1. Introduction
Apples are one of the world's most important temperate fruits with medicinal, nutritional and ornamental value. Apples are not only rich in essential nutrients for the brain such as sugar, protein, vitamins and minerals, but more importantly are rich in zinc. According to research, zinc is a component of many important enzymes in the human body and is a key element in promoting growth and development. Therefore, improving apple yield and quality can better improve human health [
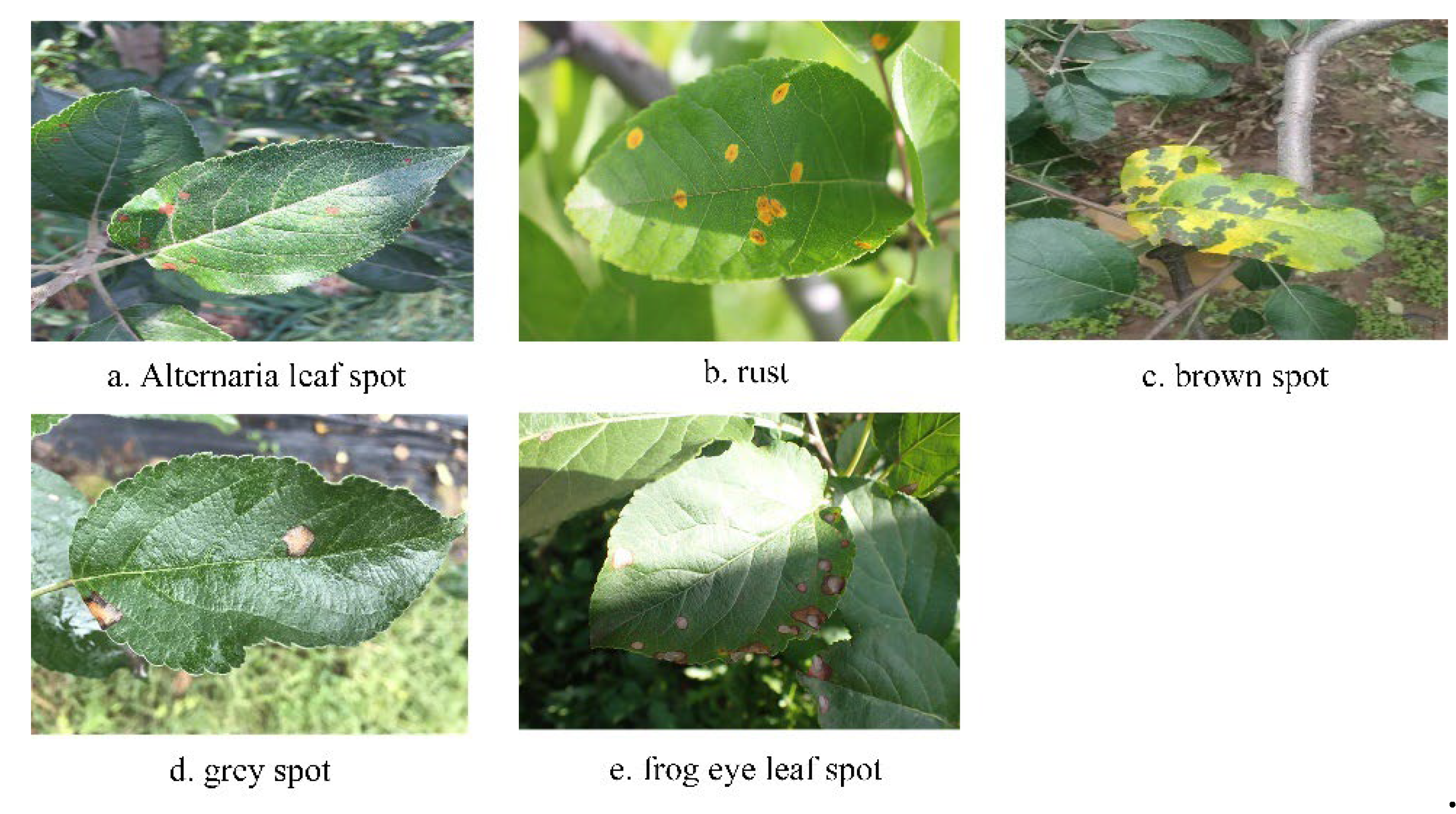
1]. The spring and summer temperatures and high humidity in northern China are favorable for the propagation and spread of pathogenic bacteria, which increases the possibility of apple tree leaves being infected with leaf spot diseases such as alternaria leaf spot, rust, brown spot, grey spot and frog eye leaf spot, etc. These diseases are the most common, coupled with the fact that artificial orchards are planted with high density and poorly ventilated, which leads to the development of these diseases. Poor ventilation has led to the frequent occurrence of these diseases in the northern region, which seriously affects the healthy development of the apple industry, and it is necessary to discover and rapidly identify leaf diseases as early as possible and carry out precise prevention and control of apple leaf diseases in order to effectively improve the yield and quality of apples [
2].
Traditional disease detection requires experienced experts to visually judge the leaves in the field. This traditional detection method is inefficient and susceptible to subjective factors resulting in leakage and misdetection. Nowadays, a large number of researches have adopted classical image processing techniques and machine learning algorithms for timely and effective detection of crop diseases [
3]. This traditional machine learning detection method recognizes some specific image features of a crop in a natural environment, but there are problems such as difficulty in feature extraction and high computational complexity [
4]. Deep learning based methods have more advantages over traditional machine learning methods in terms of detection speed, detection scale and detection accuracy. Deep learning based target detection models can be categorized into two main groups, two-stage detection models and one-stage detection models. The first category is the two-stage detection models represented by Faster R-CNN [
5] and Mask-RCNN [
6]. LEE [
7] used the Faster R-CNN model with region-based convolutional neural networks to recognize brown blight, blister blight and algal leaf spot diseases of tea, with an average precision (AP) of 63.58%, 81.08% and 64.71%, respectively. Singh [
8] used multilayer convolutional network MCNN to classify mango anthracnose leaf disease with 97.3% accuracy. He [
9] proposed a Mask-RCNN based model for detecting leaf diseases of tea tree with F1 scores of 88.3% and 95.3% for tea coal diseases and boll weevil leaf diseases, but the F1 scores was lower in distinguishing between brown leaf disease and target spot disease, with scores of only 61.1% and 66.6%. The analysis showed that the two-stage detection model needs to generate candidate frames before target classification and bounding box regression, which is difficult to apply because of high model complexity, slow speed, and low accuracy, although it can identify crop diseases.
The second category is the one-stage detection model represented by SSD [
10] and YOLO series [
11,
12,
13,
14]. Unlike the two-stage detection models, the one-stage detection models are more suitable for real-time detection tasks due to their high accuracy and fast response time. Sun [
15] et al. proposed a module for MEAN by reconfiguring the 3x3 convolutional module to detect a single diseased apple leaf in a simple environment, and the MEAN-SSD model achieved a mean average precision (mAP) value of 83.12% and an FPS value of 12.53 for apple leaf disease. Tian [
16] et al. proposed a YOLOv3-dense model for detecting apple surface anthracnose by combining the optimization of YOLOv3 model feature layer with DenseNet, which greatly improved the utilization of the features in the neural network with a detection accuracy of 95.57% and a detection speed of up to 31 FPS. Li [
17] proposed an improved model of YOLOv5s for vegetable disease detection, with a mAP0.5 of 93.1% for five disease images and a model size of 17.1M. Lin [
18] proposed an improved YOLOx-Tiny model for the detection of tobacco brown spot disease, which introduced the HMU module for information interaction and small feature extraction capability in the neck network, resulting in average precision (AP) of 80.45%. Faiza Khan [
19] obtained 99.04% average precision (AP) for three maize crop diseases using YOLOv8n model detection. The analysis showed that one-stage detection models have been effectively used to diagnose various crop diseases, but fewer studies have been applied to apple leaf disease detection. And most of the studies were realized in a simple environment using a single leaf, which could not realistically simulate the growth environment in a complex background such as different light and leaves shading each other, thus limiting their application in practical production. It is crucial to develop an apple leaf disease detection model with high detection accuracy and fast detection speed in complex backgrounds.
In order to improve the detection accuracy and speed of common apple leaf spot diseases in the apple planting environment, this paper proposes a high accuracy model and a lightweight model based on the multiscale feature extraction module ResBlock and the lightweight feature fusion module C4 together with the parameter streamlining strategy, which improves the accuracy of detecting apple leaf diseases and realizes the model lightweighting.
2. YOLOv5-Based Optimization Model
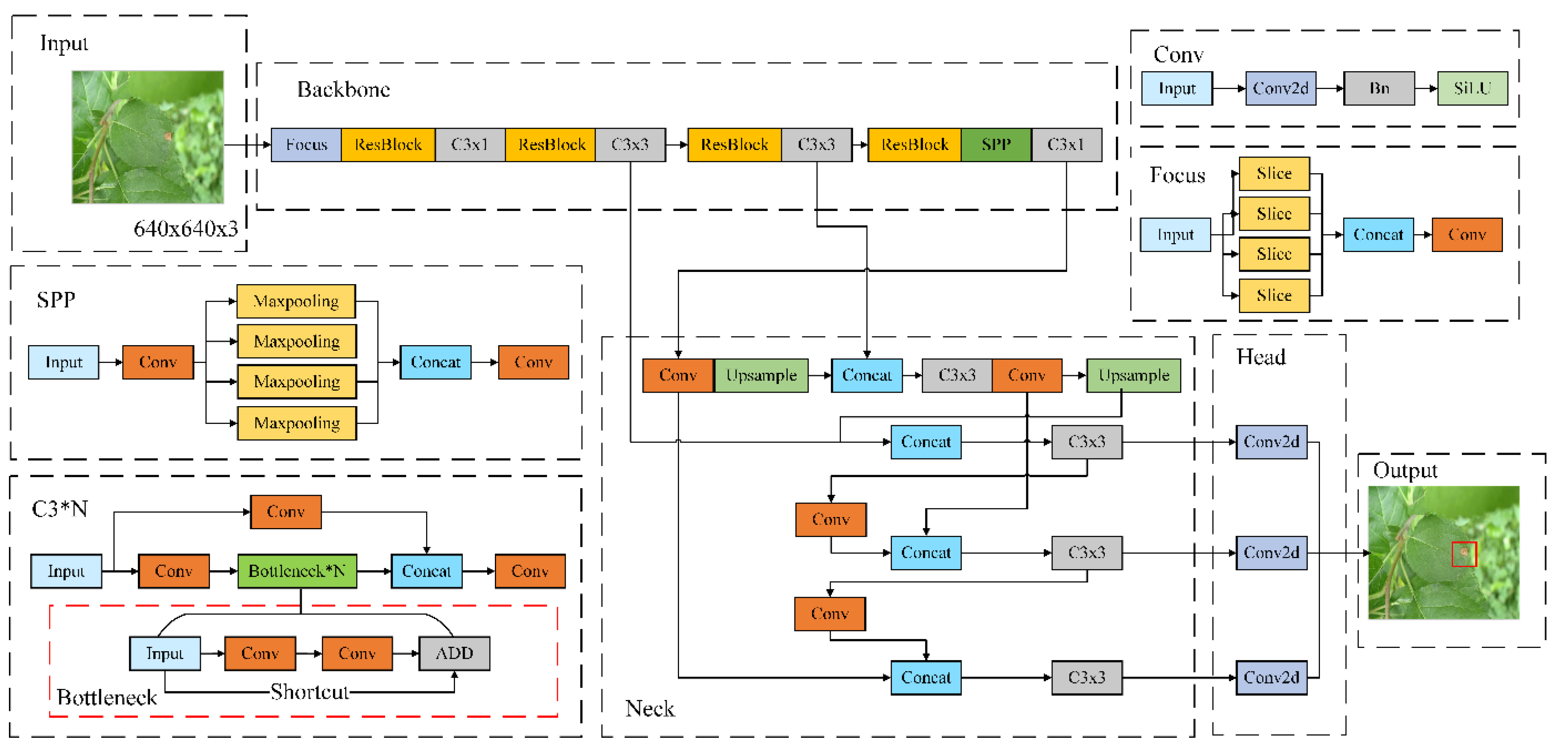
Based on the characteristics of the YOLOv5 model, the paper proposed an improved apple leaf disease detection model, YOLOv5-Res, as shown in
Figure 1, in order to improve the accuracy of the model in detecting small-scale apple leaf spot degree in a complex background. The model increases the number of Bottlenecks in the C3 module of the Backbone and Neck networks to deepen the network and improve the feature extraction ability, and adopts the lighter Focus module instead of the Conv module to balance the increase of the network parameters, and adopts the SPP module instead of the SPPF module to reduce the overfitting risk and improve the generalization ability of the model while decreasing the model parameters. generalization ability. In addition, the Resblock module is designed to adapt to input features of different scales and complexities.
Based on the YOLOv5-Res model, the C4 module was designed with the goal of model lightweighting to optimize the transfer path of the feature map information fusion process, and with the parameter streamlining strategy, the lightweight model YOLOv5-Res4 was designed.
2.1. YOLOv5 Model
The YOLOv5 [
20] model is a single-stage target recognition model proposed by Glenn Jocher in 2020. It can be categorized into YOLOv5s, YOLOv5n, YOLOv5m, YOLOv5l, and YOLOv5x versions based on the differences in network depth and network width. Among them, YOLOv5n is the lightest version of YOLOv5 series with the fastest detection speed, while YOLOv5s is the model with the best overall performance.
The YOLOv5 model structure consists of Backbone, Neck and Head. Backbone is used to extract features from the input image and consists of Conv, C3 and SPPF (Spatial Pyramid Pooling). C3 [
21] structure is used for Backbone and Neck. SPPF [
22] structure is used for maximum pooling operation of feature maps using convolution kernels of different sizes to enrich the feature representation. Neck part uses FPN [
23] (Feature Pyramid Networks) and PAN [
24] (Path Aggregation Network) to jointly enhance the semantic and spatial information. The FPN structure realizes semantic feature enhancement from top-down, and fuses the higher-order feature mapping with the lower-order feature mapping through up-sampling. The PAN structure enhances the localization features from bottom-up to achieve the purpose of feature information enrichment. The Head part generates the class probability and location information of the predicted target. The three detection heads correspond to 20 × 20, 40 × 40, and 80 × 80 feature maps, respectively. The YOLOv5 model uses CIoU [
25] to compute intersections such as the following:
.
where,, represent the centers of the predicted and real frames, respectively, stands for is to compute the Euclidean distance between two centers, represents the diagonal distance of the smallest closure region that can contain both the prediction box and the true box, is the weighting function, represent the difference between the length ratios of the predicted and real frames, respectively.
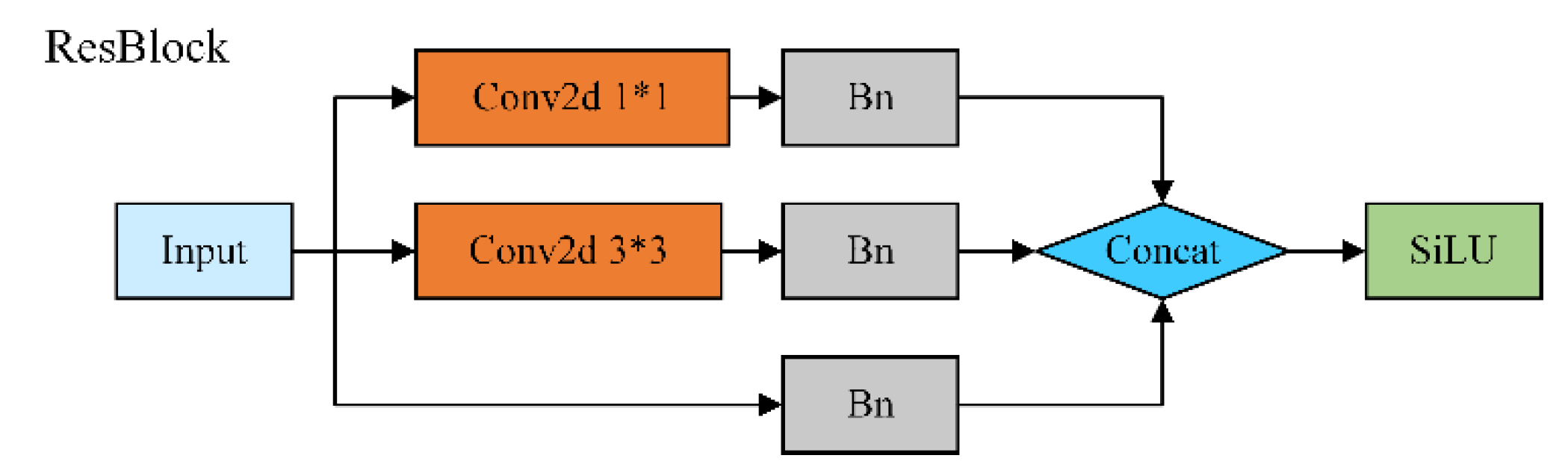
2.2. Design of ResBlock Multiscale Module
Diseases such as alternaria leaf spot, rust, brown spot, grey spot and frog eye leaf spot in artificial apple orchards, the spots of early disease are mostly small targets and the patches at the end of the stage do not have the same percentage in the leaf, so it is difficult to be detected by YOLOv5 using large convolution kernel, especially in the detection of similar diseases the ability is weaker. In order to extract on the same feature map to obtain richer feature information and improve the model's characterization and generalization ability, we designed the ResBlock module, as shown in
Figure 2.
GoogLeNet proposes the Inception [
26] model to increase the network width by introducing multiple Inception modules while reducing the number of parameters. In Inception-v[
1] [
26], multiscale feature extraction is achieved by using different sized convolutional kernels in the same layer. The core idea of the ResNet [
27] model is to learn residuals, i.e., the neural network is made to learn a residual mapping during the training process which maps the inputs directly to the outputs,
, where x denotes the inputs of the residual block and y denotes the output of the residual block, and
denotes the function that the network needs to fit. Residual connectivity makes it easier to train deeper networks.
In this paper, the ResBlock module is proposed based on the idea of Inception and ResNet, with three branch designs: 1x1 convolutional kernel, 3x3 convolutional kernel, and jump connection, and each branch contains a BN [
28] (Batch Normalization) module, which is used to improve the training stability of the model and accelerate the convergence process. Specifically, the 1x1 convolutional kernel is used to linearly transform the feature map to extract local feature information; the 3x3 convolutional kernel is used to capture a wider range of feature relationships to enhance the global expression capability; and the jump connection realizes direct information transfer and back propagation of gradient to alleviate the gradient vanishing problem. This multi-branch design utilizes different convolutional kernel feature extraction capabilities combined with jump connections to achieve feature fusion and information transfer to promote multiscale feature extraction. Meanwhile, the introduction of the BN module helps to normalize the feature distribution, accelerate the network convergence speed, and improve the model generalization ability and stability. Such a module design adapts to different scales and complexity input features to improve the model characterization ability and performance.
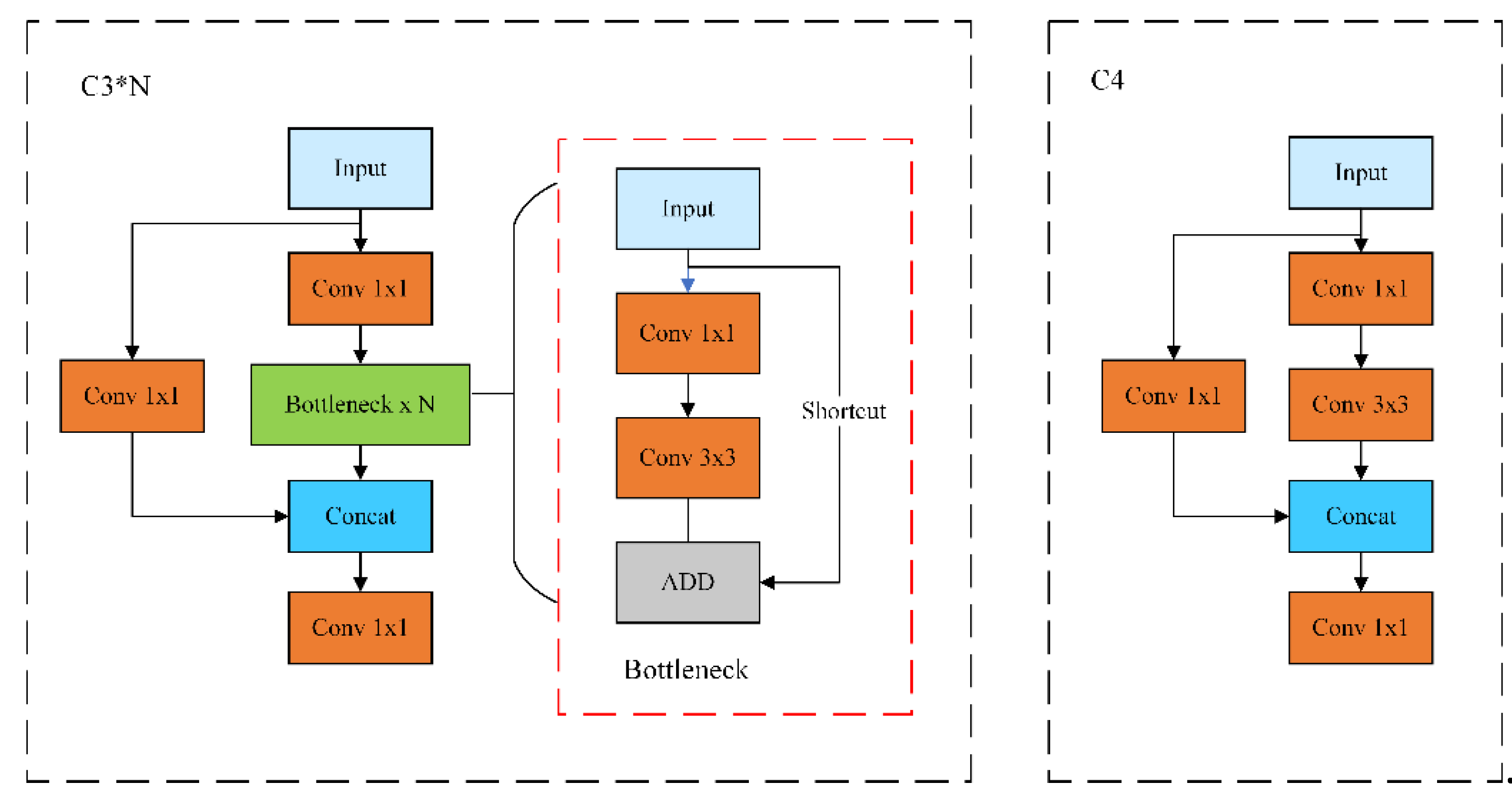
2.3. Design of the C4 Feature Fusion Module
C3 is an important building block for feature extraction in the YOLOv5s model, which consists of multiple convolutional modules (Conv), bottleneck layers (Bottleneck), BN (Batch Normalizations) layers and SiLU [
41] activation functions. Where the convolution module consists of a Conv2d convolution layer plus a BN layer plus a SiLU activation function; the bottleneck layer is a 1x1 convolution followed by a 3x3 convolution, where the 1x1 convolution halves the number of channels and the 3x3 convolution doubles the number of channels and then adds the inputs, which is set up to improve the network's characterization and performance. The C3 module as a whole is divided into three steps, which are First PW [
42] (Pointwise Convolution) to downscale the data, then convolution with regular convolution kernel, and finally PW to upscale the data. The C3 structure is shown in the left panel of Fig 3.
C3 aims to enhance the learning capability of the network by feature extraction of the input image through multi-layer operations such as convolution module, bottleneck layer, BN layer, etc., which increase the complexity and computation of the model. In this paper, on the basis of C3, considering that the first convolution of the main branch and the first convolution of the bottleneck layer are both point-by-point convolutions, the two layers are merged to form the C4 module, C4 uses 1x1 convolution to reduce the number of channels in the main branch to extract the shallow features to obtain more detailed information, 3x3 convolution is used to increase the sensory field to extract the deeper information, and the side branch introduces a 1x1 convolutional layer to realize feature fusion. With the above structure, the C4 module achieves the optimization of the information transfer path within the module, simplifies the feature transfer and interaction process, and avoids information loss and blurring to better capture local features and spatial information, and its structure is shown in the right panel of
Figure 3.
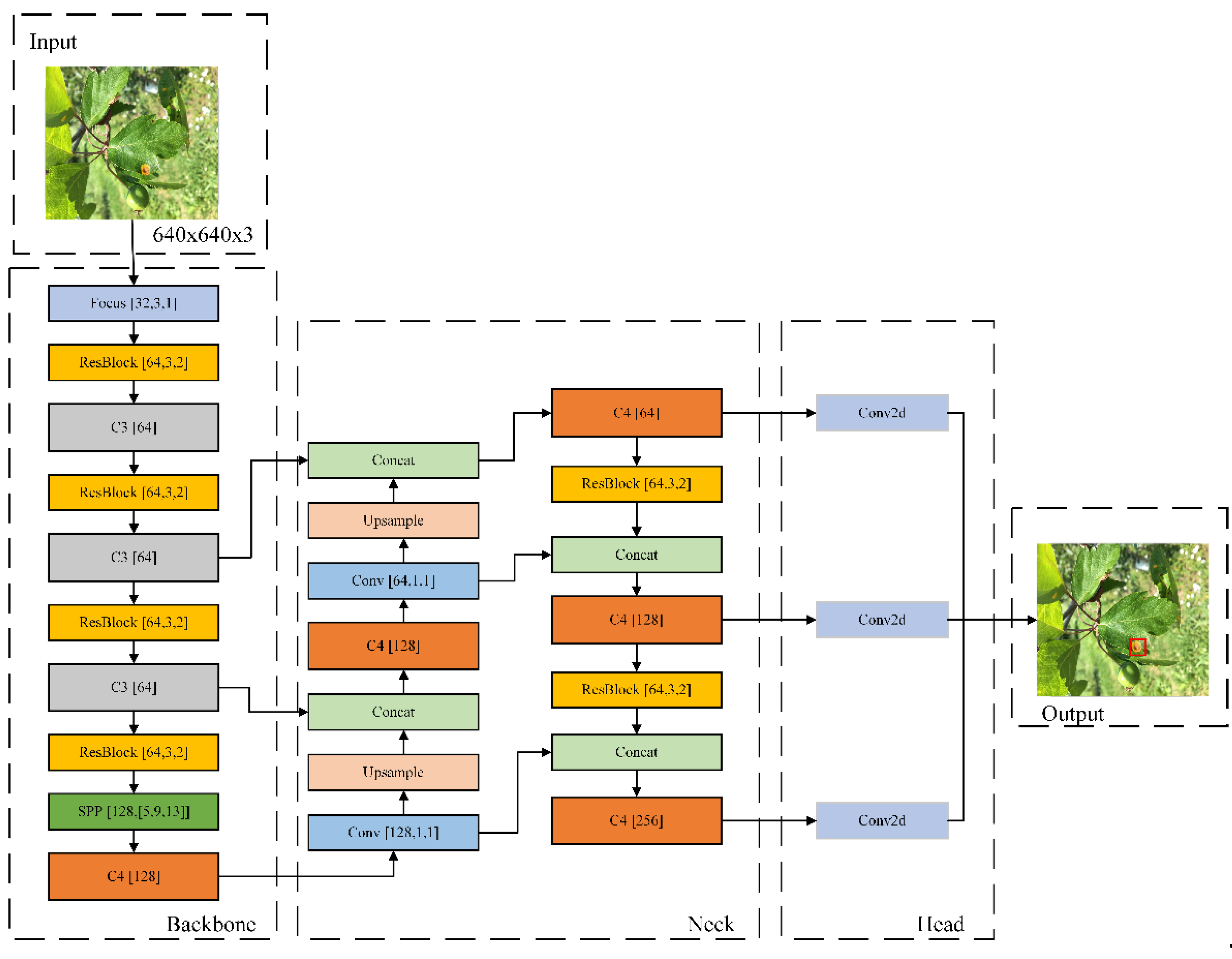
2.4. YOLOv5-Res4 Based on Parameter Refinement Strategy
Since the image feature extraction is mainly done in the Backbone part, Neck is mainly used for feature fusion of the feature map. In this paper, we use the lighter weight Focus [
31] module to replace the layer 0 Conv module in the Backbone part, reduce the Bottleneck structure coefficient in the C3 module to 1 in order to maximize the lightweight purpose, use the ResBlock multiscale module to replace the other Conv modules in the Backbone network, and introduce the SPP [
22] module to directly connect with the ResBlock module to improve the model's ability to extract features from the Backbone network. The SPP module is directly connected to the ResBlock module to improve the model's understanding of global information and reduce the risk of model overfitting to improve the model generalization ability, and the streamlined C4 module is used to optimize the feature information transfer path to further improve the model's detection performance and feature representation ability. In the Neck network, the lighter C4 module is used to replace the C3 module to accelerate the feature transfer and interaction process, which is conducive to the rapid dissemination and fusion of semantic information. The Conv module in the 18th and 21st layers mainly plays the role of downsampling, for this reason, the ResBlock multiscale module is used to replace the Conv module to improve the detection accuracy and reduce the computation volume while maintaining sufficient sensory field, so as to accelerate the model inference speed.
In summary, the YOLOv5-Res4 model adopts a lightweight module, and the strategy of channel reduction and feature fusion achieves a significant reduction in the number of model parameters and computational complexity under the premise of maintaining high detection accuracy, and the model realizes a shallower compact structure, as shown in
Figure 4.
3. Analysis of Experimental Results
3.1. Dataset and Experimental Environment
To validate the effectiveness of the apple leaf disease recognition model proposed in this study, a new ALPP dataset was composed using a fusion of the AppleLeaf9 dataset [
32] and the Ai Studio dataset [
36].The AppleLeaf9 dataset fuses PlantVillage [
33], ATLDSD [
34], PPCD2020 and PPCD[2021] [
35]. PlantVillage contains only single leaf images of simple environments. The images of ATLDSD, PPCD2020 and PPCD2021 were collected from orchards under different weather conditions, which included a variety of situations such as sunny, cloudy, after rain, with light, against light, near, far, pitch and elevation. In this paper, five types of leaf images of alternaria leaf spot; rust; brown spot; grey spot; frog eye leaf spot were selected, and in order to increase the diversity of the images as much as possible and to improve the generalization ability of the model, the alternaria leaf spot; rust; brown spot and grey spot leaf images were examined by using the Ai Studio dataset. rust; brown spot; grey spot; and frog eye leaf spot using the Ai Studio dataset, as shown in
Figure 5.
The images were labeled using Labeling software, and the label of each target feature corresponded to the target feature, and redundant information was ignored during image labeling. A total of 4863 images were labeled, which were randomly divided into training set, testing set, and validation set in the ratio of 6:2:2, and the specific number distribution of each disease category is shown in
Table 1.
The main parameters of the computer platform used in this experiment are: Intel Core i9-13900K CPU main frequency 3.00 GHz, running memory 64 GB and NVIDIA GeForce RTX3090 24GB GPU. The PyTorch2.0.1 deep learning framework is used to build and evaluate the model. The training parameters were set with an image input size of 640x640, the training epoch was 150, the image batch size was set to 2, Adam optimizer, initial learning rate setting of 0.01, momentum value of 0.937, and weight decay parameter of 0.0005.
3.2. Model Evaluation Indicators
To evaluate the performance of the improved YOLOv5s model, the evaluation metrics used in this study were categorized into precision (P), recall (R), single-category average precision (AP), mean average precision (mAP), number of parameters (Parameters) and floating point operations (FLOPs). Where mAP, Parameters and FLOPs denote the mean average precision of disease detection, the number of network parameters and the real-time processing speed of the model, respectively. The formula for model performance evaluation metrics is shown in Equations (1)-(6):
where,
denotes the number of correctly detected positive samples,
denotes the number of incorrectly detected negative samples,
denotes the number of undetected positive samples.
denotes the convolutional kernel size,
and
denote the number of input and output channels, respectively,
and
denote the height and width of the output feature map, respectively.。
3.3. Experimental Analysis of the ResBlock Module
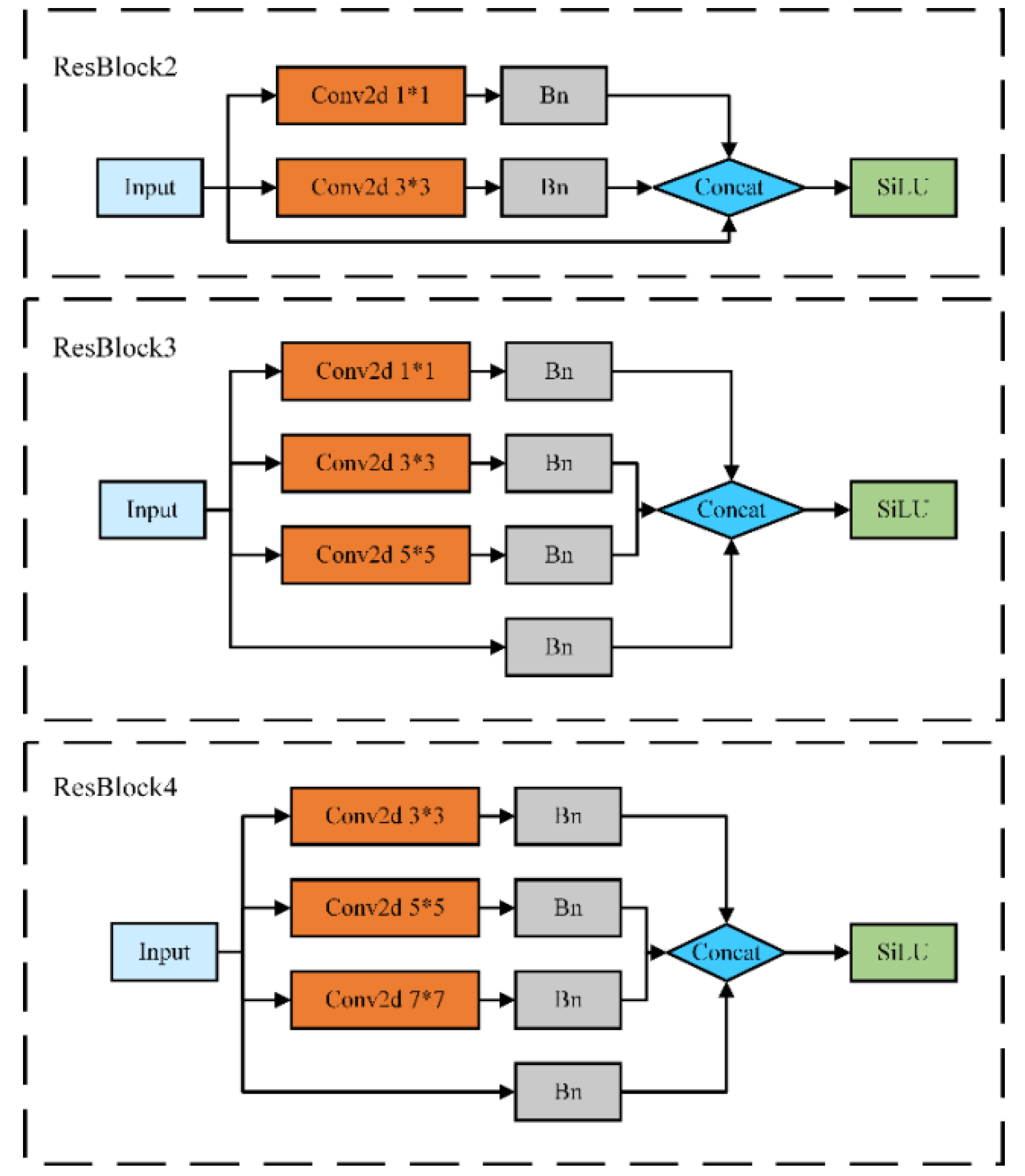
In order to verify the effectiveness of the ResBlock module, this paper reconstructs the ResBlock2, ResBlock3 and ResBlock4 modules as shown in
Figure 6. Keeping the other structures in the YOLOv5-Res model unchanged, ResBlock is replaced with ordinary Conv, ResBlock2, ResBlock3 and ResBlock4 for comparison experiments.
Leaf disease features vary greatly at different stages of development, and 1x1 and 3x3 parallel convolution kernels are used to ensure that the detailed information in the image can be adequately captured. Different sensory fields facilitate the extraction of local features of different sizes, which can better capture the detailed information in the image and improve the robustness of the network. As shown in
Table 2 of the experimental results, the mAP@0.5 values of ResBlock are 1.88%, 2.18% and 2.28% higher than those of ResBlock2, ResBlock3 module and ResBlock4 module, respectively. In addition, ResBlock and ResBlock2 models have the same number of parameters or model size, but there is a large difference in accuracy, which is mainly because the BN layer plays a role in speeding up the training speed to improve the model accuracy. Comparing the FLOPs values of the three improved modules, the ResBlock module is also more advantageous. The experimental results show that using the ResBlock module to replace the Conv module in the Backbone structure of YOLOv5s is an effective way to improve the model, and although the ResBlock module increases the number of parameters, this multiscale voxel extraction module adapts to the needs of apple leaf disease detection and improves the accuracy of the detection model.
3.4. Experiments with the C4 Module
In order to evaluate the effectiveness of the C4 feature fusion module, several different feature fusion strategies were compared in this experiment. The YOLOv5-Res4 model was used as the base model, and other mainstream feature fusion module replacements, e.g., CSP [
37], C3, and C2f [
38] modules, were used sequentially in place of the C4 module for comparison.
The experimental results, as seen in
Table 3, show that the mAP@0.5 values of the CSP, C3 and C2f modules are 77.5%, 77.8% and 77.2%, which are 2.4%, 2.1% and 2.7% lower than that of the YOLOv5-Res4 network model with the addition of the C4 feature fusion module in terms of the mAP@0.5 values, respectively. The C3 module introduces the idea of constant mapping of ResNet residual networks on top of the strategy of channel separation of the CSP module by stacking multiple BottleNeck [
39] modules on the main branch for tuning the model depth. And C2f references the ELAN [
40] network's idea of using short gradient paths to stack more Blocks, which greatly enriches the gradient flow information. In contrast, C4 feature fusion module streamlines the network structure, which only consists of three 1x1Convs with a step size of 1 and one 3x3Conv with a step size of 1. Finally, the low-level and high-level feature maps are spliced by Concat, which retains more spatial and semantic information, and is conducive to the enhancement of the model's expressive ability.
In addition, the C4 feature fusion module obtains the lowest Parameters and FLOPs while maintaining high detection accuracy. And the introduction of the C4 module enables the network to have better sensing capability and information fusion ability to improve the accuracy and robustness of target detection.
3.5. Experiments with Model Streamlining Strategies
In order to verify the effectiveness of the parameter streamlining scheme, this paper designs several models for exploratory validation, and the specific streamlining methods of each model are shown in
Table 4:
As shown in
Table 5 of the experimental results, both the C4 module and the Resblock module play a positive effect in simplifying the model complexity and recognition accuracy. Since the feature extraction is mainly in the shallow network, model D has 1.7% less model parameters than model C, while mAP@0.5 is improved by 0.5%. The effect of the SPP module was verified using model D, model E and model F. The results show that the mAP@0.5 value gradually increases and reaches the highest value of 80.8% in model F. The mAP@0.5 value of the YOLOv5-Res4 model is 79.9%, with the parameters only 22.89% and 61.7% of those of the F and YOLOv5n models, and the model size is 2.4MB, which significantly reduces the model size and computational cost.
3.6. YOLOv5-Res Comparative Analysis Experiment
In order to further validate the model performance, and considering the differences between the YOLOv5-Res accuracy model and the YOLOv5-Res4 lightweight model, this experiment compares YOLOv5-Res and YOLOv5-Res4 separately with the current mainstream single-target detection models.
The experimental results of the YOLOv5-Res model compared with DT-DETR [
41], YOLOv5s [
20], YOLOv5s-TR [
20], YOLOv5s6 [
20], YOLOv8s [
42], and YOLOv9 [
43] are shown in
Table 6. The YOLOv5-Res precision is improved by 1.6%, 1.3%, 3.8%, 5.2%, and 1.96%, respectively, 4.3%, 3.8%, 5.2%, and 1.96%, recall is improved by 7.5%, -0.3%, 2.3%, 2.6%, 2.6%, and 4.3%, and mAP@0.5 is improved by 7.5%, 2.8%, 3.7%, 4.3%, and 3.5%, respectively. Compared to the latest YOLOv9, YOLOv5-Res has improved 5.2, -1.7%, and 2% in precision, recall, and mAP@0.5, respectively, and YOLOv5-Res still has the highest detection performance with fewer number of parameters.
Table 7 shows the mAP@0.5 values of the mainstream models for detecting various categories of disease characteristics, among which the best performing models for detecting alternaria leaf spot, rust, brown spot, grey spot, and frog eye leaf spot were YOLOv9, YOLOv5s6, YOLOv9, RTDETR-L, and YOLOv5-Res. Comparing the detection results of the other models, YOLOv5-Res has a more balanced performance, and only grey sport has a mAP@0.5 lower than 80%.
3.7. YOLOv5-Res4 Comparative Analysis Experiment
Since the YOLOv5-Res4 model focuses more on lightweight, this paper compares it with lightweight YOLO models such as YOLOv3-tiny [
44], YOLOv5n [
20], YOLOv5n6 [
20] and YOLOv8n [
42]. The experimental results are shown in
Table 8, where the YOLOv5-Res4 model improves the precision by 8.1%, 0.9%, 1.3%, and -0.2%, the recall by 13.2%, 3.7%, 7.9%, and 3.2%, and the mAP@0.5 by 11.7%, 2.2%, and 5.3%, respectively, with the lowest number of parameters and model sizes, 2.9%, but FLOPs are only 5.3, providing better performance.
The experimental data of mainstream lightweight algorithmic models on various categories of diseases are shown in
Table 9, and the detection performance is higher than that of other algorithmic models in all categories, except that the mAP@0.5 value of Grey sport is lower than that of the YOLOv8n model by 3.4%.
The YOLOv5-Res model and YOLOv5-Res4 model were validated using the test set, and the detection results are shown in
Figure 7. The apple leaf disease targets were small, and the YOLOv5s model and YOLOv5n model had misdetection and omission in detecting alternaria leaf spot and grey spot, and had a slightly lower confidence level compared with YOLOv5-Res and YOLOv5-Res4. In summary, the YOLOv5-Res and YOLOv5-Res4 models were able to accomplish the task of accurate and efficient apple leaf spot detection.
4. Conclusions
Aiming at the problem of difficulty in accurately detecting lesion targets due to small-scale lesions and complex backgrounds in apple leaf images, this study proposes YOLOv5-Res and YOLOv5-Res4. Among them, the proposed ResBlock multiscale module helps to extract features at different scales and preserve image detail information. And the feature fusion module C4 enhances the ability to extract feature information of small targets while being lightweight. By compressing the depth and width of the model, the lightweight model YOLOv5-Res4 is obtained. The improved algorithm model YOLOv5-Res has mAP@0.5 of 82.1%, FLOPs of 15.6G, parameters of 5486055, and model size of 10.8MB. The YOLOv5-Res4 model has mAP0.5 of 79.9%, FLOPs of 5.3G, parameter count of 1094471, and model size of 2.4MB. Overall, YOLOv5-Res was able to achieve high accuracy in detecting apple leaf diseases, while YOLOv5-Res4 still had high detection accuracy while being lightweight, and it also had certain advantages in comparison with mainstream models.
References
- Bi C, Wang J, Duan Y, et al. MobileNet based apple leaf diseases identification. Mobile Networks and Applications, 2022: 1-9. [CrossRef]
- Abbaspour-Gilandeh Y, Aghabara A, Davari M, et al. Feasibility of using computer vision and artificial intelligence techniques in detection of some apple pests and diseases. Applied Sciences, 2022, 12(2): 906. [CrossRef]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: a review. Plant Methods, 2021, 17: 1-18. [CrossRef]
- Fuentes, A.; Yoon, S.; Park, D.S. Deep learning-based techniques for plant diseases recognition in real-field scenarios//Advanced Concepts for Intelligent Vision Systems: 20th International Conference, ACIVS 2020, Auckland, New Zealand, February 10–14, 2020, Proceedings 20. Springer International Publishing, 2020: 3-14.
- Ren S, He K, Girshick R, et al. Faster r-cnn: Towards real-time object detection with region proposal networks. Advances in neural information processing systems, 2015, 28.
- He K, Gkioxari G, Dollár P, et al. Mask r-cnn//Proceedings of the IEEE international conference on computer vision. 2017: 2961-2969.
- Lee, S.H.; Wu, C.C.; Chen, S.F. Development of image recognition and classification algorithm for tea leaf diseases using convolutional neural network//2018 ASABE Annual International Meeting. American Society of Agricultural and Biological Engineers, 2018: 1.
- Singh U P, Chouhan S S, Jain S, et al. Multilayer convolution neural network for the classification of mango leaves infected by anthracnose disease. IEEE access, 2019, 7: 43721-43729. [CrossRef]
- Li H, Shi H, Du A, et al. Symptom recognition of disease and insect damage based on Mask R-CNN, wavelet transform, and F-RNet. Frontiers in Plant Science, 2022, 13: 922797. [CrossRef]
- Liu W, Anguelov D, Erhan D, et al. Ssd: Single shot multibox detector//Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part I 14. Springer International Publishing, 2016: 21-37.
- Redmon J, Divvala S, Girshick R, et al. You only look once: Unified, real-time object detection//Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 779-788.
- Redmon J, Farhadi A. YOLO9000: better, faster, stronger//Proceedings of the IEEE conference on computer vision and pattern recognition. 2017: 7263-7271.
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767, 2018.
- Bochkovskiy, A.; Wang, C.Y.; Liao HY, M. Yolov4: Optimal speed and accuracy of object detection. arXiv preprint arXiv:2004.10934, 2020.
- Sun H, Xu H, Liu B, et al. MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neural networks. Computers and Electronics in Agriculture, 2021, 189: 106379. [CrossRef]
- Tian Y, Yang G, Wang Z, et al. Detection of apple lesions in orchards based on deep learning methods of cyclegan and yolov3-dense. Journal of Sensors, 2019, 2019. [CrossRef]
- Li J, Qiao Y, Liu S, et al. An improved YOLOv5-based vegetable disease detection method. Computers and Electronics in Agriculture, 2022, 202: 107345. [CrossRef]
- Lin J, Yu D, Pan R, et al. Improved YOLOX-Tiny network for detection of tobacco brown spot disease. Frontiers in Plant Science, 2023, 14: 1135105. [CrossRef]
- Khan F, Zafar N, Tahir M N, et al. A mobile-based system for maize plant leaf disease detection and classification using deep learning. Frontiers in Plant Science, 2023, 14: 1079366. [CrossRef]
- Jocher G, Chaurasia A, Stoken A, et al. ultralytics/yolov5: v6. 0-yolov5 classification models, apple m1, reproducibility, clearml and deci. ai integrations. Zenodo, 2022.
- Park H, Yoo Y, Seo G, et al. C3: Concentrated-comprehensive convolution and its application to semantic segmentation. arXiv preprint arXiv:1812.04920, 2018.
- He K, Zhang X, Ren S, et al. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE transactions on pattern analysis and machine intelligence, 2015, 37(9): 1904-1916. [CrossRef]
- Lin T Y, Dollár P, Girshick R, et al. Feature pyramid networks for object detection//Proceedings of the IEEE conference on computer vision and pattern recognition. 2017: 2117-2125.
- Liu S, Qi L, Qin H, et al. Path aggregation network for instance segmentation//Proceedings of the IEEE conference on computer vision and pattern recognition. 2018: 8759-8768.
- Zheng Z, Wang P, Liu W, et al. Distance-IoU loss: Faster and better learning for bounding box regression//Proceedings of the AAAI conference on artificial intelligence. 2020, 34(07): 12993-13000.
- Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions//Proceedings of the IEEE conference on computer vision and pattern recognition. 2015: 1-9.
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition//Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 770-778.
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift//International conference on machine learning. pmlr, 2015: 448-456.
- Hua, B.S.; Tran, M.K.; Yeung, S.K. Pointwise convolutional neural networks//Proceedings of the IEEE conference on computer vision and pattern recognition. 2018: 984-993.
- Ang G, Han R, Yuepeng S, et al. Construction and verification of machine vision algorithm model based on apple leaf disease images. Frontiers in Plant Science, 2023, 14: 1246065. [CrossRef]
- Lucic A, Oosterhuis H, Haned H, et al. FOCUS: Flexible optimizable counterfactual explanations for tree ensembles//Proceedings of the AAAI Conference on Artificial Intelligence. 2022, 36(5): 5313-5322.
- Yang, Q.; Duan, S.; Wang, L. Efficient identification of apple leaf diseases in the wild using convolutional neural networks. Agronomy, 2022, 12(11): 2784. [CrossRef]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv preprint arXiv:1511.08060, 2015.
- Jingze Feng, Xiaofei Chao. Apple Tree Leaf Disease Segmentation Dataset[DS/OL]. V1. Science Data Bank, 2022[2024-02-26]. https://cstr.cn/31253.11.sciencedb.01627. CSTR:31253.11.sciencedb.01627.
- Thapa R, Zhang K, Snavely N, et al. The Plant Pathology Challenge 2020 data set to classify foliar disease of apples. Applications in plant sciences, 2020, 8(9): e11390. [CrossRef]
- Ai Studio poublic datasets. (2024). “Pathological images of apple leaves.” Available at: https://aistudio.baidu.com/aistudio/datasetdetail/11591/0.
- Wang C Y, Liao H Y M, Wu Y H, et al. CSPNet: A new backbone that can enhance learning capability of CNN//Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops. 2020: 390-391.
- Sun Y, Chen G, Zhou T, et al. Context-aware cross-level fusion network for camouflaged object detection. arXiv preprint arXiv:2105.12555, 2021.
- Sandler M, Howard A, Zhu M, et al. Mobilenetv2: Inverted residuals and linear bottlenecks//Proceedings of the IEEE conference on computer vision and pattern recognition. 2018: 4510-4520.
- Zhang X, Zeng H, Guo S, et al. Efficient long-range attention network for image super-resolution//European Conference on Computer Vision. Cham: Springer Nature Switzerland, 2022: 649-667.
- LV, Wenyu, et al. Detrs beat yolos on real-time object detection. arXiv preprint arXiv:2304.08069, 2023.
- Reis D, Kupec J, Hong J, et al. Real-Time Flying Object Detection with YOLOv8. arXiv preprint arXiv:2305.09972, 2023.
- Wang C Y, Yeh I H, Liao H Y M. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. arXiv preprint arXiv:2402.13616, 2024.
- Adarsh, P., Rathi, P., & Kumar, M. (2020, March). YOLO v3-Tiny: Object Detection and Recognition using one stage improved model. In 2020 6th international conference on advanced computing and communication systems (ICACCS) (pp. 687-694). IEEE.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).