Introduction
This study investigates the presence and potential health benefits of Bifidobacteria within a Swiss fermented milk and colostrum product with a history of use in chronic condition management (Ward et al. 2014; Ruggiero et al. 2014). Prior research has characterized the product’s overall microbial composition, focusing on plasmids, Lactobacillaceae, Lactococcaceae (Pacini and Ruggiero 2019), and bacteriophages (Pacini and Ruggiero 2020). In the present study, the Bifidobacteriaceae family within this product is examined as well as the role of Bifidobacterial enzymes in the endogenous production of GcMAF (Gc protein-derived Macrophage Activating Factor).
Bifidobacteria are a unique group of bacteria. They belong to the order Bifidobacteriales, which interestingly contains only one family—Bifidobacteriaceae (Biavatti and Mattarelli 2018). This is supported by findings from “The All-Species Living Tree” Project (Yarza et al. 2008). Bifidobacteria themselves are gram-positive, non-motile, and anaerobic, thriving in oxygen-free environments. Primarily found in the mammalian gastrointestinal tract (Schell et al. 2002), Bifidobacteria have also been isolated from other locations like the vagina (Albert et al. 2015) and mouth (Bifidobacterium dentium; Inchingolo et al. 2022). Notably, they are a dominant genus contributing to a healthy gut microbiota (Hidalgo-Cantabrana et al. 2017; Schell et al. 2002). Interestingly, before the 1960s, Bifidobacteria were classified as “Lactobacillus bifidus”; advancements in microbiology led to their reclassification as a distinct genus, Bifidobacterium.
Various strains of Bifidobacteria have been linked to several health-supporting properties. Following is a breakdown of some of the potential benefits associated with Bifidobacteria:
Nutrient Absorption: Bifidobacteria can help break down complex carbohydrates and produce short-chain fatty acids (SCFAs) that support gut cell metabolism and promote nutrient absorption (Duranti et al. 2020).
Immune System Support: A healthy gut microbiome rich in Bifidobacteria contributes to a balanced immune response by reducing inflammation and modulating the immune system’s activity (Gavzy et al. 2023).
Diarrhea Prevention: Certain Bifidobacteria strains might help prevent diarrhea caused by rotavirus or antibiotic use by competing with harmful pathogens and supporting a healthy gut barrier. (Bodke and Jogdand 2022).
Improved Bowel Movements: Bifidobacteria can aid digestion and promote regular bowel movements by softening stool and increasing stool frequency (Okada et al. 2023).
Lactose Tolerance: Some Bifidobacteria strains can help break down lactose, the sugar found in milk, potentially easing symptoms in individuals with lactose intolerance (Chen et al. 2021).
Inflammatory Bowel Disease: Studies suggest certain Bifidobacteria strains might play a role in reducing inflammation associated with ulcerative colitis (Yao et al. 2021).
Atopic Dermatitis (Eczema): Bifidobacteria supplementation in infants might be linked to a reduced risk of developing eczema (Kim et al. 2010).
Cancer: Bifidobacteria show anti-cancer effects in colon cancer cells (Faghfoori et al. 2021); studies using preclinical mouse models and human clinical trials have revealed a link between Bifidobacteria and the efficacy of tumor-targeting immunotherapy (Longhi et al. 2020).
Alzheimer’s disease: Treatment with Bifidobacteria can suppress amyloid β accumulation and neuro-inflammation (Wu et al. 2020).
Materials and Methods
This study investigates the microbial composition of a commercially available product called “Freeze-Dried Bravo—Fermented Colostrum and Probiotic Complex” manufactured in Switzerland by Silver Spring Sagl (Arzo-Mendrisio, Ticino). The analysis was performed using the Axiom Microbiome Array; the analysis was carried out by Eurofins Microbiology Laboratories Inc. (New Berlin, Wisconsin, USA). The composition of plasmids, Lactobacillaceae, Lactococcaceae and bacteriophages in this product was previously described elsewhere (Pacini and Ruggiero 2019; Pacini and Ruggiero 2020). For the product under investigation, the manufacturing process involves a 48-hour fermentation of bovine milk and colostrum at room temperature, followed by freeze drying. A separate publication details the product’s nutritional facts and overall microbial composition (Antonucci et al. 2019).
The product object of these studies is manufactured in Switzerland in a Demeter-certified facility. The Demeter biodynamic certification is the oldest ecological certification for agriculture products worldwide. It signifies that a product comes from a facility adhering to the principles of biodynamic agriculture. Biodynamic agriculture has an holistic approach; it goes beyond organic farming by treating the farm or the facility as a self-contained ecosystem and emphasizes a balanced relationship between soil, plants, animals, and workers (Vaish et al. 2024). Biodynamic workers utilize unique practices like composting methods that involve specific preparations made from manure and other natural materials; planting calendars based on lunar cycles; biodynamic sprays made from fermented plant and animal materials.
Demeter certification builds upon and surpasses the requirements for organic certification. It has more stringent regulations around imported fertility sources (manure, compost, etc.) that is encouraging on-site solutions. Weed, pest, and disease control that is emphasizing preventative measures and natural solutions. Water conservation and biodiversity practices. Demeter certification provides a guarantee that the product comes from a facility committed to environmental responsibility, animal welfare, and potentially higher quality produce.
The Axiom Microbiome Array used in this study utilizes a microarray platform, where each probe on the array is designed to detect specific DNA sequences from various microbes (Thissen et al. 2019). The Axiom Microbiome Array works by extracting DNA from the sample and this DNA is then applied to the microarray chip. If the DNA from the sample matches the sequence on a specific probe, it will bind to that probe, indicating the presence of that particular microbe in the sample. Compared to traditional methods like PCR, the Axiom Microbiome Array offers several advantages such as high-throughput: It can analyze a large number of samples simultaneously, making it efficient for large-scale studies.
Comprehensiveness: It can detect a wide range of microorganisms, including known and potentially novel species. Standardized platform: The use of a standardized microarray platform ensures consistency and reproducibility in results across different laboratories. The Axiom Microbiome Array used in this study includes a total of approximately 12,000 species and uses a software program designed to analyze the composition of a microbiome in a sample, the Microbiome Detection Analysis Software (MiDAS). This program works by comparing two values for each organism: Probes Observed and Probes Expected. Probes Observed indicates the number of probes that successfully detected the target organism in the sample. Probes Expected indicates the total number of probes designed to detect that specific organism. MiDAS calculates an Initial Score based on this ratio. A higher score indicates a greater likelihood of the organism being present. MiDAS then uses an iterative process to refine its analysis. MiDAS analyzes the Initial Scores and identifies the organism with the highest score as the most likely to be present. The probes specific to this chosen organism are then removed from the analysis. MiDAS then repeats the process with the remaining probes and organisms. The process of identifying the most likely organism, removing its probes, and re-analyzing continues until adding new organisms no longer improves the accuracy of the analysis is called Iterations. Finally, MiDAS determines positive detection based on probe intensity. A probe is considered positive if its signal is stronger than 99% of random control probes, and detected in at least 20% of the probes designed for that specific target organism. In addition to the Initial Score, the program calculates the Conditional Score because the presence of other microbes in the sample could influence the detection of a particular target. The Conditional Score considers this by taking into account the observed probe data for all targets. It reflects the likelihood of the target being present given the context of the entire microbial community identified in the sample. The Axiom Microbiome Array demonstrates high accuracy in identifying species within mock microbial communities used for testing. It achieved a perfect 100% success rate in these controlled environments (Thissen et al. 2019).
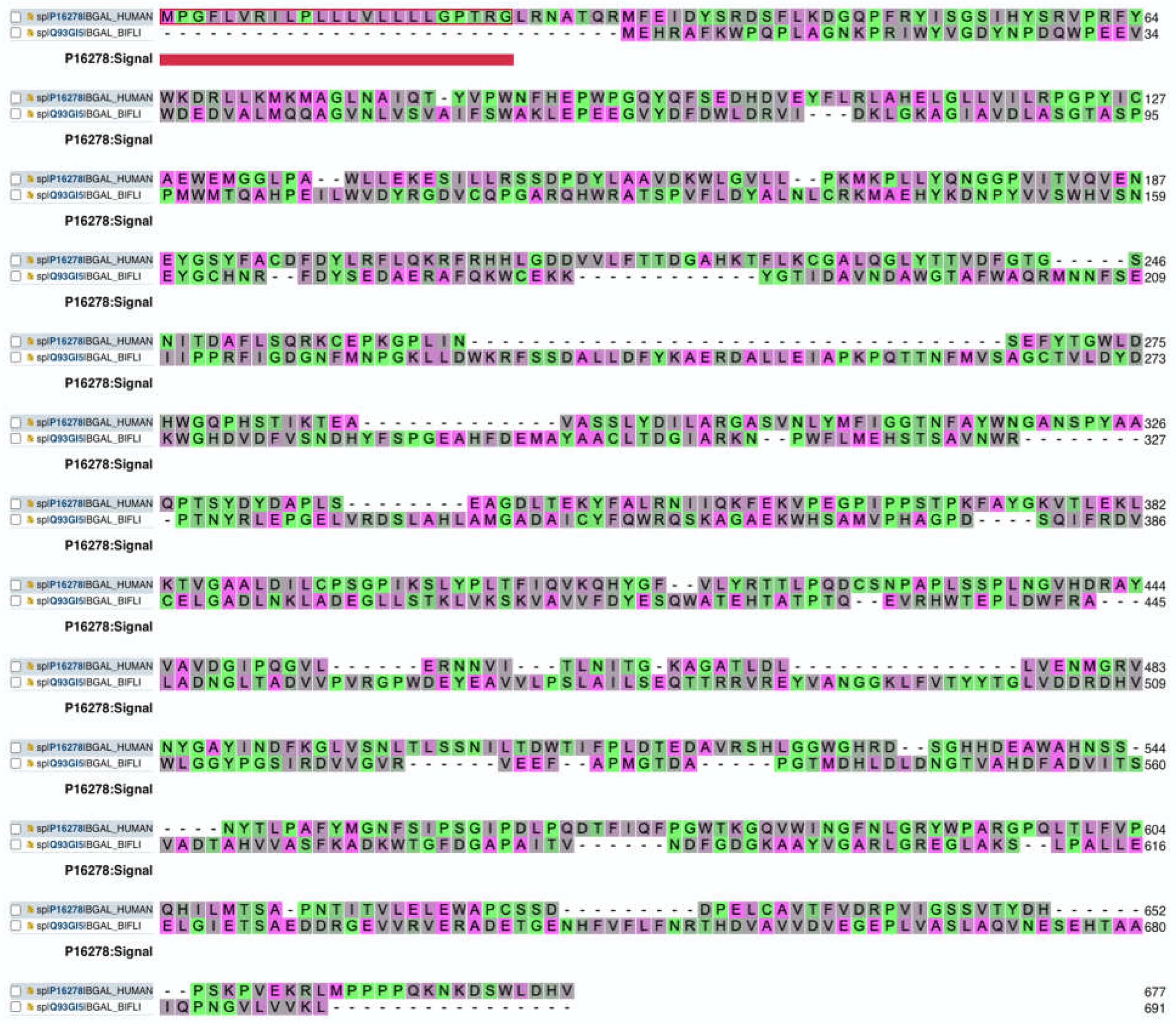
In addition, this study investigates the characteristics of two enzymes synthesized by Bifidobacteria, beta-galactosidase (Hsu et al. 2005) and sialidase (Nishiyama et al. 2018) and compares the amino acid sequences of these enzymes with those of the corresponding human counterparts. Sequences and structures of proteins were studied using the database UniProt and, more specifically, the function designated “align” (UniProt Consortium 2023).
Results
The Axiom Microbiome Array demonstrated the presence of a high number of targets corresponding to microbial species that fit into the definition of probiotics and are members of the families of Bifidobacteriaceae.
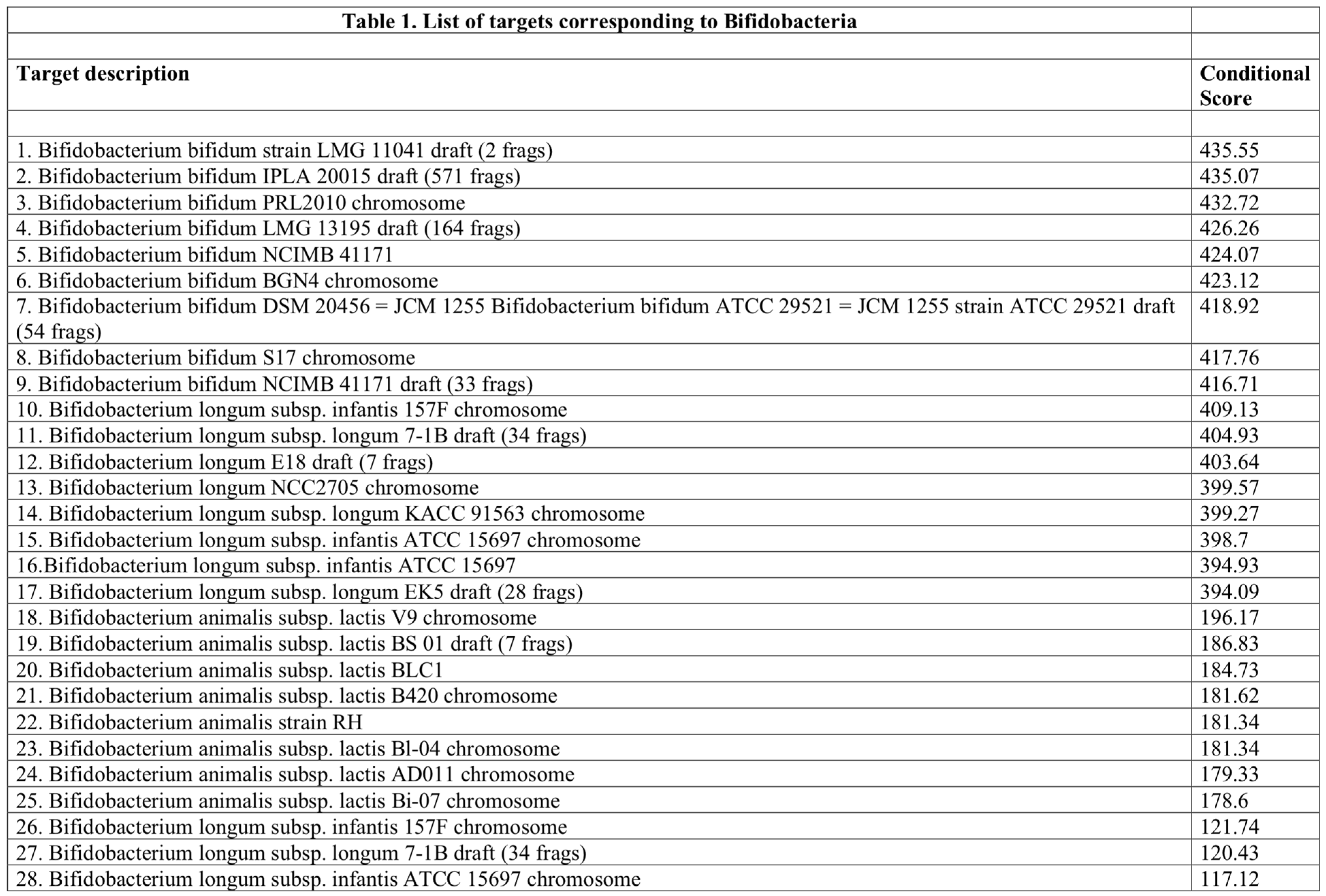
Table 1, shows the composition of the product as far as Bifidobacteriaceae are concerned.
The array provided 28 iterations, each one corresponding to a unique target. The targets are ordered in descending sequence with those showing the highest Conditional Score at the top of the list. Although the assay is not quantitative, it can be deduced that higher Conditional Score corresponds to higher number of copies (Thissen et al. 2019). A higher Conditional Score indicates a stronger likelihood of the target microbe being present in the sample, even if other microbes are also present. A low Conditional Score might suggest the target microbe is less likely to be present, even if the Initial Score was high. This could be due to interference from other microbes or limitations in the assay. The assay uses a threshold for the ratio of Conditional Score to Initial Score. If this ratio is above the threshold, it increases confidence in considering the target microbe as “present” in the sample. As far as other information is concerned, in the context of the Axiom Microbiome Array, “frags” refers to DNA fragments since these probes target specific fragments of the microbial DNA, rather than the entire genome. This is because microarray technology typically works best with shorter DNA fragments for efficient hybridization with the probes and targeting specific informative fragments allows the assay to identify a wider range of microbes within a single test.
The assay is capable of precise microbe identification. For example, entry n. 7
“Bifidobacterium bifidum DSM 20456 = JCM 1255 Bifidobacterium bifidum ATCC 29521 = JCM 1255 strain ATCC 29521 draft (54 frags)”
contains the following information. Bifidobacterium bifidum: This confirms the presence of this particular type of bacteria. DSM 20456 = JCM 1255 = ATCC 29521: This indicates that the assay identified DNA fragments matching multiple reference strains of Bifidobacterium bifidum. These reference strains are identified by their collection numbers in different culture collections: DSM (German Collection of Microorganisms and Cell Cultures); JCM (Japan Collection of Microorganisms); ATCC (American Type Culture Collection). Multiple Matches: The fact that the assay result lists multiple reference strains suggests that the detected DNA fragments match characteristics of all three strains to a significant degree. This is possible because these reference strains are very closely related. However, due to the limitations of targeting specific DNA fragments, the assay might not be able to definitively distinguish between these highly similar strains. Strain ATCC 29521 (draft 54 frags): This clarifies that while the assay detected fragments matching all three reference strains, it provides additional information for the ATCC 29521 strain. Draft (54 frags): Indicates the assay didn’t sequence the entire genome. “54 frags” signifies that the assay detected a higher number of DNA fragments (54) matching the ATCC 29521 strain. This result strongly suggests the presence of a Bifidobacterium bifidum strain very similar to, if not exactly, Bifidobacterium bifidum ATCC 29521 in the sample. The high number of detected fragments (54) strengthens the confidence in this identification.
Another example is provided by entry n. 23
“Bifidobacterium animalis subsp. lactis Bl-04 chromosome”.
This refers to the complete set of genetic material (DNA) for a specific strain of bacteria: Bifidobacterium animalis subspecies lactis Bl-04. Animalis subsp. lactis indicates a specific subspecies within the Bifidobacterium animalis species. Subspecies are closely related groups within a species that have some unique characteristics. Bl-04 refers to a specific strain of Bifidobacterium animalis subsp. lactis. Different strains within a subspecies can have slight variations in their genetic makeup and functional properties. Chromosome refers to the single, circular DNA molecule that contains the complete genetic information for this particular strain (Bl-04) of Bifidobacterium animalis subsp. lactis. By studying the complete chromosome of Bifidobacterium animalis subsp. lactis Bl-04, it is possible to gain valuable insights into genes, unique characteristics, and evolutionary relationships. Identifying the genes present on the chromosome can reveal the potential functionalities of this strain; comparing the chromosome of Bl-04 with other Bifidobacterium strains can help identify the genetic basis of any unique features of this strain, such as probiotic properties; studying the chromosome can shed light on the evolutionary history and relationships between different Bifidobacterium strains. The complete chromosome sequence of Bifidobacterium animalis subsp. lactis Bl-04 is publicly available in online databases like the National Center for Biotechnology Information (NCBI) or GenBank.
Next, the amino acid sequences of beta-galactosidase and sialidase from Bifidobacteria (Hsu et al. 2005; Nishiyama et al. 2018) and humans were compared. These two enzymes were chosen because they are responsible for the production of GcMAF, a potent activator of macrophages, from the Gc protein that is abundant in colostrum (Ruggiero et al. 2016; Nabeshima et al. 2020); it had been previously demonstrated that the product object of this study has a very high GcMAF activity (Carter et al. 2020), possibly due to the enzymatic conversion of the Gc protein by these two enzymes released by Bifidobacteria.
Figure 1, shows the degree of identity and helix propensity of human and Bifidobacterial sialidase in the region where the two sequences were aligned.
Figure 2, shows the results of alignment of human and Bifidobacterial sialidase where positively charged amino acids are highlighted. The rationale for the choice of positively charged amino acids lays in the interaction with the negatively charged sialic acid of GcMAF.
Figure 3, shows the results of alignment of human beta-galactosidase (Oshima et al. 1988) and beta-galactosidase from Bifidobacterium longum (Hsu et al. 2005).
The degree of identity and helix propensity in the region where the two sequences were aligned are highlighted.
Figure 4, shows the results of alignment where negatively charged amino acids are highlighted. The rationale for the choice of negatively charged amino acids lays in the interaction with the positively charged hydroxyl group of the terminal galactose residue that remains exposed once the sialic acid is removed from GcMAF.
Discussion
Bifidobacteria have been known for decades for a plethora of health-supporting properties with potential therapeutic impact in a number of diseases (Chen et al. 2021). It is reasonable to state that the unequalled abundance of Bifidobacteria targets in the product that is the object of this study contributes to its health-promoting effects observed in a number of conditions ranging from cancer to viral infection (Ward et al. 2014; Ruggiero et al. 2014; Branca et al. 2015; Schwalb et al. 2016; Blythe et al. 2017; Carter et al. 2020; Blythe and Ruggiero 2020; Zunaid et al. 2020). It is also reasonable to postulate that a major factor contributing to the health-supporting effects of the Bifidobacteria present in the product, is constituted by the ability of these bacteria to promote the endogenous production of GcMAF in milk and colostrum.
As thoroughly reviewed by Nabeshima et al. (2020), the potent immune stimulant, anti-cancer, anti-angiogenetic factor GcMAF, derives from the Gc protein that is present in blood, milk and colostrum. The concentration of Gc protein in bovine colostrum, that is in the colostrum used to manufacture the product, is close to that of human blood/plasma; 300-600 mg/L in human blood (Nabeshima et al. 2020); 250 mg/L in bovine colostrum (Ena et al. 1992). In human blood, the variant Gc1 of Gc protein is converted to GcMAF through deglycosylation operated by beta-galactosidase of activated B lymphocytes, and sialidase of activated T lymphocytes. These passages of deglycosylation uncover the N-acetylgalactosamine (GalNAc) that is linked to the amino acid Threonine420 (Thr420 or T420) present in domain III of Gc protein, thus permitting the electrostatic interaction between the positively-charged GalNAc of GcMAF and the negatively charged pocket of the GcMAF receptor. The rationale for such a molecular mechanism of action based on electrostatic interactions is reinforced by the observation that the Gc2 variant of human Gc protein, a variant that has Lysine at position 420 instead of Threonine (T420K mutation), and is not glycosylated, functions as GcMAF thanks to the positive charge of Lysine. In the product that is the object of this study, the enzymatic mechanisms responsible for GcMAF generation are operated by the enzymes released by Bifidobacteria, beta-galactosidase and sialidase, which have significant functional similarities with their human counterparts. In other words, during the process of fermentation that is performed to manufacture the product, the enzymes produced by the Bifidobacteria, convert the Gc protein of bovine milk and colostrum into the active GcMAF. Interestingly, however, the overall GcMAF activity of the products is higher than that observed with human blood-derived GcMAF (Carter et al. 2020).
The reason for this greater activity is to be found in the presence of chondroitin sulfate in milk and colostrum together with the ability of Bifidobacteria and other microbes of the product to metabolize it, thus conferring GcMAF-like activity to chondroitin sulfate. We previously hypothesized, in an article quoted by Nabeshima et al. (2020), that chondroitin sulfate has GcMAF-like activity thanks to the presence of GalNAc as one of its two constituent sugars (Ruggiero et al. 2016). It was also hypothesized that chondroitin sulfate could have been responsible for the anticancer effects observed with Coley’s vaccine (Ruggiero 2017) as well as for the observed effects of GcMAF in autism (Ruggiero 2016). In normal conditions, endogenous chondroitin sulfate in plasma, milk, and colostrum is complexed with proteins (Coppa et al. 2011; Ruggiero 2021) and, therefore, its GalNAc moiety is unable to interact with the negatively charged pocket of the GcMAF receptor. However, during the process of fermentation that is performed to manufacture the product, the combined action of acidity and proteolysis from microbial proteases (Ruggiero 2021), leads to the unmasking of GalNAc that is thus able to interact with the active site of the GcMAF receptor in a manner functionally superimposable to that of the GalNAc attached to Thr420 of GcMAF.
The GcMAF-like activity of chondroitin sulfate is further supported by the similarities of the physical properties between the sequence of alternating sugars constituting chondroitin sulfate (GalNAc and glucuronic acid) and the active site of GcMAF, that is the amino acid sequence TPT420ELAKLVNKRSE where GalNAc is attached to T420 (Schneider et al. 2003; Gregory et al. 2010). The sequence TPT420ELAKLVNKRSE exhibits a tendency to alternate between polar and aliphatic amino acids; likewise, the repeating disaccharide units of chondroitin sulfate involve one sugar unit with aliphatic properties—glucuronic acid has a certain aliphatic features due to its non-aromatic ring structure—and a polar sugar (GalNAc). The similarity is even more striking in the core of the active sequence, that is PT420E. From the point of view of surface charge distribution, Proline (P) and glucuronic acid both possess aliphatic features and have a cyclic structure in their molecules. T420 has GalNAc that is identical to the GalNAc of chondroitin sulfate. Glutamic acid (E) and glucuronic acid have some aliphatic features, possess a carboxylic acid group at one end of the molecule, and the basic carbon backbone and the placement of the carboxylic acid group are similar in both molecules. Therefore, the sequence PT420E has physical properties superimposable to the sequence glucuronic acid-GalNAc-glucuronic acid that constitutes chondroitin sulfate. However, unlike GcMAF, where there is one active site (T420-GalNAc or K420) per molecule, chondroitin sulfate has 50 to 100 repeated units of alternating sugars, with a resulting significantly higher probability of interaction with the GcMAF receptor. In addition, domain III of Gc protein has several negatively charged amino acids that may be implicated in its binding to the GcMAF receptor (Nabeshima et al. 2020); likewise, chondroitin sulfate is a negatively charged macromolecule and such a physical property, together with the propensity to bind to proteins (Ruggiero 2021), might favor its binding to the GcMAF receptor. Given the uniform distribution of negative charges on the surface of chondroitin sulfate as well as its inherent flexibility, such a binding would not require site specificity at variance with what occurs with GcMAF.
Conclusions
This study provides compelling evidence for the Bifidobacteria-rich product’s potential health benefits observed in various conditions. The high concentration of Bifidobacteria strains likely contributes to these effects through two main mechanisms:
Enhanced endogenous GcMAF production: The Bifidobacteria in the product express beta-galactosidase and sialidase enzymes, functionally similar to their human counterparts. During fermentation, these enzymes facilitate deglycosylation of the Gc protein present in bovine milk and colostrum, converting it into the active GcMAF molecule.
Activation of chondroitin sulfate: The presence of chondroitin sulfate in milk and colostrum, along with its metabolization by Bifidobacteria and other microbes present in the product, contributes to the product’s efficacy. Chondroitin sulfate, with its GalNAc sugar moiety, exhibits GcMAF-like activity after being freed from protein complexes by the acidic and proteolytic environment during fermentation. This process allows GalNAc to interact with the GcMAF receptor, thus amplifying the overall immune-stimulatory effects.
A limitation of this study consists in the fact that it relies on a targeted microarray technique, potentially missing uncharacterized microbes within the product’s complex microbial community. Additionally, in silico analysis of enzymes may not fully represent their functionality within the dynamic gut environment. These limitations notwithstanding, the present findings suggest that the Bifidobacteria-rich product may promote health benefits through a combination of enhanced GcMAF generation and activation of chondroitin sulfate, warranting further investigation into its therapeutic potential in various disease models.
Corresponding Author: Marco Ruggiero, MD, PhD, National Coalition of Independent Scholars. 125 Putney Rd Battleboro, VT 05301, United States of America. email: marco.ruggiero@ncis.org
Funding
The author did not receive any funding for this study.
Acknowledgements
MR thanks the Colleagues at Silver Spring Sagl for sharing the results of the Axiom Microbiome Array for the purpose of writing this article. In addition, MR acknowledges the great work of Dr. Aldo Ruggiero, MD (1923-2006), pioneer of radiology in Prato, Italy, founder of the Studio Radiologico Ruggiero, source of boundless inspiration for this and many other scientific articles.
Conflicts of interest
MR consults for Silver Spring Sagl, the company producing the fermented product described in this study. However, he had no prior knowledge of the results of the Axiom Microbiome Array that was independently performed by Eurofins Microbiology Laboratories Inc. (New Berlin Wisconsin, USA) on a commercially available lot as part of the company’s quality controls. He did not receive any remuneration for writing this article. MR is Editor-in-Chief of the American Journal of Immunology and is waived from the Article Processing fee for this contribution; he receives no remuneration for his editorial work.
Ethics
This article is original and contains material that has not been submitted or published in any scientific journal. A preprint of the first submission of this article had been posted in a multidisciplinary preprint platform (Ruggiero, M. Genetic Analysis of Bifidobacteriaceae in a Fermented Milk and Colostrum Product in Relation to Immune Function. Preprints 2024, 2024051503.
https://doi.org/10.20944/preprints202405.1503.v1. https://
www.preprints.org/manuscript/202405.1503/v1).
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Advisory
No information in this paper is intended or implied to be a substitute for professional medical advice, diagnosis or treatment.
References
- Albert AY, Chaban B, Wagner EC, Schellenberg JJ, Links MG, van Schalkwyk J, Reid G, Hemmingsen SM, Hill JE, Money D; VOGUE Research Group. A Study of the Vaginal Microbiome in Healthy Canadian Women Utilizing cpn60-Based Molecular Profiling Reveals Distinct Gardnerella Subgroup Community State Types. PLoS One. 2015 Aug 12;10(8):e0135620. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4534464/. [CrossRef]
- Antonucci N, Pacini S, Ruggiero M. Use of an Extremely Biodiverse Probiotic and a Supplement Based on Microbial Chondroitin Sulfate is Associated with a Significant Decrease of Serum Free Kappa Light Chains as well as a Trend Toward Normalization of Kappa/Lambda Ratio and of Plasma Cell Bone Marrow Infiltration in a Case of Multiple Myeloma. Am J Immunol. 2019, 15; 5-9. https://thescipub.com/abstract/10.3844/ajisp.2019.5.9. [CrossRef]
- Biavatti B, Mattarelli P. Chapter 3 - Related Genera Within the Family Bifidobacteriaceae. The Bifidobacteria and Related Organisms.
- Biology, Taxonomy, Applications. 2018; 49-66 https://doi.org/10.1016/B978-0-12-805060-6.00003-X. [CrossRef]
- Blythe J, Ruggiero M, Pacini S. Intermittent fasting and probiotic yogurt consumption are associated with reduction of serum alpha- N-acetylgalactosaminidase and increased urinary excretion of lipophilic toxicants. Madridge J Immunol. 2017; 1(1): 23-27 https://madridge.org/journal-of-immunology/mjim-1000107.php. [CrossRef]
- Blythe, J. & Ruggiero, M. (2020). Effects on the Immune System of a Three-Month Consumption of an Extremely Diverse Probiotic Yogurt: Decrease of Serum Alpha-N-Acetylgalactosaminidase Activity, Detoxification and Gut Microbiota Normalization. American Journal of Immunology, 16(1), 31-41 https://thescipub.com/abstract/10.3844/ajisp.2020.31.41. [CrossRef]
- Bodke H, Jogdand S. Role of Probiotics in Human Health. Cureus. 2022 Nov 9;14(11):e31313. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9733784/. [CrossRef]
- Bonten E, van der Spoel A, Fornerod M, Grosveld G, d’Azzo A. Characterization of human lysosomal neuraminidase defines the molecular basis of the metabolic storage disorder sialidosis. Genes Dev. 1996 Dec 15;10(24):3156-69. https://pubmed.ncbi.nlm.nih.gov/8985184/. [CrossRef]
- Branca JJ, Pacini S, Ruggiero M. Effects of Pre-surgical Vitamin D Supplementation and Ketogenic Diet in a Patient with Recurrent Breast Cancer. Anticancer Res. 2015 Oct;35(10):5525-32. https://pubmed.ncbi.nlm.nih.gov/26408720/. [PubMed]
- Carter, M., Pacini, S. & Ruggiero, M. (2020). Consumption of an Extremely Biodiverse Probiotic and a Supplement based on Microbial Chondroitin Sulfate is Associated with Very Low Serum Alpha-N-acetylgalactosaminidase (Nagalase) Activity and Decrease of C-reactive Protein Values. American Journal of Immunology, 16(1), 8-18 https://thescipub.com/abstract/10.3844/ajisp.2020.8.18. [CrossRef]
- Chen J, Chen X, Ho CL. Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front Bioeng Biotechnol. 2021 Dec 22;9:770248. https://pubmed.ncbi.nlm.nih.gov/35004640/. [CrossRef]
- Coppa GV, Gabrielli O, Buzzega D, Zampini L, Galeazzi T, Maccari F, Bertino E, Volpi N. Composition and structure elucidation of human milk glycosaminoglycans. Glycobiology. 2011 Mar;21(3):295-303. https://pubmed.ncbi.nlm.nih.gov/21030540/. [CrossRef]
- Duranti S, Longhi G, Ventura M, van Sinderen D, Turroni F. Exploring the Ecology of Bifidobacteria and Their Genetic Adaptation to the Mammalian Gut. Microorganisms. 2020 Dec 22;9(1):8. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7822027/. [CrossRef]
- Ena, JM Pérez MD, Aranda P, Sánchez L, Calvo M. Presence and changes in the concentration of vitamin D-binding protein throughout early lactation in human and bovine colostrum and milk. J. Nutritional Biochem. 1992 3(10):498-502. ISSN 0955-2863. [CrossRef]
- Faghfoori Z, Faghfoori MH, Saber A, Izadi A, Yari Khosroushahi A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021 May 12;21(1):258. https://pubmed.ncbi.nlm.nih.gov/33980239/. [CrossRef]
- Gavzy SJ, Kensiski A, Lee ZL, Mongodin EF, Ma B, Bromberg JS. Bifidobacterium mechanisms of immune modulation and tolerance. Gut Microbes. 2023 Dec;15(2):2291164. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10730214/. [CrossRef]
- Gregory KJ, Zhao B, Bielenberg DR, Dridi S, Wu J, Jiang W, Huang B, Pirie-Shepherd S, Fannon M. Vitamin D binding protein-macrophage activating factor directly inhibits proliferation, migration, and uPAR expression of prostate cancer cells. PLoS One. 2010 Oct 18;5(10):e13428. https://pubmed.ncbi.nlm.nih.gov/20976141/. [CrossRef]
- Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and Their Health-Promoting Effects. Microbiol Spectr. 2017 Jun;5(3). https://pubmed.ncbi.nlm.nih.gov/28643627/. [CrossRef] [PubMed]
- Hsu CA, Yu RC, Chou CC. Production of beta-galactosidase by Bifidobacteria as influenced by various culture conditions. Int J Food Microbiol. 2005 Oct 15;104(2):197-206. https://pubmed.ncbi.nlm.nih.gov/15985305/. [CrossRef]
- Inchingolo AD, Malcangi G, Semjonova A, Inchingolo AM, Patano A, Coloccia G, Ceci S, Marinelli G, Di Pede C, Ciocia AM, Mancini A, Palmieri G, Barile G, Settanni V, De Leonardis N, Rapone B, Piras F, Viapiano F, Cardarelli F, Nucci L, Bordea IR, Scarano A, Lorusso F, Palermo A, Costa S, Tartaglia GM, Corriero A, Brienza N, Di Venere D, Inchingolo F, Dipalma G. Oralbiotica/Oralbiotics: The Impact of Oral Microbiota on Dental Health and Demineralization: A Systematic Review of the Literature. Children (Basel). 2022 Jul 8;9(7):1014. https://pubmed.ncbi.nlm.nih.gov/35883998/. [CrossRef]
- Kim JY, Kwon JH, Ahn SH, Lee SI, Han YS, Choi YO, Lee SY, Ahn KM, Ji GE. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: a double-blind, randomized, placebo-controlled trial. Pediatr Allergy Immunol. 2010 Mar;21(2 Pt 2):e386-93. https://pubmed.ncbi.nlm.nih.gov/19840300/. [CrossRef]
- Longhi G, van Sinderen D, Ventura M, Turroni F. Microbiota and Cancer: The Emerging Beneficial Role of Bifidobacteria in Cancer Immunotherapy. Front Microbiol. 2020 Sep 8;11:575072. https://pubmed.ncbi.nlm.nih.gov/33013813/. [CrossRef]
- Nabeshima Y, Abe C, Kawauchi T, Hiroi T, Uto Y, Nabeshima YI. Simple method for large-scale production of macrophage activating factor GcMAF. Sci Rep. 2020 Nov 5;10(1):19122. https://pubmed.ncbi.nlm.nih.gov/33154460/. [CrossRef]
- Nishiyama K, Nagai A, Uribayashi K, Yamamoto Y, Mukai T, Okada N. Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe. 2018 Aug;52:22-28. https://pubmed.ncbi.nlm.nih.gov/29787815/. [CrossRef]
- Okada K, Takami D, Makizaki Y, Tanaka Y, Nakajima S, Ohno H, Sagami T. Effects of Bifidobacterium longum CLA8013 on bowel movement improvement: a placebo-controlled, randomized, double-blind study. Biosci Microbiota Food Health. 2023;42(3):213-221. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10315193/. [CrossRef]
- Oshima A, Tsuji A, Nagao Y, Sakuraba H, Suzuki Y. Cloning, sequencing, and expression of cDNA for human beta-galactosidase. Biochem Biophys Res Commun. 1988 Nov 30;157(1):238-44. https://pubmed.ncbi.nlm.nih.gov/3143362/. [CrossRef]
- Pacini S, Ruggiero M. Natural Plasmids in a Swiss Fermented Milk and Colostrum Product assessed by Microbiome Array. Madridge J Immunol. 2019; 3(2): 100-108 https://madridge.org/journal-of-immunology/mjim-1000123.pdf. [CrossRef]
- Pacini S, Ruggiero M. Phage composition of a fermented milk and colostrum product assessed by microbiome array; putative role of open reading frames in reference to cell signaling and neurological development. J Neurol Stroke. 2020; 10(2):80‒90 https://medcraveonline.com/JNSK/JNSK-10-00416.pdf. [CrossRef]
- Ruggiero, M. (2016). Gc Protein-Derived Macrophage Activating Factor (GcMAF) and Autism: Do Clinical Results Require a Novel Interpretation?. American Journal of Immunology, 12(4), 77-82 https://thescipub.com/abstract/ajisp.2016.77.82. [CrossRef]
- Ruggiero, M. (2017). Is Rerum® the New Coley’s Vaccine?. American Journal of Immunology, 13(2), 91-98 https://thescipub.com/abstract/ajisp.2017.91.98. [CrossRef]
- Ruggiero, M. (2021). Release of Endogenous Chondroitin Sulfate and Heparin as Consequence of Dysregulated Proteolysis in COVID-19. American Journal of Immunology, 17(1), 40-46 https://thescipub.com/abstract/10.3844/ajisp.2021.40.46. [CrossRef]
- Ruggiero M, Ward E, Smith R, Branca JJ, Noakes D, Morucci G, Taubmann M, Thyer L, Pacini S. Oleic Acid, deglycosylated vitamin D-binding protein, nitric oxide: a molecular triad made lethal to cancer. Anticancer Res. 2014; Jul;34(7):3569-78. https://pubmed.ncbi.nlm.nih.gov/24982371/. [PubMed]
- Ruggiero M, Reinwald H, Pacini S. Is chondroitin sulfate responsible for the biological effects attributed to the GC protein-derived Macrophage Activating Factor (GcMAF)? Med Hypotheses. 2016 Sep;94:126-31. https://pubmed.ncbi.nlm.nih.gov/27515218/. [CrossRef]
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14422-7. https://pubmed.ncbi.nlm.nih.gov/12381787/. [CrossRef]
- Schneider GB, Grecco KJ, Safadi FF, Popoff SN. The anabolic effects of vitamin D-binding protein-macrophage activating factor (DBP-MAF) and a novel small peptide on bone. Crit Rev Eukaryot Gene Expr. 2003;13(2-4):277-84. https://pubmed.ncbi.nlm.nih.gov/14696974/. [CrossRef]
- Schwalb, M., Taubmann, M., Hines, S., Reinwald, H. & Ruggiero, M. (2016). Clinical Observation of a Novel, Complementary, Immunotherapeutic Approach based on Ketogenic Diet, Chondroitin Sulfate, Vitamin D3, Oleic Acid and a Fermented Milk and Colostrum Product. American Journal of Immunology, 12(4), 91-98 https://thescipub.com/abstract/ajisp.2016.91.98. [CrossRef]
- Thissen JB, Be NA, McLoughlin K, Gardner S, Rack PG, Shapero MH, Rowland RRR, Slezak T, Jaing CJ. Axiom Microbiome Array, the next generation microarray for high-throughput pathogen and microbiome analysis. PLoS ONE 2019, 14(2): e0212045. https://pubmed.ncbi.nlm.nih.gov/30735540/. [CrossRef] [PubMed] [PubMed Central]
- UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023 Jan 6;51(D1):D523-D531. https://pubmed.ncbi.nlm.nih.gov/36408920/. [CrossRef]
- Vaish S, Soni SK, Singh B, Garg N, Zareen Ahmad I, Manoharan M, Trivedi AK. Meta-analysis of biodynamic (BD) preparations reveal the bacterial population involved in improving soil health, crop yield and quality. J Genet Eng Biotechnol. 2024 Mar;22(1):100345. https://pubmed.ncbi.nlm.nih.gov/38494258/. [CrossRef]
- Ward E, Smith R, Branca JJ, Noakes D, Morucci G, Thyer L. (2014). Clinical experience of cancer immunotherapy integrated with oleic acid complexed with de-glycosylated vitamin D-binding protein. American Journal of Immunol. 2014; 10(1), 23-32 https://thescipub.com/abstract/ajisp.2014.23.32. [CrossRef]
- Wu Q, Li Q, Zhang X, Ntim M, Wu X, Li M, Wang L, Zhao J, Li S. Treatment with Bifidobacteria can suppress Aβ accumulation and neuroinflammation in APP/PS1 mice. PeerJ. 2020 Oct 28;8:e10262. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7602682/. [CrossRef]
- Yao S, Zhao Z, Wang W, Liu X. Bifidobacterium Longum: Protection against Inflammatory Bowel Disease. J Immunol Res. 2021 Jul 23;2021:8030297. https://pubmed.ncbi.nlm.nih.gov/34337079/. [CrossRef]
- Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Rosselló-Móra R. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008 Sep;31(4):241-50. https://pubmed.ncbi.nlm.nih.gov/18692976/. [CrossRef]
- Zunaid IR, Pacini S, Ruggiero M. Significance of hydrophobic and charged sequence similarities in sodium-bile acid cotransporter and vitamin D-binding protein macrophage activating factor. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.03.03.975524v1. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).