1. Introduction
Enveloped viruses, characterized by a lipid bilayer derived from the host cell membrane, represent a diverse group of pathogens [
1]. In particular, enveloped viruses such as human immunodeficiency virus (HIV) and coronavirus (CoV) pose a significant threat to human. The process of entry of the enveloped virus genome into the target cell involves two primary steps, initial envelope binding to cell surface receptor and subsequent fusion with the cell membranes [
2,
3,
4]
. As a type of CoV, the COVID-19 represents a class of RNA viruses characterized by an enveloped structure with a diameter of approximately 80-160 nm [
5]
. The enveloped structure of COVID-19 is composed of a lipid membrane with three types of glycoproteins on its surface: the spike protein (S), which functions in receptor recognition, cell lysis, and primary antigen; the small envelope glycoprotein (E) that facilitates attachment to the cell membrane; and the membrane glycoprotein (M), which is responsible for the formation of the envelope and the budding of new viruses [
6,
7,
8]
. The virus achieves its invasion primarily by binding to host cell receptors through the S portion [
9,
10]
. The S portion contains two subunits, S1 and S2, and forms a cloverleaf-shaped trimer with three S1 heads and a trimeric S2 stalk. S1 can be further subdivided into N-terminal domain (NTD), receptor binding domain (RBD) and C-terminal domain (CTD) [
11]
. The RBD locates at the tip of S1 head [
12]
. The S1 subunit of COVID-19 is primarily responsible for recognizing and binding to the angiotensin converting enzyme 2 (ACE2) on the host cell, facilitating the virus's entry into the host cell [
13]. The RBD is also the main part of antigen variation and immune escape, for example, the COVID-19 Omicron mutant, which has been widely spread in the world, has 17 amino acid mutations in the RBD region
. Meanwhile
, the RBD also provides a target for the development of preventive COVID-19 vaccines and drugs [
14]. The frequent ineffectiveness of vaccines or acquisition of drug resistance due to the high variability of RNA viruses is a common occurrence. To address such issues, the discovery of antiviral compounds that act through different mechanisms, like compounds that target differentially glycosylated binding of the RBD, or compounds that produce protein-protein interactions with conserved domains of the RBD, are crucial.
Lectins are diverse proteins that bind carbohydrates and agglutinate erythrocytes [
15]. Since lectins can specifically recognize and bind to carbohydrate structures, they are classified into different lectin families [
16]. Numerous lectins were identified to have antiviral properties, with a particular attention on lectins that target densely glycosylated RBD of enveloped viruses. The OAAH lectin exhibits robust anti-influenza virus activity through specific binding to high-mannose glycans present on the envelope glycoprotein hemagglutinin, effectively preventing viral entry into host cells [
17]. Griffithsin (GRFT), a lectin isolated from the red algae, shows remarkable broad-spectrum antiviral activity. Previous studies have demonstrated that the ability of GRFT to inhibit HIV infection at a picomolar concentration, along with its ability to induce resistance against the Severe Acute Respiratory Syndromes (SARS) and Middle East Respiratory Syndrome (MERS) viruses [
18,
19,
20]. As for the CV-N lectin, in addition to its inhibitory effect on HIV virus, it also has potential for the treatment of the filovirus Ebola through sugar-specific recognition [
21]. Undoubtedly, algae lectins have a vast potential as natural antiviral compounds, suggesting promising applications in various fields. Microcystis viridis lectin (MVL), is one of algae lectins isolated from M. viridis NIES-102 strain, a unicellular freshwater bloom-forming cyanobacterium [
22]. MVL consisted of 113 amino acid residues including two tandemly repeated homologous
54-residue carbohydrate binding domains [
22]
, consist homodimer stabilized by the boomerang-shaped monomers interlock, and each domain from one monomer contacts both domains from the second monomer [
23]
. MVL also showed the inhibition of HIV-1 envelope-mediated cell fusion at nanomolar concentrations by binding to high mannose N-linked carbohydrate on the surface of the envelope glycoprotein gp120 [
24]
.
In this study, our research focuses on the screening and mechanistic exploration of blue-green algae Microcystis viridis lectin MVL with anti-COVID-19 potential against four COVID-19 variants. For this purpose, we prepared recombinant MVL, and assessed its binding properties against spike protein RBDs of COVID-19 variants, comparing with different type of lectins such as Jacalin, galactose-specific lectin from Artocarpus altilis [
25], mannose-specific lectin from Dioscoria batata (DB1) [
26], Concanavalin A (Con A) from Canavalia ensiformis [
27], PHA-M legume lectin from Phaseolus vulgaris [
28], and CSL3 from eggs of the chum salmon (Oncorhynchus keta) [
29].
2. Results
2.1. Expression, Purification and Characterization of the Recombinant MVL
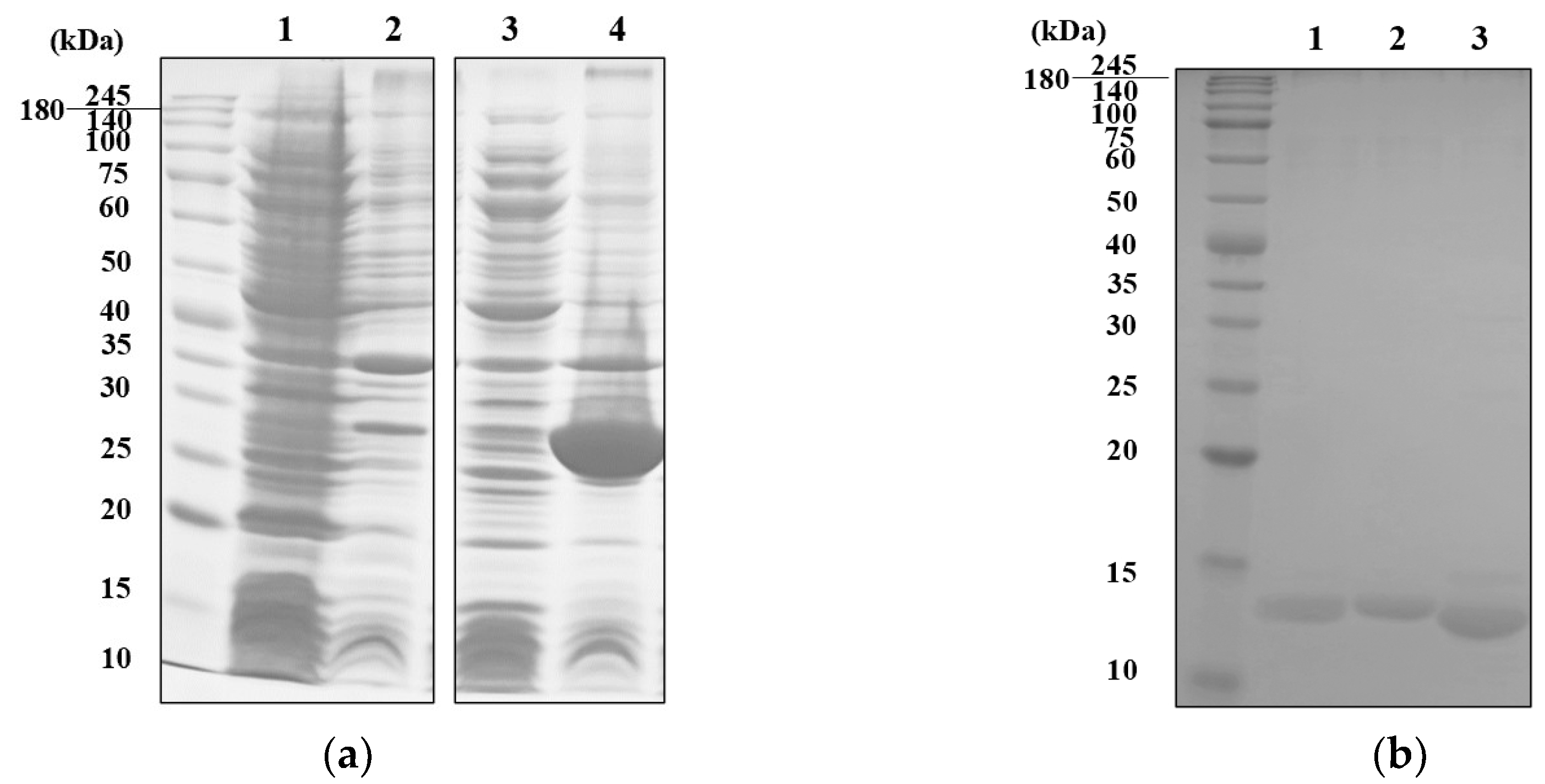
The recombinant MVL (rMVL) was successfully expressed as a TrxA (thioredoxin) fusion protein by using a pET32a vector (
Figure 1a and
Figure S1). Although the optimization included varying host cell types, culture temperature and conditions were attempted, rMVL-TrxA was expressed in the inclusion bodies under all tested conditions (
Supplementary Figure S2).
In order to obtain the soluble rMVL, we conducted the purification under a denaturing condition and refolding of rMVL-TrxA. Briefly, the inclusion bodies were denatured in 8 M urea and then purified by a nickel affinity chromatography (
Figure S2). Subsequently, the rMVL-TrxA was refolded by dialysis at 4°C with 20 mM Tris buffer (pH 7.8) to remove urea. After removing the TrxA tag (13.9 kD) by thrombin digestion, purified rMVL (15.6 kDa) was successfully obtained (
Figure 1b and
Figure S3). The activity of rMVL and other lectins was confirmed by the hemagglutination assays with 2% rabbit red blood cells. The minimum hemagglutinating concentration of rMVL was 0.008 mg/mL, showing a slightly lower activity than the reported native MVL (0.0015 mg/mL) (
Table 1 and
Figure S4).
2.2. Interaction and Binding Properties of rMVL with COVID-19 Spike Protein RBDs
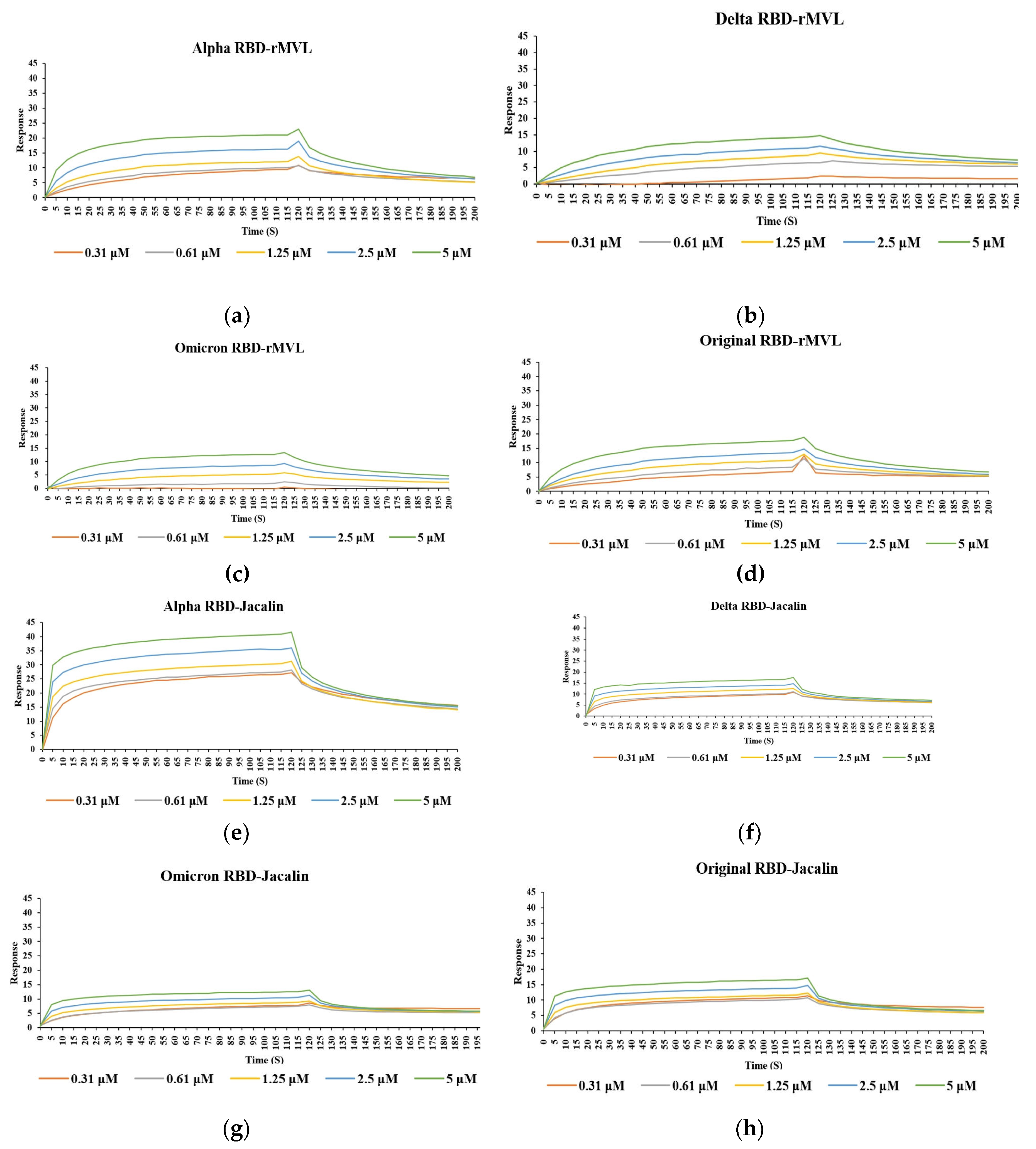
The binding properties of six lectins including rMVL (
Table 1) against COVID-19 spike protein RBDs from four variants were assessed by surface plasmon resonance (SPR) technology (
Figure 2). As a result, MVL and Jacalin lectin showed the binding ability with spike protein RBD of various COVID-19 mutants, while other lectins showed no or very weak binding abilities to the spike protein RBDs (
Figure 2).
Table 2 summarizes the binding properties of lectins against RBDs, including kinetic parameters. In particular, Jacalin, which is a galactose-specific lectin, can effectively interact with spike proteins RBDs from four mutants, (alpha, delta, omicron and original RBDs) with similar
KD values ranging from 3.20 to 4.78 x 10
-7 M (
Figure 2a and
Table 2). On the other hand, MVL lectin, which was reported as a mannose -specific lectin [
23], also showed interactions with the various mutant spike protein RBDs. However, its binding ability was lower than that of Jacalin, with
K
D values ranging from 1.29×10
-6 to 2.73×10
-6 M (
Figure 2b,
Table 2). The binding ability of MVL to each COVID-19 spike protein RBD followed this order from highest to lowest: alpha, delta, original and omicron RBDs. While the other mannose-specific lectins, Con A, PHA-M, DB1, and CSL3, did not interact with the spike protein RBD.
2.3. Carbohydrate Binding Specificity of MVL
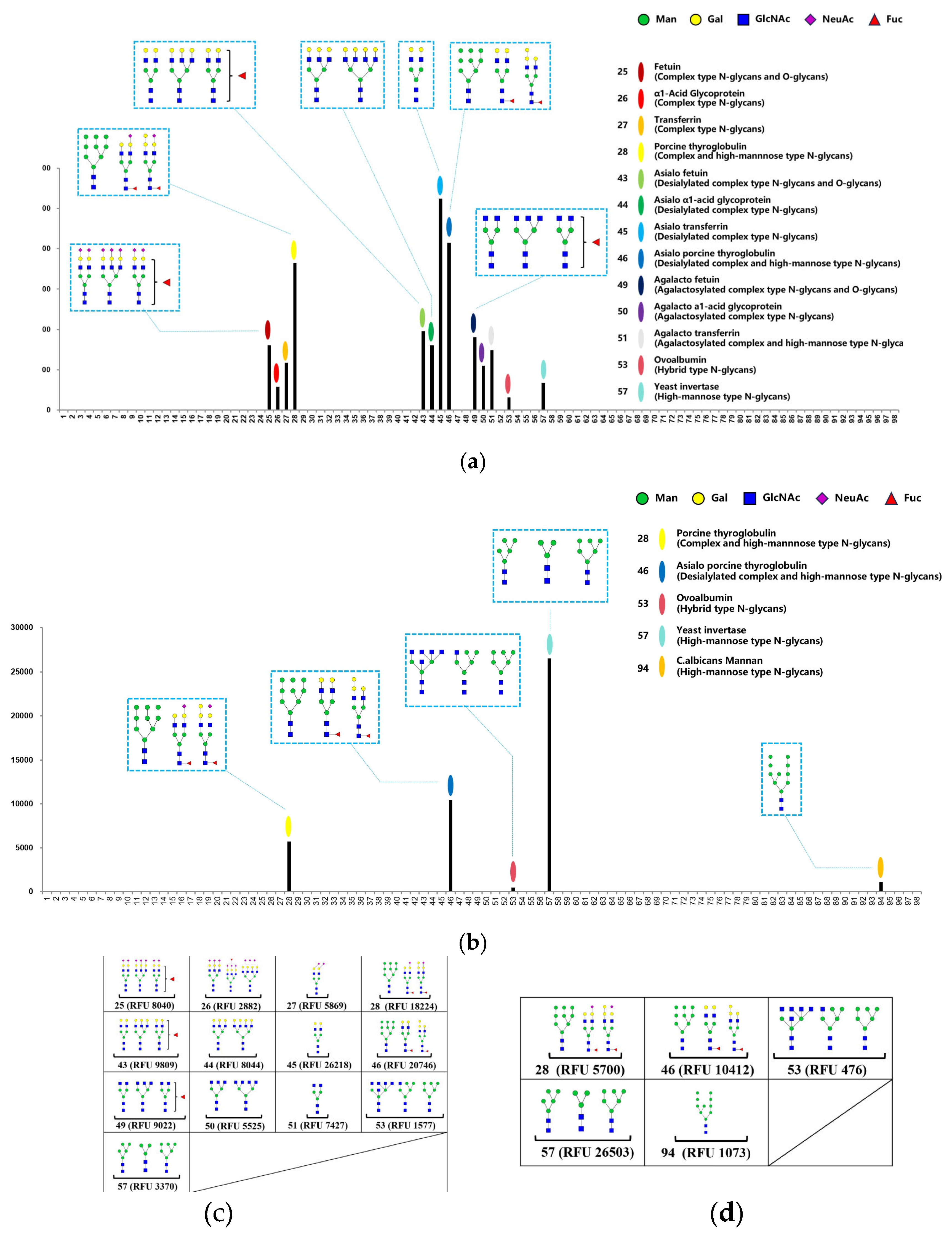
MVL lectins are considered to be mannose-binding lectins [
23], but their detailed glycan specificities are not known. Therefore, we analyzed the glycan specificity of rMVL using glycan array analysis technology and compared it with mannose-binding DB1. As shown in
Figure 3, rMVL recognized 13 types of glycans in 4 categories, including complex-type, desialylated complex-type, agalactosylateds complex-type and high mannose-type N-glycans. In contrast, DB1 recognized 5 types of glycans in 3 categories, including complex-type, desialylated complex-type, and high-mannose type glycans.
rMVL binds to four glycoproteins, i.e., Fetuin, α1-acid glycoprotein, transferrin, and thyroglobulin with different sugar terminus (
Figure 3a,c). rMVL showed the highest binding to glycans containing desialylated complex-type N-glycans with galactose terminus (No.43-46 in
Figure 3a and 3c). The highest binding was observed for asialo transferrin at the relative fluorescence units (RFU) of 26218 (No.45 in
Figure 3a,c) and asialo porcine thyroglobulin at RFU of 20746 (No.46 in
Figure 3a,c), respectively. The rMVL binding to glycans containing complex-type N-glycans and agalactosylated complex-type N-glycans were lower than that of desialylated complex-type N-glycans (No.25-28 or 49-51 in comparison with No. 43-46 in
Figure 3a,c). This suggests that the galactose terminus enhances the binding. Therefore, the binding to glycans containing complex-type N-glycans varied between glycoproteins, RFU ranging from 2882 (α1-acid glycoprotein) to 18224 (porcine thyroglobulin glycan), suggesting relatively low binding to NeuAc terminus. Conversely, the binding to glycans containing agalactosylated complex-type N-glycans did not exceed 1000, indicating relatively low binding to GlcNAc terminus. The binding effects of MVL lectin to high mannose type N-glycans were also low, RFU 1577 (ovalbumin) and RFU 3370 (invertase), respectively. These results suggest that rMVL preferentially binds to the galactose terminus comparing to the mannose, GlcNAc, and NeuAc terminus.
In addition, the DB1 lectin exhibited recognition exclusively towards five tested glycans. These glycans share the sugar chain of high mannose type N-glycan (No. 28, 46, 53, 57 and 94 in
Figure 3b,d), confirming that DB1 lectin recognizes the mannose-terminus. This lectin showed the highest binding affinity with the yeast invertase glycan at the RFU of 26503, and the lowest binding with the hybrid type N-glycan of ovoalbumin at the RFU of 476 (No. 57 and No. 53 in
Figure 3b,d). These results indicate that MVL exhibits sugar-binding specificity distinct from that of DB1 and suggest that this specificity is related to their differential interaction with the spike protein RBDs. Consequently, these results indicate that rMVL is a mannose/galactose-specific binding lectin rather than a mannose-binding lectin.
2.4. Galactose Inhibits The Interaction between Lectins and RBDs
The inhibitory effects of specific glycans on the lectin binding to the spike protein were examined. The results showed that untreated Jacalin bound well to the alpha and omicron spike protein RBDs. However, after galactose treatment (30 min), the binding of Jacalin was inhibited almost completely in a concentration-dependent manner, even at the lowest concentration of galactose (90% at 12.5 mM for alpha and 80% at 12.5 mM for omicron) (
Figure 4c,d). These findings suggest that Jacalin and spike protein RBDs interact via glycan-specific recognition. The effect of galactose on the MVL lectin was quite different. The untreated MVL could bind to alpha and omicron spike protein RBDs. However, the galactose treated MVL exhibited interesting SPR profiles: although the binding of MVL to the spike protein RBDs was inhibited by galactose, as with jacalin lectin, treatment with the highest concentration of 100 mM galactose did not completely inhibit the interaction of MVL lectin with alpha or omicron spike protein RBDs, and approximately 40% of SPR signal was retained. Furthermore, there was no significant difference between the response at the lowest sugar concentration of 12.5 mM and the highest concentration of 100 mM (
Figure 4a,b). These results indicate that the interaction between MVL and RBDs are only partially dependent on galactose-specific interactions.
When the carbohydrate recognition sites of MVL were blocked with excessive (1000-times more) amount of galactose, nearly half MVL still retained binding ability to RBDs. Therefore, we hypothesized that the interaction between MVL lectin and spike protein RBDs were not only glycan-specific recognition, but also protein-protein interaction between MVL lectin and spike protein RBD.
2.5. The Protein-Protein Interaction between MVL Lectin and RBDs
To explore the protein-protein interaction between MVL and RBDs, we used
in silico method to predict the binding sites and modes between them. According to the structural comparison of alpha-fold2 predicted RBD mutants (
Figure 5), these structures are almost identical, even though Omicron RBD contains more than a dozen mutation sites compared with other RBDs.
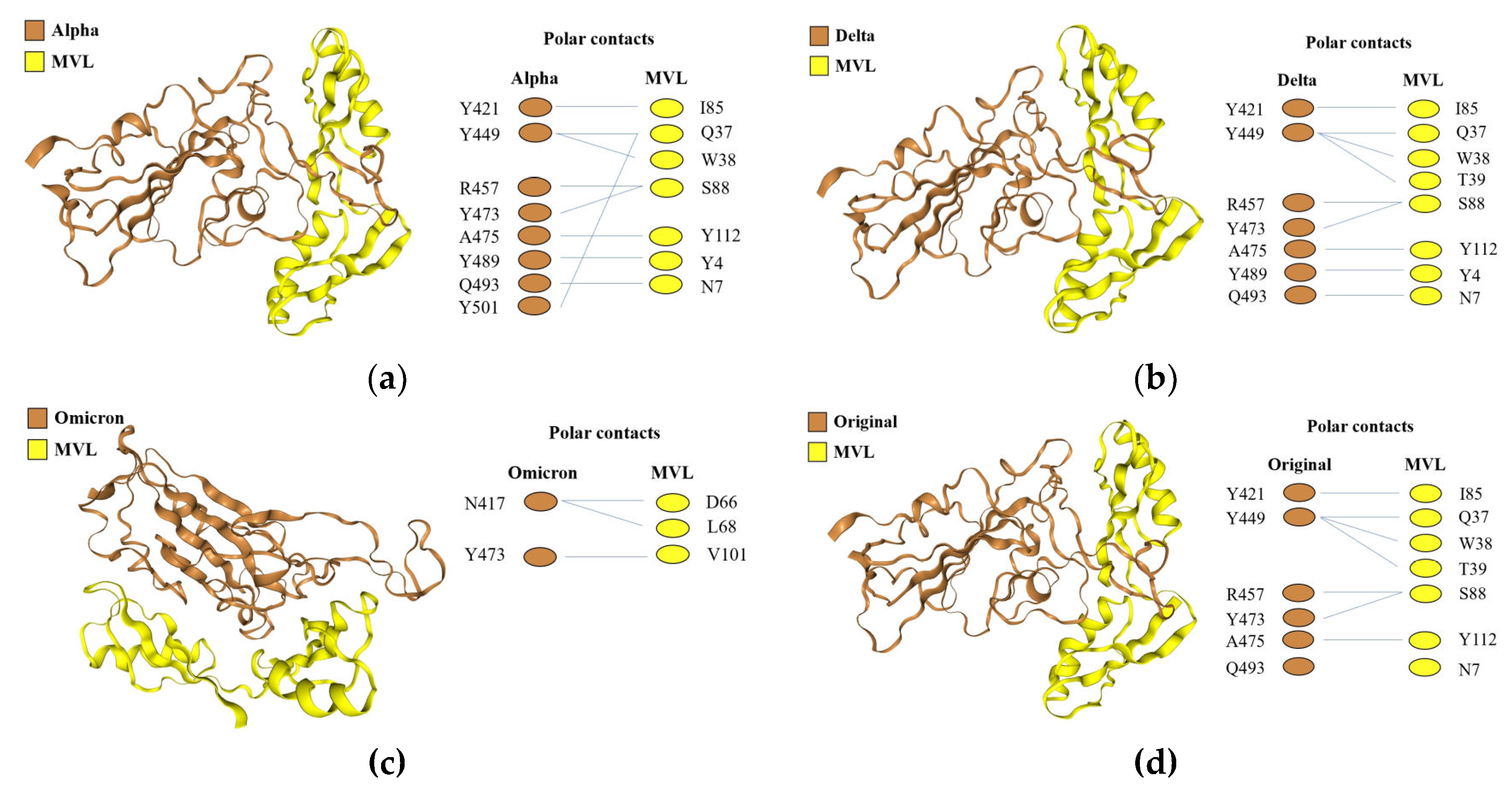
Subsequently, the docking analysis between four spike protein RBDs and rMVL was performed using HDOCK software, and the complex structure with the lowest docking energy score was selected, alpha -293.37 kcal/mol, delta -296.37 kcal/mol, omicron -307.74 kcal/mol, and original -294.01 kcal/mol, respectively (
Figure 6)
Compared with omicron, which has 16 mutations compared to other RBDs, the protein-protein interaction sites of alpha, delta, original RBDs and MVL were close. Alpha RBD has 8 amino acid sites binding to MVL (
Figure 6a), one more Y501 than the delta RBD, and the rest are completely consistent (
Figure 6b). Y501 was the only mutation site of RBD of alpha variant, while the R452 and K478 mutation sites of RBD of delta variant did not bind to MVL. Delta has one more Y489 than original RBD, and the rest were completely same (
Figure 6d). As for omicron, only two amino acids bound to MVL, and Y473 also exists in the other three RBDs, while N417 was the unique RBD mutation amino acid site of omicron variant (
Figure 6c). Another more interesting phenomenon is that tyrosine was most predominant in the binding site of RBDs to MVL, occupying almost half of the position.
However, it should be noted that although the lowest energy score with the highest confidence showed that MVL bound to the C-terminal amino acids of RBDs, which contribute to binding to the ACE2 receptor (
Figure 5b). The docking results of the lowest energy score from the second to the tenth showed that MVL was combined with RBDs in a different manner (
Figure S5). These results suggest that MVL interacts with Covid-19 S-protein RBDs at the binding site of RBDs to ACE2 through protein-protein interaction.
3. Discussion
In this study, the interaction of six lectins with four COVID-19 spike protein RBDs was analyzed using the SPR. As a result, two galactose-specific lectins MVL and Jacalin showed high affinity to COVID -19 RBDs. Although many mannose-binding lectins were reported to recognize high mannosylated COVID-19 RBDs [
30], ConA, PHA-M and DB1 lectins, the mannose-binding lectins, were unable to interact with the RBDs in this study. The most interesting finding in this study is that MVL can bind to the spike protein RBD through protein-protein interaction. When the sugar chain recognition was blocked by galactose, jacalin binding to the COVID-19 RBD was lost, but MVL (almost half) was able to bind to the spike protein RBDs, suggesting the protein-protein interaction.
In Silico docking analysis suggested the presence of protein-protein interaction between MVL lectin and spike protein RBDs. The complex structures showed that MVL lectin interacted with COVID-19 spike protein RBDs mainly through polar contacts. Lan et al. performed X-ray crystallography analysis with a complex composed of recombinant COVID-19 RBD and ACE2 expressed in Hi5 insect cells [
31]. The analysis showed that a total of 17 residues of the RBD were in contact with 20 residues of ACE2 and that these 17 residues were mainly located at the C-terminal end of the amino acid sequence of the RBD, from K417 to Y505 (
Figure 5b) [
31].
In Silico docking results demonstrated that the binding sites of the four RBDs to MVL were also located at the C-terminal end from amino acid 417, and three of them, Y449, Y489, and Q493, were associated with the amino acid position of the RBD binding to hACE2. The other sites, although not identical, were also in the vicinity of the binding site of RBD and hACE2. This result implies that MVL prevents the binding of RBD to hACE2 by masking the amino acid binding site between RBD and hACE2. It is worth noting that many π - π stacks in MVL were also involved to varying degrees in the binding with RBDs. The π - π stack refers to the attractive, non-covalent interactions between aromatic rings, which is strongly associated with the broad-spectrum antiviral properties of lectins [
32,
33].
The number of polar interaction sites of the four mutant RBDs with MVL was 8 for alpha, 7 for delta, 6 for original, and 2 for omicron, respectively. This number of interactions was consistent with the order of affinity between MVL and mutant RBDs as measured by SPR. In other words, more binding sites have higher affinity than fewer.
In addition, MVL can recognize complex-type N-glycans, desialylated complex-type N-glycans, and agalactosylated complex-type N-glycans whose termini are galactose. It has been reported that the RBD expressed in HEK293 cells has galactose groups at the terminal N331 and N343 of the N-glycan chain [
34,
35]. These glycosylation properties of spike protein RBDs were thought to be the main reason why jacalin and MVL, but not other lectins such as Con A and DB1, bind to spike protein RBDs. However, the current lectins that recognize the spike protein or RBD of COVID-19 by carbohydrates are mostly high-mannose binding lectins. For example, Griffithsia lectin derived from the red marine alga
Griffithsia sp. and BanLec lectin isolated from
Musa acuminata are both high-mannose lectins that can effectively inhibit the binding of COVID-19 spike protein to the hACE2 receptor at nM concentration [
36,
37]. COVID-19 spike protein contains a total of 22 glycosylation sites, including high-mannose, hybrid and complex-type N-glycosylation sites, with high-mannose-type glycosylation sites accounting for the majority [
38]. Therefore, the above two high-mannose-type lectins can inhibit COVID-19 infection by recognizing the high-mannose glycosylation sites and binding to the S1 or S2 region of the COVID-19 spike protein. However, few studies have also reported the interaction between some lectins that specifically recognize the complex-type N-glycan with galactose moiety and the COVID-19 RBD. The FRIL lectin isolated from
Edible Lablab Beans is a complex-type N-glycan binding lectin that can recognize the sugar chain with galactose groups on the outermost side, and has a weak affinity with mannose carbohydrate simultaneously, which is similar to MVL lectin. FRIL lectin can effectively recognize COVID-19 galactose glycoproteins, interfering with COVID-19 enter cells [
39].
The MVL shows strong resistance to COVID-19, while its carbohydrate specificity is galactose rather than the previously reported mannose. Although MVL also shows a weak mannose recognition, it was only about 10% of galactose. This may be the reason why MVL can bind well to RBD compared to other mannose lectins. Therefore, to explain this difference, it is necessary to analyze the interaction between the MVL lectin and RBDs at the atomic level by cryo-electron microscopy in the future study.
Research on lectin anti-envelope virus has mainly focused on lectin binding to virus RBD through carbohydrate-specific recognition and inhibition of virus invasion [
40,
41,
42]. There has been limited research on lectin inhibition of binding of virus RBD to its receptor through protein-protein interaction, except for C-type lectin fold [
43]. Thus, MVL lectin has dual antiviral potent activity via protein-protein interaction inhibitor and carbohydrate-specific recognition.
4. Materials and Methods
4.1. Materials
PHA-M (Cat# L-8902) and ConA (Cat# C-2010) lectins were purchased from Sigma, and Jacalin lectin (Cat# L-1150) was purchased from Vector Laboratories Co. The lectins were obtained as lyophilized powders and stored at -30°C. During use, the lectin powders were dissolved in 100 mM Tris-HCl buffer at a concentration of 1 mg/ml. DB1 lectin was purified according to our previous work [
26]. Briefly, the homogenized sample from
Dioscorea batatas yam tubers was first purified by ammonium sulfate precipitation method, followed by hydrophobic interaction chromatography on a Phenyl-Toyopearl 650 M column, and finally purified by anion-exchange chromatography on a HiTrap Q column with a NaCl gradient. CSL3 was also purified from chum salmon (
Oncorhynchus keta) eggs as previously described [
29] and stored at -30˚C until use. All other reagents were of the purest grade commercially available. The anti-EGFP of western blot detection was purchased from Fujifilm, anti-mouse IgG secondary antibody was obtained from Roche.
4.2. Expression And purification of Recombinant MVL
For the expression of recombinant
Microcystis viridis NIES-102 lectin (rMVL), cDNA encoding MVL subcloned into pVT118N vector in our previous work [
22] was used. After introducing the restriction enzyme sites, NcoI and HindIII, by polymerase chain reaction (PCR) using MVL forward and reverse primers, the PCR fragment of MVL was introduced into the pET32a vector by ligation with T4 DNA ligase, then the resulting plasmids were transformed into DH5α competent cells. Next day seeded single positive colony into LB medium (0.1% ampicillin) and cultured at 37°C overnight to extract plasmids using the extraction kit (Cat# FAPDE 001-1). The successfully constructed plasmid confirmed by sequencing analysis was temporarily stored at -30°C.
The constructed plasmids were transformed into BL21 (DE3) Escherichia coli competent cells and selected by LB plates supplemented with ampicillin. Meanwhile, the empty vector pET32a was also transformed into BL21 (DE3) as a negative control. Single colony was inoculated into 3.5 ml of liquid LB (0.1% ampicillin) and pre-cultured at 37°C overnight. Then the bacteria were inoculated into 200 ml LB (0.1% ampicillin) at 1:100 ratio and shaking incubated at 37°C until OD600 reached 0.4-0.6. After approaching target concentration, 0.5 mM of IPTG was added to induce expression at 23°C for further 4 h. Subsequently, the Escherichia coli cells were harvested by centrifugation at 8000 xg for 10 min, removed supernatant and recovered precipitate was stored at -30°C.
The precipitate was resuspended in sonication buffer (50 mM Tris, 100 mM NaCl, pH 8.0) and sonicated on ice for 30 min. (30 s on/30 s off). The debris was removed by centrifugation at 18,000 xg for 15 min. The supernatant and insoluble precipitate were analyzed by 12.5% SDS–PAGE. The inclusion bodies, existing in a precipitated form, were initially washed three times with a cleaning buffer to remove cellular debris in wash buffer (50 mM Tris, 100 mM NaCl, 10 mM EDTA, 0.5% (w/w) TritonX-100, pH 8.0) at 4°C, each time 30 min and collected the precipitate by centrifugation at 15 000 xg for 10 min. Subsequently, the inclusion bodies were subjected to overnight treatment at 4°C with denaturation buffer (50 mM Tris, 100 mM NaCl, 1 mM 2-mercaptoethanol, 8 M Urea, pH 7.8). Next day, the supernatant was recovered after centrifugation at 15 000 xg for 10 min, and purified by nickel affinity chromatography (ÄKTA Pure Protein Purification System) at denaturation conditions with loading buffer (50 mM Tris, 100 mM NaCl, 1 mM 2-Mercaptoethanol, 5 mM Imidazole, 8 M Urea, pH 7.8). The sample was eluted at 20 mM, 100 mM, 250 mM imidazole concentration, and fractions were immediately analyzed by 12.5% SDS–PAGE electrophoresis.
The fractions containing the target protein were mixed and dialyzed into thrombin cleavage buffer (20 mM Tris, 30 mM NaCl, 2 mM, CaCl2, pH 8.0). Then, 10U thrombin was added to the sample to remove the TrxA-tag at RT for 2 h reaction, terminated by adding PSMF until 2 mM final concentration. Following this, the lectin and TrxA-tag were separated through further nickel affinity chromatography. Pure lectin obtained at flow through (FT), and TrxA tag recovered at 100 mM imidazole concentration fraction. Similarly, the digested lectin-TrxA samples, FT fraction and 100 mM imidazole fraction were detected by SDS-PAGE.
4.3. Construction of COVID-19 Mutants Spike Protein RBD Expression System
This experiment involved four types of spike protein RBDs of COVID-19, including the original (wild-type) and three mutants, alpha, delta, and omicron. The original COVID-19 spike protein RBD plasmid, pcDNA3-SARS-CoV-2-S-RBD-sfGFP, was kindly obtained from Prof. Erik Procko laboratory via addgene [
44]. Alpha and delta variants undergo site-directed mutagenesis on the N501Y single mutation site for the alpha variant, as well as the L452R and T478K dual mutation sites for the delta variant using the quick-change method.
For the omicron variant, due to numerous mutation sites in its RBD, the In-fusion method was employed to ligate the synthetic omicron spike protein RBD gene fragment (Vector Laboratories company) with the pcDNA3-sfGFP vector fragment, and constructed the pcDNA3-omicron-RBD-sfGFP plasmid. In simple terms, the process began by designing primers for the amplification of the Omicron RBD and pcDNA3-sfGFP vector fragment, then amplified through PCR. The resulting products were identified and recovered by 1.2% agarose gel electrophoresis. Subsequently, the insert fragment and linearized vector fragment were ligated together by 5X In-Fusion Snap Assembly Master Mix. Because the selection resistance of plasmid is also to ampicillin, the amplification and extraction methods for the constructed COVID-19 mutants spike protein RBD plasmid were consistent with the lectin plasmid construction method mentioned earlier.
The successfully constructed plasmid was transfected into HEK293T cells for secretion expression using polyethylenimine (PEI). Firstly, HEK293T cells were thawed and seeded in 10 cm tissue culture dishes with DMEM medium containing 10% fetal bovine serum (FBS). The cells were cultured for 3 days at 37°C with 5% CO2 concentration. After successful cell recovery, the cells underwent two times passages before the PEI transfection procedure, and the confluence of cells before transfection was about 90%. Mixed 10 μg of plasmid to be transfected with 250 μl of Opi-EME medium, then added 25 μl of PEI, mixed well again, and reacted at RT for 15 min. The resulting mixture was transferred to HEK293T cells and incubated for 24 h. The next day, the culture medium was replaced with an FBS-free DMEM medium, and the cells were cultured for 72 h before harvesting.
The harvested culture medium was purified through nickel column affinity chromatography. The culture medium was loaded onto a 1 mL pre-packed nickel column using binding buffer (50 mM Tris, 100 mM NaCl, pH 8.0). The elution was performed with the elution buffer (50 mM Tris, 100 mM NaCl, 500 mM Imidazole, pH 8.0). The fractions were collected at 20 mM, 50 mM, and 250 mM imidazole concentrations, then subjected to Western blotting analysis.
4.4. SDS-PAGE and Western Blotting
The purified MVL lectin fractions were subsequently analyzed using 12.5% SDS-PAGE electrophoresis to evaluate their expression and purity and stained with Coomassie Brilliant Blue for 1 h and then destained with acetic acid-methanol solution.
The Spike protein RBDs were also separated using SDS-PAGE and then transferred onto Polyvinylidene-Fluoride membranes. The membranes were blocked with 5% skim milk powder for 1 hour at RT and then incubated with anti-EGFP primary antibody at 4°C overnight. The next day, the membranes were subjected to secondary incubation with an anti-mouse IgG secondary antibody, followed by photographic detection.
4.5. Hemagglutination Assays
The biological activity of commercial lectin, recombinant lectin, and naturally extracted lectin was evaluated based on their aggregation effect on 2% rabbit erythrocytes (purchased from Japan Bio Serum Co., Hiroshima, Japan). The experiment was conducted in a U-bottom 96-well plate. Initially, 50 μL of PBS were dispensed into each well, followed by the addition of 50 μL of lectin into the wells of the first column. After thorough mixing, the mixture was subjected to two-fold serial dilution into the subsequent columns. The 96 well plate was stand at RT for 30 min, and observed for hemagglutination. Subsequently, the minimum concentration of lectins required to induce hemagglutination was determined.
4.6. Surface Plasmon Resonance (SPR) Detection of the Interaction between Lectin and COVID-19 Spike Protein RBDs.
The binding affinity (KD) of 6 kinds of lectins and 4 kinds of spike protein RBDs was measured at RT by the Biocore T200 molecular interaction instrument. Firstly, the spike protein RBDs and the lectin samples were dialyzed into HBS-EP+ buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.05% surfactant P 20, pH 7.4). Then, spike protein RBDs were diluted to 100 μg/ml by 10 mM sodium acetate (pH 4.5) and immobilized on a CM5 chip after biotinylating according to the instrument manual reaching a target level of 200 response units (RU), done by the amine coupling Kit (BR-1000-50) provide from GE company. Besides the sample flow cell, a reference flow cell that does not capture any protein molecular was used to correct the response contribution like the bulk shifts. The lectin samples to be analyzed were twofold diluted from 5 μM to 0.31 μM with HBS-EP buffer, and flowed through the sensor chip at 30 μl/min for 120 s, subsequently. To correct baseline drift or other disturbances, two blank cycles were performed before injecting lectin samples into the sensor chip. Between sample injections, the sensor chip was regenerated with pH 2.5 glycine at 30 μl/min for 60 sec to remove any bound analytes and clean the chip surface for the next analysis cycle. The entire experiment was conducted at 25°C. After the determination, the two-state reaction model of Biacore T200 evaluation software was used for interaction analysis, and the analysis parameter Rmax was set to ‘local’.
The interaction between lectins and spike protein RBDs was analyzed using SPR to determine if it depends on carbohydrate-specific recognition binding or protein-protein interaction binding. The specific carbohydrates corresponding to the lectins were prepared with HBS-EP+ buffer. They were then mixed with the lectin samples and allowed to react for 30min at RT. The final concentration of lectin was 5 μM, and the final concentrations of carbohydrate were 100 mM, 50 mM, 25 mM, 12.5 mM and 0 mM in gradient. Alpha and omicron spike protein RBDs, which have the strongest and weakest affinity to Jacalin and MVL, were selected for interaction verification. The rest of the experimental procedures were consistent with the section 2.10.
4.7. Carbohydrate-Specificity Analysis of MVL and DB1 Lectins
MVL (10 μg) and DB1 (10 μg) were labeled with Cy3-N-hydroxysuccinimide ester (GE Healthcare), and excess Cy3 was removed with Sephadex G-25 desalting columns (GE Healthcare), respectively. Cy3-labeled lectin was diluted with probing buffer [25 mM Tris-HCl (pH 7.5), 140 mM NaCl, 2.7 mM KCl, 1 mM CaCl2, 1 mM MnCl2, and 1% Triton X-100] to 10 μg/mL and was incubated with the glycan microarray at 20°C overnight. The glycan array was washed three times with probing buffer, and fluorescence images were captured using a Bio-Rex scan 200 evanescent-field-activated fluorescence scanner (Rexxam Co. Ltd., Kagawa, Japan).
4.8. In Silico Analysis
The spike protein RBD structure of original COVID-19 was obtained from the RDB database under the ID 6w41, and used as a template to predict the three-dimensional structure of RBDs of Alpha, Delta and Omicron mutants through SWISS-MODEL (
https://swissmodel.expasy.org/). The three-dimensional structure modeling of MVL lectin was predicted by alphafold2 (
https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb). The HDOCK molecular docking web server (
http://hdock.phys.hust.edu.cn/) was used for visual analysis to analyze the protein-protein interaction surface characteristics between MVL lectin and various RBDs. The docking result with the lowest energy score was selected, and PyMOL software was used for visual analysis to analyze the protein-protein interaction surface characteristics between MVL lectin and various RBDs.
5. Conclusions
In this study, the anti-COVID-19 abilities of six lectins from different sources was screened, and two lectins, Jacalin and MVL, with broad-spectrum anti-COVID-19 were successfully verified. The affinity order of these lectins to four types of COVID-19 spike protein RBDs was identical, which was alpha>delta>original>omicron. Jacalin and COVID-19 RBD were combined by galactose-specific recognition, while MVL and COVID-19 RBD were interacted by galactose-specific recognition and protein-protein interaction. According to the results of in silico analysis, MVL mainly bound with COVID-19 RBD through polar contact and mainly combined with the N-terminal of RBD. The number of amino acid binding sites between MVL and RBD is alpha>delta>original>omicron, which is consistent with the affinity order determined by SPR. In addition, π - π stack may also play a role in the antiviral effect of MVL.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Schematic diagram of an open reading frame (ORF) composed of lectin and thioredoxin (a) and amino acid sequence of TrxA-MVL fusion protein (b); Figure S2: Optimization of culture conditions for MVL-TrxA expression; Figure S3: The purification profiles of rMVL-Trx fusion protein (a) and thrombin-digested MVL-TrxA (b) on His-trap HP column; Figure S4: Hemagglutination assays of lectins using rabbit erythrocyte; Figure S5: Docking simulation of MVL and spike protein RBDs variants.
Author Contributions
Conceptualization, Z.W. and T.O.; designed experiments and performed analyses, Z.W., Z.Y., M.S., H.T. and T.O.; validation, Z.W., K.D., M.H., E.F. and T.O.; data curation, Z.W., Z.Y., M.S., and T.O.; writing—original draft preparation, Z.W. and T.O.; writing—review and editing, Z.W., K.D., E.F. and T.O..; visualization, Z.W. and T.O.; supervision, K.D., E.F. and T.O.; funding acquisition, Z.W. and T.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS Open Partnership Joint Research Projects, grant number JPJSBP120239904 (T.O) and Tohoku University's Advanced Graduate School Pioneering Research Support Project for PhD Students (Z.W.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Acknowledgments
This work is part of the Marie Skłodowska-Curie Actions, Research and Innovation Staff Exchange (RISE) project ’Algae4AV’ (
http://algaenet4av.eu/), and we thank the project leader Prof. Panagiotis Madesis (INAB/CERTH, Greece) and Prof. Nikolaos Labrou (Agricultural University of Athens, Greece) for
their support and guidance in our project. We thank Ms. Jinko Murakami for technical assistance in glycan array analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Más, V.; Melero, J.A. Entry of enveloped viruses into host cells: membrane fusion. Sub-cellular biochemistry 2013, 68, 467–487. [Google Scholar] [PubMed]
- Fenouillet, E.; Barbouche, R.; Jones, I. Cell entry by enveloped viruses: redox considerations for HIV and SARS-coronavirus. Antioxidants & redox signaling 2007, 9, 1009–1034. [Google Scholar]
- Mas, V.; Melero, J. Entry of Enveloped Viruses into Host Cells: Membrane Fusion. Structure and Physics of Viruses 2013, 68, 467–487. [Google Scholar]
- Cross, K.J.; Burleigh, L.M.; Steinhauer, D.A. Mechanisms of cell entry by influenza virus. Expert Rev. Mol. Med. 2001, 3, 1–18. [Google Scholar] [CrossRef]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Aqeel, M.; Khalid, N.; Hashem, M.; Alamari, S.; Zafar, S.; Qasim, M.; Irshad, M.K.; Qari, S.H. Spike glycoproteins: Their significance for corona viruses and receptor binding activities for pathogenesis and viral survival. Microb. Pathog. 2020, 150, 104719–104719. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.; Lavillette, D.; Denolly, S. The SARS-CoV-2 Envelope and Membrane proteins modulate maturation and retention of the Spike protein, allowing assembly of virus-like particles. The Journal of biological chemistry 2020, 100111. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xiang, Y. Spike Glycoprotein-Mediated Entry of SARS Coronaviruses. Viruses 2020, 12, 1289. [Google Scholar] [CrossRef] [PubMed]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? International journal of molecular sciences 2021, 22, 992. [Google Scholar] [CrossRef]
- Nassar, A.; Ibrahim, I.M.; Amin, F.G.; Magdy, M.; Elgharib, A.M.; Azzam, E.B.; Nasser, F.; Yousry, K.; Shamkh, I.M.; Mahdy, S.M.; et al. A Review of Human Coronaviruses’ Receptors: The Host-Cell Targets for the Crown Bearing Viruses. Molecules 2021, 26, 6455. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Current opinion in virology 2021, 50, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; et al. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Balbín, Y.; Santana-Mederos, D.; Paquet, F.; Fernandez, S.; Climent, Y.; Chiodo, F.; Rodriguez, L.; Ramírez, B.; Leon, K.; Hernández, T.; Castellanos-Serra, L.; Garrido, R.; Chen, G.; García-Rivera, D.; Rivera, D.; Verez-Bencomo, V. Molecular Aspects Concerning the Use of the SARS-CoV-2 Receptor Binding Domain as a Target for Preventive Vaccines. ACS Central Science 2021, 7, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Sharon, N. Lectin-carbohydrate complexes of plants and animals: an atomic view. Trends Biochem. Sci. 1993, 18, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Pang, X.; Liu, T.; Ning, Z.; Cheng, G. The Roles of Direct Recognition by Animal Lectins in Antiviral Immunity and Viral Pathogenesis. Molecules 2015, 20, 2272–2295. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Hirayama, M.; Morimoto, K.; Hori, K. The OAAH Family: Anti-Influenza Virus Lectins. Methods in molecular biology 2020, 2132, 683–693. [Google Scholar] [PubMed]
- Micewicz, E.D.; Cole, A.L.; Jung, C.-L.; Luong, H.; Phillips, M.L.; Pratikhya, P.; Sharma, S.; Waring, A.J.; Cole, A.M.; Ruchala, P. Grifonin-1: A Small HIV-1 Entry Inhibitor Derived from the Algal Lectin, Griffithsin. PLOS ONE 2010, 5, e14360. [Google Scholar] [CrossRef]
- Millet, J.K.; Séron, K.; Labitt, R.N.; Danneels, A.; Palmer, K.E.; Whittaker, G.R.; Dubuisson, J.; Belouzard, S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antivir. Res. 2016, 133, 1–8. [Google Scholar] [CrossRef]
- Férir, G.; Palmer, K.E.; Schols, D. Synergistic activity profile of griffithsin in combination with tenofovir, maraviroc and enfuvirtide against HIV-1 clade C. Virology 2011, 417, 253–258. [Google Scholar] [CrossRef]
- Barrientos, L.G.; Gronenborn, A.M. The Highly Specific Carbohydrate-Binding Protein Cyanovirin-N: Structure, Anti-HIV/Ebola Activity and Possibilities for Therapy. Mini-Reviews Med. Chem. 2005, 5, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Ogawa, T.; Muramoto, K.; Kamio, Y.; Jimbo, M.; Kamiya, H. Isolation and Characterization of a Mannan-Binding Lectin from the Freshwater Cyanobacterium (Blue-Green Algae) Microcystis viridis. Biochem. Biophys. Res. Commun. 1999, 265, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.C., Jr.; Lee, J.Y.; Cai, M.; Bewley, C.A.; Clore, G.M. Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2. J. Biol. Chem. 2005, 280, 29269–29276. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A.; Cai, M.; Ray, S.; Ghirlando, R.; Yamaguchi, M.; Muramoto, K. New carbohydrate specificity and HIV-1 fusion blocking activity of the cyanobacterial protein MVL: NMR, ITC and sedimentation equilibrium studies. Journal of molecular biology 2004, 339, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S. Jacalin: a jackfruit (Artocarpus heterophyllus) seed-derived lectin of versatile applications in immunobiological research. J. Immunol. Methods 1998, 212, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Gaidamashvili, M.; Ohizumi, Y.; Iijima, S.; Takayama, T.; Ogawa, T.; Muramoto, K. Characterization of the Yam Tuber Storage Proteins from Dioscorea batatas Exhibiting Unique Lectin Activities. J. Biol. Chem. 2004, 279, 26028–26035. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.; Yariv, J.; Helliwell, J.; Kalb, A.; Dodson, E.; Papiz, M.; Wan, T.; Campbell, J. The structure of the saccharide-binding site of concanavalin A. EMBO J. 1989, 8, 2189–2193. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, Y.; Whittier, R.F.; Yamanaka, H.; Carredano, E.; Gotoh, M.; Sota, H.; Hasegawa, Y.; Shinohara, Y. The high specificities of Phaseolus vulgaris erythro-and leukoagglutinating lectins for bisecting GlcNAc or β1–6-linked branch structures, respectively, are attributable to loop B. Journal of Biological Chemistry 2002, 277, 16928–16935. [Google Scholar] [CrossRef] [PubMed]
- Shiina, N.; Tateno, H.; Ogawa, T.; Muramoto, K.; Saneyoshi, M.; Kamiya, H. Isolation and characterization of L-rhamnose-binding lectins from chum salmon (Oncorhynchus keta) eggs. Fish. Sci. 2002, 68, 1352–1366. [Google Scholar] [CrossRef]
- Barre, A.; Damme, E.; Simplicien, M.; Poder, S.; Klonjkowski, B.; Benoist, H.; Peyrade, D.; Rougé, P. Man-Specific Lectins from Plants, Fungi, Algae and Cyanobacteria, as Potential Blockers for SARS-CoV, MERS-CoV and SARS-CoV-2 (COVID-19) Coronaviruses: Biomedical Perspectives. Cells 2021, 10. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Ziółkowska, N.E.; O'Keefe, B.R.; Mori, T.; Zhu, C.; Giomarelli, B.; Vojdani, F.; Palmer, K.E.; McMahon, J.B.; Wlodawer, A. Domain-Swapped Structure of the Potent Antiviral Protein Griffithsin and Its Mode of Carbohydrate Binding. Structure 2006, 14, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Swanson, M.D.; Boudreaux, D.M.; Salmon, L.; Chugh, J.; Winter, H.C.; Meagher, J.L.; André, S.; Murphy, P.V.; Oscarson, S.; Roy, R.; et al. Engineering a Therapeutic Lectin by Uncoupling Mitogenicity from Antiviral Activity. Cell 2015, 163, 746–758. [Google Scholar] [CrossRef]
- Gstottner, C.; Zhang, T.; Resemann, A.; Ruben, S.; Pengelley, S.; Suckau, D.; Welsink, T.; Wuhrer, M.; Domínguez-Vega, E. Structural and functional characterization of SARS-CoV-2 RBD domains produced in mammalian cells. Analytical chemistry 2021, 93, 6839–6847. [Google Scholar] [CrossRef] [PubMed]
- Lenza, M.P.; Oyenarte, I.; Diercks, T.; Quintana, J.I.; Gimeno, A.; Coelho, H.; Diniz, A.; Peccati, F.; Delgado, S.; Bosch, A.; et al. Structural Characterization of N-Linked Glycans in the Receptor Binding Domain of the SARS-CoV-2 Spike Protein and their Interactions with Human Lectins. Angew. Chem. 2020, 132, 23971–23979. [Google Scholar] [CrossRef]
- Cai, Y.; Xu, W.; Gu, C.; Cai, X.; Qu, D.; Lu, L.; Xie, Y.; Jiang, S. Griffithsin with a broad-spectrum antiviral activity by binding glycans in viral glycoprotein exhibits strong synergistic effect in combination with a pan-coronavirus fusion inhibitor targeting SARS-CoV-2 spike S2 subunit. Virologica Sinica 2020, 35, 857–860. [Google Scholar] [CrossRef]
- Chan, J.F.W.; Oh, Y.J.; Yuan, S.; Chu, H.; Yeung, M.L.; Canena, D.; Chan, C.C.S.; Poon, V.K.M.; Chan, C.C.Y.; Zhang, A.J.; Cai, J.P. A molecularly engineered, broad-spectrum anti-coronavirus lectin inhibits SARS-CoV-2 and MERS-CoV infection in vivo. Cell Reports Medicine 2022, 3. [Google Scholar] [CrossRef]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N-and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef]
- Liu, Y.M.; Shahed-Al-Mahmud, M.; Chen, X.; Chen, T.H.; Liao, K.S.; Lo, J.M.; Wu, Y.M.; Ho, M.C.; Wu, C.Y.; Wong, C.H.; Jan, J.T. A carbohydrate-binding protein from the edible lablab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell reports 2020, 32. [Google Scholar] [CrossRef] [PubMed]
- Garrison, A.R.; Giomarelli, B.G.; Lear-Rooney, C.M.; Saucedo, C.J.; Yellayi, S.; Krumpe, L.R.H.; Rose, M.; Paragas, J.; Bray, M.; Olinger, G.G., Jr.; et al. The cyanobacterial lectin scytovirin displays potent in vitro and in vivo activity against Zaire Ebola virus. Antivir. Res. 2014, 112, 1–7. [Google Scholar] [CrossRef]
- Shahzad-Ul-Hussan, S.; Gustchina, E.; Ghirlando, R.; Clore, G.M.; Bewley, C.A. Solution Structure of the Monovalent Lectin Microvirin in Complex with Manα(1–2)Man Provides a Basis for Anti-HIV Activity with Low Toxicity. Journal of Biological Chemistry 2011, 286, 20788–20796. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Damme, E.; Simplicien, M.; Benoist, H.; Rougé, P. Man-Specific, GalNAc/T/Tn-Specific and Neu5Ac-Specific Seaweed Lectins as Glycan Probes for the SARS-CoV-2 (COVID-19) Coronavirus. Marine Drugs 2020, 18. [Google Scholar] [CrossRef] [PubMed]
- Dohnálek, J.; Skálová, T. C-type lectin-(like) fold – Protein-protein interaction patterns and utilization. Biotechnology Advances 2022, 58, 107944. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.K.; Dorosky, D.; Sharma, P.; Abbasi, S.A.; Dye, J.M.; Kranz, D.M.; Herbert, A.S.; Procko, E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 1261. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
SDS-PAGE profiles of rMVL-TrxA expressed by pET32a vector (a) and purified rMVL after removal of TrxA tag (b). (a) Supernatants and pellets of bacterial lysates were analyzed. Lanes represent the supernatant (lanes 1, 3) and the pellet (lanes 2, 4) from cells with pET32a vector (lanes 1, 2) or pET32a-MVL (lanes 3, 4). (b) After the digestion of rMVL-TrxA by α-thrombin, rMVL was recovered by nickel affinity. The digested sample with α-thrombin (lane 1), rMVL in flowthrough (lane 2), and the TrxA tag eluted with 250 mM imidazole (lane 3) were analyzed.
Figure 1.
SDS-PAGE profiles of rMVL-TrxA expressed by pET32a vector (a) and purified rMVL after removal of TrxA tag (b). (a) Supernatants and pellets of bacterial lysates were analyzed. Lanes represent the supernatant (lanes 1, 3) and the pellet (lanes 2, 4) from cells with pET32a vector (lanes 1, 2) or pET32a-MVL (lanes 3, 4). (b) After the digestion of rMVL-TrxA by α-thrombin, rMVL was recovered by nickel affinity. The digested sample with α-thrombin (lane 1), rMVL in flowthrough (lane 2), and the TrxA tag eluted with 250 mM imidazole (lane 3) were analyzed.
Figure 2.
SPR profiles of the interaction between spike protein RBDs and lectins. Spike protein RBDs of different COVID-19 strains, alpha (a, e), delta (b, f), omicron (c, g), and original (d, h), were used as ligands attached on a CM5 sensor chip in Biacore T200. rMVL(a-d) and Jacalin (e-h) samples were used as analytes at different concentrations (0.31, 0.62, 1.25, 2.5 and 5 µM; traces with orange, gray, yellow, blue, and green, respectively) to record responses to determine interaction between RBDs and lectins.
Figure 2.
SPR profiles of the interaction between spike protein RBDs and lectins. Spike protein RBDs of different COVID-19 strains, alpha (a, e), delta (b, f), omicron (c, g), and original (d, h), were used as ligands attached on a CM5 sensor chip in Biacore T200. rMVL(a-d) and Jacalin (e-h) samples were used as analytes at different concentrations (0.31, 0.62, 1.25, 2.5 and 5 µM; traces with orange, gray, yellow, blue, and green, respectively) to record responses to determine interaction between RBDs and lectins.
Figure 3.
The glycan array analysis profiles of recombinant MVL (a) and DB1 (b) lectins and list of the glycans that bound to MVL (c) and DB1 (d). Symbol Nomenclature for Glycans (SNFG) is used to represent oligosaccharides on the graph (green circle for mannose, yellow circle for galactose, blue square for GlcNAc, purple diamond for NeuAc, and red triangle for fucose).
Figure 3.
The glycan array analysis profiles of recombinant MVL (a) and DB1 (b) lectins and list of the glycans that bound to MVL (c) and DB1 (d). Symbol Nomenclature for Glycans (SNFG) is used to represent oligosaccharides on the graph (green circle for mannose, yellow circle for galactose, blue square for GlcNAc, purple diamond for NeuAc, and red triangle for fucose).
Figure 4.
Interaction between lectins and alpha or omicron spike protein RBDs after specific carbohydrate treatment. MVL (a, b) and jacalin (c, d) were treated with galactose (0 mM, 12.5 mM, 25 mM, 50 mM, 100 mM) at RT for 30 min before SPR. SPR performed as
Figure 2.
Figure 4.
Interaction between lectins and alpha or omicron spike protein RBDs after specific carbohydrate treatment. MVL (a, b) and jacalin (c, d) were treated with galactose (0 mM, 12.5 mM, 25 mM, 50 mM, 100 mM) at RT for 30 min before SPR. SPR performed as
Figure 2.
Figure 5.
Comparison of predicted three-dimensional structure of four COVID-19 spike protein RBDs (a) and RBD (brown)-ACE2 (green) complex of SARS-Covid-2 (PDB ID 6M0J) [
31](b). The alpha-fold predicted structure of COVID-19 alpha, delta and omicron RBDs were displayed with cartoon presentation with green, blue, magenta, and yellow colors.
Figure 5.
Comparison of predicted three-dimensional structure of four COVID-19 spike protein RBDs (a) and RBD (brown)-ACE2 (green) complex of SARS-Covid-2 (PDB ID 6M0J) [
31](b). The alpha-fold predicted structure of COVID-19 alpha, delta and omicron RBDs were displayed with cartoon presentation with green, blue, magenta, and yellow colors.
Figure 6.
The docking results of MVL and spike protein RBDs by HDOCK molecular docking web server. The complex strcutures of MVL docking with the alpha RBD (a), the delta RBD (b), the omicron RBD (c), and the original RBD (d). RBDs and MVL are displayed in cartoon presentation with brown and yellow color, respectively. Residues for polar interaction are shown.
Figure 6.
The docking results of MVL and spike protein RBDs by HDOCK molecular docking web server. The complex strcutures of MVL docking with the alpha RBD (a), the delta RBD (b), the omicron RBD (c), and the original RBD (d). RBDs and MVL are displayed in cartoon presentation with brown and yellow color, respectively. Residues for polar interaction are shown.
Table 1.
The hemagglutination activities of recombinant and natural lectins.
Table 1.
The hemagglutination activities of recombinant and natural lectins.
| Lectin Name |
Minimum Agglutination
Concentration (mg/mL) |
Carbohydrate Specificity |
| rMVL |
0.008 |
D-mannose/D-galactose [23, this study] |
| Jacalin |
0.005 |
D-galactose/GalNAc [25] |
| Con A |
0.008 |
D-mannose [27] |
| PHA-M |
0.002 |
complex-type N-glycans [28] |
| DB1 |
0.03 |
D-mannose [26] |
| CSL3 |
0.0002 |
L-rhamnose [29] |
Table 2.
SPR binding kinetics of lectins to the spike protein RBDs.
Table 2.
SPR binding kinetics of lectins to the spike protein RBDs.
| Name |
Interaction
with RBDs |
Affinity with RBD [KD (M)] |
| Alpha |
Delta |
Omicron |
Original |
| MVL |
Yes |
1.29×10-6
|
1.65×10-6
|
2.73×10-6
|
2.08×10-6
|
| Jacalin |
Yes |
3.20×10-7
|
3.39×10-7
|
4.78×10-7
|
4.28×10-7
|
| DB1 |
No |
― |
― |
― |
― |
| Con.A |
No |
― |
― |
― |
― |
| PHA-M |
No |
― |
― |
― |
― |
| CSL3 |
No |
― |
― |
― |
― |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).