept is a Turing machine. And a Turing machine or Computer that is able to consider the whole and describe the sum of everything in it absolutely in unambiguous terms is a useful tool because it can tell you about things and not how things appear to be but how things really are according to the information available.
Further Considerations and Implications
Further interesting follow on ideas from this might be the use of a component known to generate an XO signature, and then process this type of control through the context using drivers with different mutually exclusive relationships until you find further XO or vice versa combinations. Another interesting question that springs to mind is could it be that there are certain combinations of mutually exclusive relationships for interrogating components in the context able to provide enough information with respect to any or at least many newly interesting targets or functional entities: for example, if we examine results for known approved medicines or their targets, that the results positively correlate i.e. with time, from this type of test and the data generated we might realize that the fly or a similar two-part test provides enough information to fast-track a candidate and potential therapeutic for drug development and ultimately clinical trials?
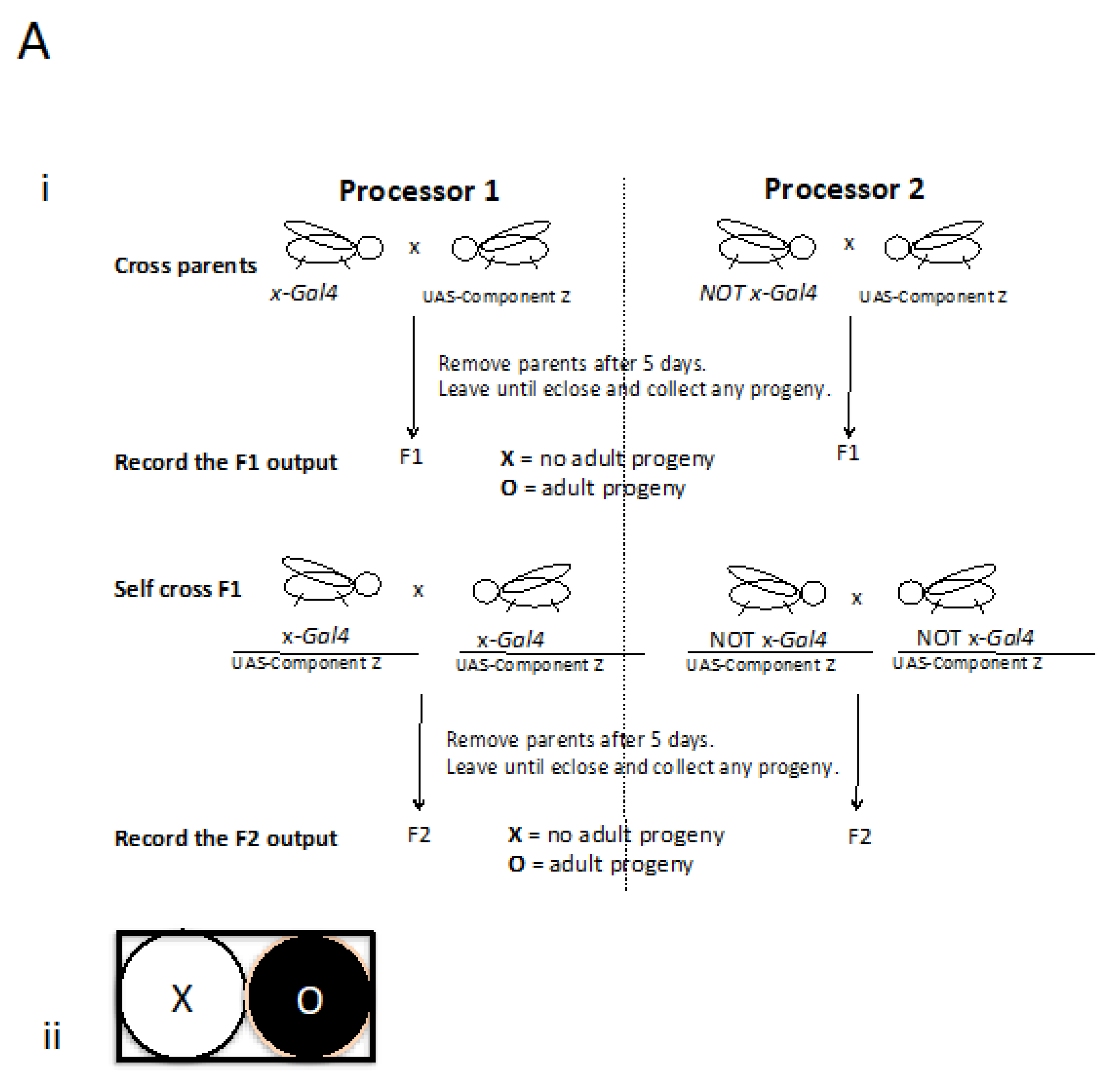
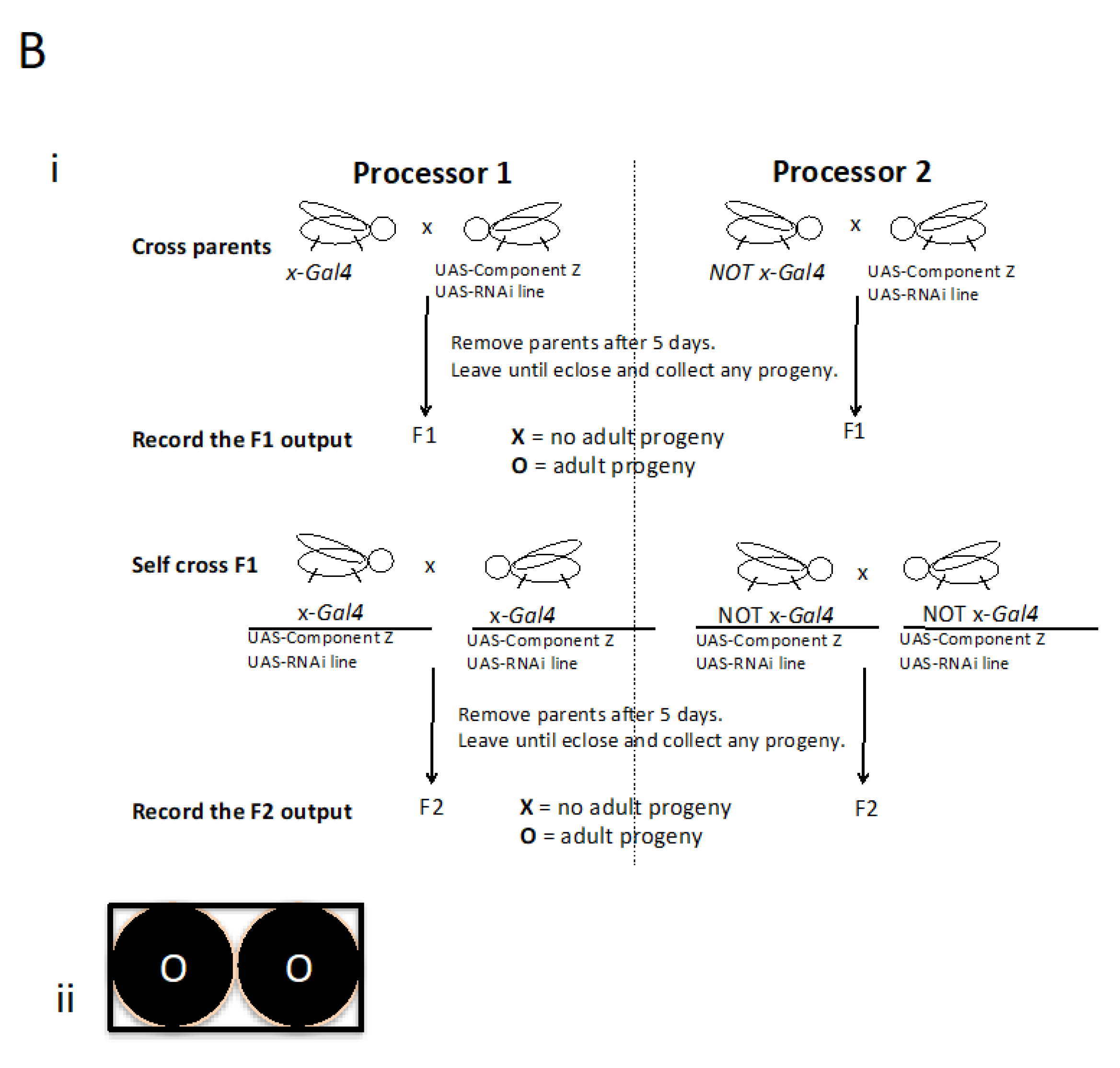
Recently, COVID-19 (Hu et al., 2020; Sekhawat et al., 2020) has highlighted the time it takes to find a solution and, together with the anticipation of emerging disease, questioned our security. Therefore with this imaginable Computer in mind, I would like to outline a hypothetical scenario: let`s imagine part of a virus e.g. SARS-CoV-2 is a timely example, but also any gene or mutation causing disease. Using molecular biology, this could be any part or combination, let`s call this component Z and imagine with the use of appropriate drivers this might generate an XO or vice versa output (
Figure 2a: please allow for this conjecture based on prior knowledge and first principles that give us good reason to believe that an example may exist). The context can now be searched for e.g. a potential nucleotide candidate medicine that can bring about a change in the output (
Figure 2b). Concerning potential medicines the use of nucleotides have progressed (for informative reviews concerning the state of the art please see e.g. Yoshioka et al., 2019; Yamakawa et al., 2019) so these, in addition to vaccines, should be considered. Interestingly, I must add that a number of viruses modulate Hedgehog signaling (Smelkinson et al., 2017).
To end with. We are told, circa 330 BC, Aristotle said something like “To say that that which is, is not OR that which is not, is – is a falsehood and to say that that which is, is AND that which is not, is not – is true”.
Figure 2.
A hypothetical or imaginable Fly Computer. A) i: Mutually exclusive GAL4 drivers described by x and NOT x are used to interrogate a component Z to identify a relevant context. ii: If a computation represented in Ai generates an XO output, it can now be interrogated further. B) i: Component Z could now be investigated together with the directed expression of another functional entity, in this example an RNAi. ii: The idea being that a change in the XO output shown in Ai to OO would be of high interest.
Figure 2.
A hypothetical or imaginable Fly Computer. A) i: Mutually exclusive GAL4 drivers described by x and NOT x are used to interrogate a component Z to identify a relevant context. ii: If a computation represented in Ai generates an XO output, it can now be interrogated further. B) i: Component Z could now be investigated together with the directed expression of another functional entity, in this example an RNAi. ii: The idea being that a change in the XO output shown in Ai to OO would be of high interest.
Acknowledgements
I would like to thank everybody for making all this possible. Special mentions for Michael Ebenazer Kwadjo Omari Owuo Jr., Edward Christopher Sheeran, Damini Ebunoluwa Ogulu, Own it (Atlantic). Ariana Grande-Butera, Into you (Republic). Adam Richard Wiles and Samuel Frederick Smith, Promises (Columbia), Calvin Cordozar Broadus Jr. & Terius Youngdell Nash, Gangsta Luv (Priority), Andreas Kleerup and Titiyo Yambalu Felicia Jah, Trouble (U OK?), Lisa Fischer and the Rolling Stones, Gimme Shelter (Decca), Gordon Matthew Thomas Sumner, Andy Summers and Stewart Copeland, Roxanne (A&M), Pharrell Williams, Freedom (Columbia), Kate Bush, Running Up That Hill (EMI), Peroulakis Nikos & Zinos, Olos o Kosmos Thalassa (Independent), Ichiro Yamaguchi and Sakanaction, Music (サカナクション - ミュージック) (Victor), Benito Antonio Martinez Ocasio, Where She Goes (Rimas) and with Jhay Cortez, Dakiti (Rimas), Abel Makkonen Tesfaye, Blinding Lights (XO), Beyonce Giselle Knowles-Carter, XO (Parkwood), Barry Gibb and to the late Ivor Neville Kamen, Each Time You Break My Heart (Sire), Robin & Maurice Gibb, Staying Alive (RSO), Leonard Cohen, Anthem (Columbia) and Johann Holzel, Out of the Dark (EMI Electrola).
References
- Cicek, I. O., Karaca, S., Brankatschk, M. Eaton, S., Urlaub, H. and Shcherbata, H. R. (2016). Hedgehog signaling strength is orchestrated by the mir-310 cluster of microRNAs in response to diet. Genetics 202(3): 1167-1183. [CrossRef]
- Garen, A. and Gehring, W. (1972). Repair of the lethal developmental defect in deep orange embryos of Drosophila by injection of normal egg cytoplasm. Proc. Natl. Acad. Sci. USA. 69(10): 2982-5. [CrossRef]
- Hao, L., Sakurai, A., Watanabe, T., Sorensen, E., Nidom, C. A., Newton, M. A., Ahlquist, P. and Kawaoka, Y. (2008). Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454(7206): 890-3. Epub 2008 Jul 9 . [CrossRef]
- Hu, B., Guo, H., Zhou, P. and Shi, Z. (2020). Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews Microbiology 6: 1-14. [CrossRef]
- Ingham, P. W. (2018). From Drosophila segmentation to human cancer therapy. Development 145(21): dev168898. [CrossRef]
- Lee, T. and Luo, L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22(3): 451-461. [CrossRef]
- Ma, J. and Ptashne, M. (1987). The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 50(1): 137-142. [CrossRef]
- Merkling, S. H., Riahi, H., Overheul, G. J., Schenck, A. and van Rij, R. P. (2019). Peroxisome-associated Sgroppino links fat metabolism with survival after RNA virus infection in Drosophila. Sci. Rep. 9(1): 2065. [CrossRef]
- Oxford English Dictionary (OED), 1989. Second Edition revised. Oxford University Press.
- Parmantier, E., Lynn, B., Lawson, D., Turmaine, M., Namini, S. S., Chakrabarti, L., McMahon, A. P., Jessen, K. R. and Mirsky, R. (1999). Schwann cell-derived desert hedgehog controls the development of peripheral nerve sheath. Neuron 23:713-724. [CrossRef]
- Sekhawat, V., Green, A. and Mahadeva, U. (2020). COVID-19 autopsies: conclusions from international studies. Diagn Histopathol (Oxf). [CrossRef]
- Smelkinson, M. G., Guichard, A., Teijaro, J. R., Malur, M., Loureiro, M. E., Jain, P., Ganesan, S., Zuniga, E. I., Krug, R. M., Oldstone, M. B. and Bier E. (2017). Influenza NS1 directly modulates Hedgehog signalling during infection. PLoS Pathog. 13(8):e1006588. 1006588. eCollection 2017 Aug. [CrossRef]
- Spratt, S. J. (2013). A computation using mutually exclusive processing is sufficient to identify specific Hedgehog signaling components. Front. Genet. 4:284. eCollection 2013. [CrossRef]
- Tan, C. Y., Virtue, S. Bidault, G., Dale, M., Hagen, R., Griffin, J. L. and Vidal-Puig, A. (2015). Brown Adipose Tissue Thermogenic Capacity is Regulated by ELOVL6. Cell Rep. 13: 2039-2047. [CrossRef]
- Turing, A. M. (1936). On computable numbers, with an application to the Entscheidungsproblem. Proc. Lond. Math. Soc. 42: 230-265. [CrossRef]
- Turing, A. M. (1950). I.-Computing machinery and Intelligence. Mind 236: 433-460. [CrossRef]
- Umehara, F., Tate, G., Itoh, K., Yamaguchi, N., Douchi, T., Mitsuya, T. and Osame, M. (2000). A Novel Mutation of Desert Hedgehog in a Patient with 46, XY Partial Gonadal Dysgenesis Accompanied by Minifascicular Neuropathy. Am. J. Hum. Genet. 67(5): 1302-1305. [CrossRef]
- Yao, P. J., Manor, U., Petralia, R. S., Brose, R. D., Wu, R. T. Y., Ott, C., Wang, Y-X., Charnoff, A., Lippincott-Schwartz, J. and Mattson, M. P. (2017). Sonic hedgehog pathway activation increases mitochondrial abundance and activity in hippocampal neurons. Mol. Biol. Cell 28: 387-395. [CrossRef]
- Yamaguchi, M. and Takashima, H. (2018). Drosophila Charcot-Marie-Tooth Disease Models. Adv. Exp. Med. Biol. 1076:97-117. [CrossRef]
- Yamakawa, K., Nakano-Narusawa, Y., Hashimoto, N., Yokohira, M. & Matsuda, Y. (2019). Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment. Int. J. Mol. Sci. 20(17). Pii: E4224. [CrossRef]
- Yoshioka, K., Kunieda, T., Asami, Y., Guo, H., Miyata, H., Yoshida-Tanaka, K., Sujino, Y., Piao, W., Kuwahara, H., Nishina, K., Hara, R. I., Nagata, T., Wada, T, Obika, S. and Yokota, T. (2019). Highly Efficient Silencing of microRNA by Heteroduplex Oligonucleotides. Nucleic Acids Res. 47(14): 7321-7332. [CrossRef]
- Zalokar, M. (1971). Transplantation of nuclei in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 68(7): 1539-41.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).