Submitted:
24 May 2024
Posted:
27 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Cell Types in Hepatic Fibrosis
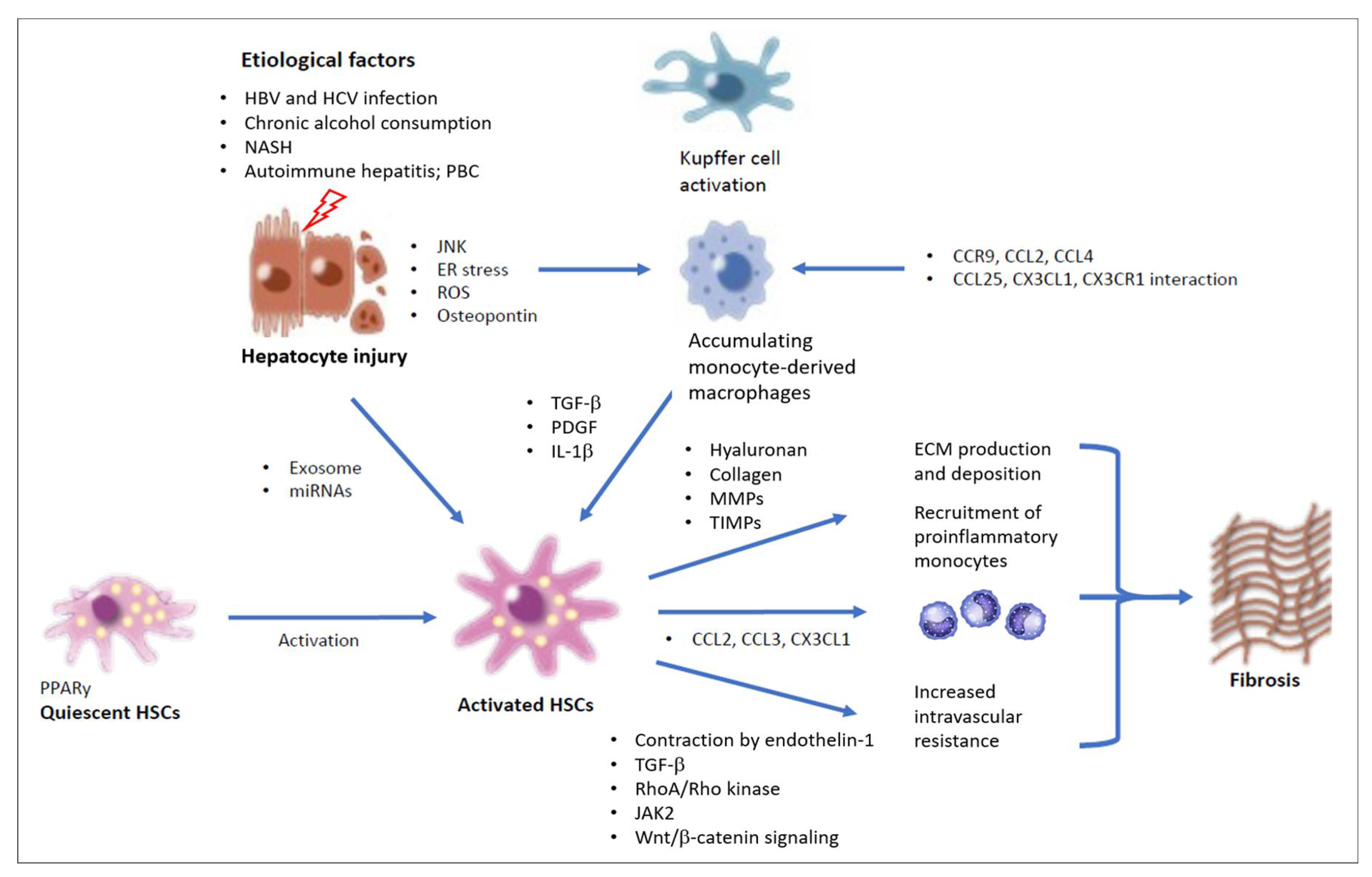
2.1. Hepatic Stellate Cells and Myofibroblasts
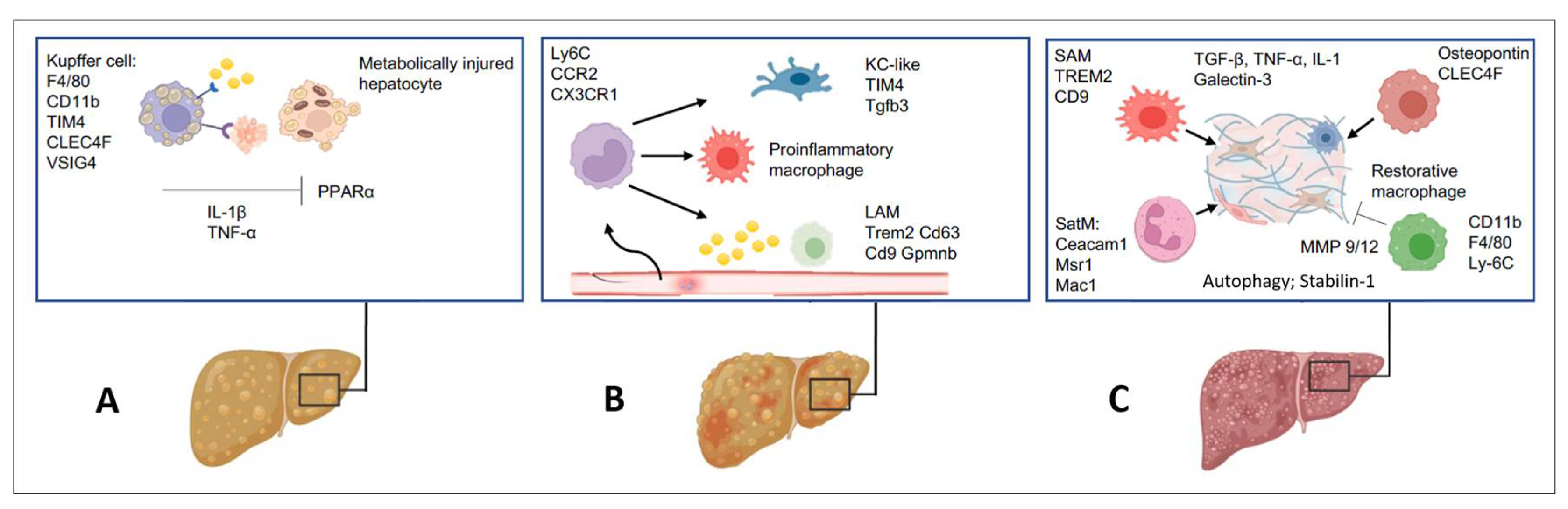
2.2. Inflammatory Cell Species Driving Hepatic Fibrogenesis
2.3. Hepatocytes
2.4. Liver Sinusoidal Endothelial Cells
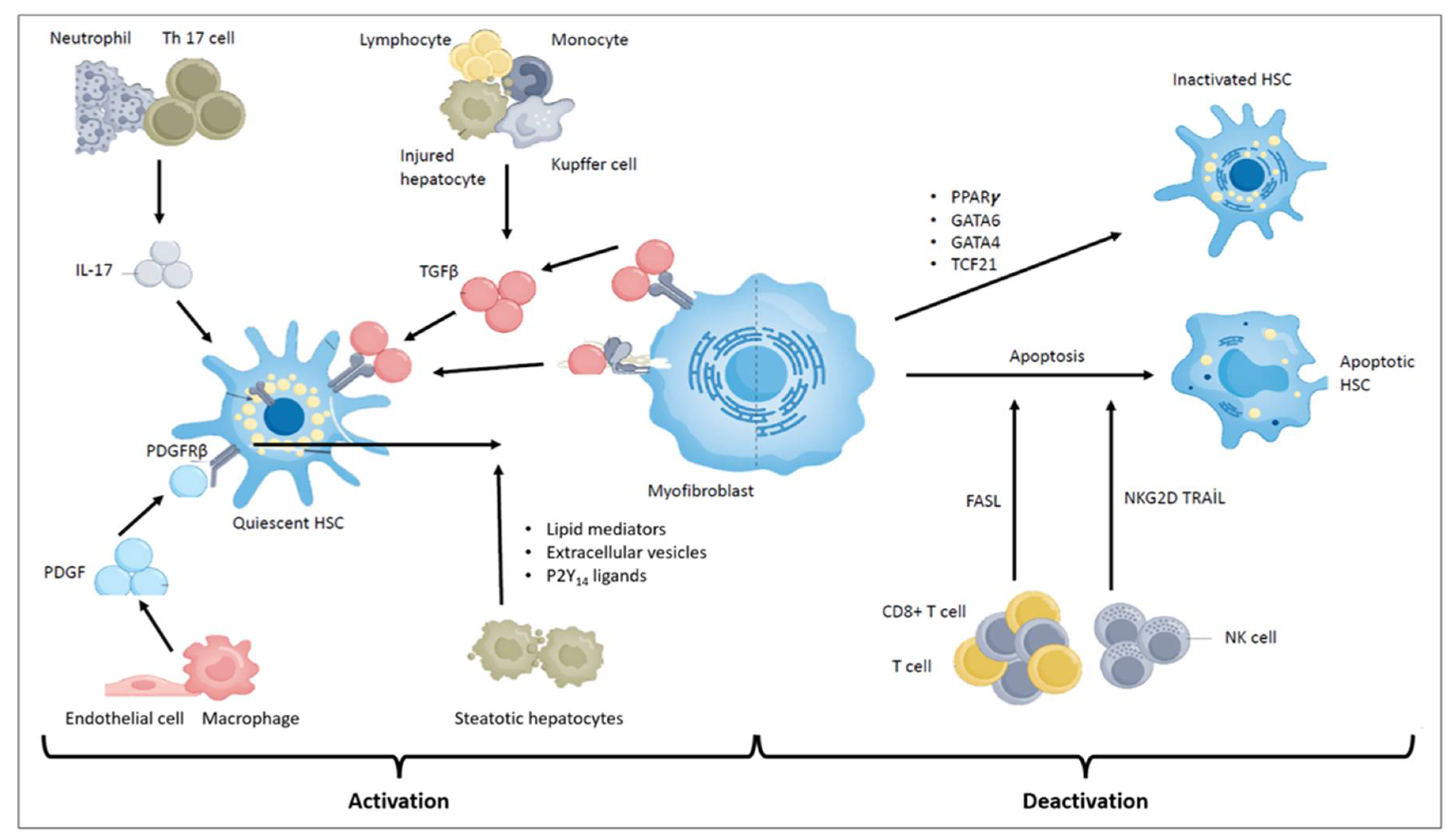
2.5. Portal Fibroblasts
2.6. Other Immune Cells in the Pathogenesis of Liver Fibrosis
2.7. Metabolic Reprogramming of HSCs in Liver Fibrogenesis
2.8. Metabolic Regulation of Liver Fibrosis
2.9. Epigenetic Regulation of HSCs
3. Mechanisms Driving Regression of Liver Fibrosis
4. The Fate of Fibrogenic Myofibroblasts
4.1. HSC Apoptosis
4.2. HSC Senescence
4.3. HSC Inactivation
5. Potential Novel Therapeutic Targets for Treating Liver Fibrosis
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| αSMA | α-smooth muscle actin |
| APCs | antigen-presenting cells |
| ATP | adenosine triphosphate |
| BAMBI | BMP activin membrane-bound inhibitor |
| CEACAM1 | carcinoembryonic antigen-related adhesion molecule 1 |
| CLD | chronic liver disease |
| COL1A | collagen 1A |
| DAMPs | danger-associated molecular patterns |
| DNMT | DNA methyltransferase |
| mDCs | myeloid dendritic cells [also dubbed “classical” or “conventional”] |
| ECM | extracellular matrix |
| EGF | epidermal growth factor |
| EMT | epithelial-to-mesenchymal transition |
| FXR | farnesoid X receptor |
| GFAP | glial fibrillary acidic protein |
| HAND2 | heart and neural crest derivatives 2 |
| HBV | hepatitis B virus |
| HCC | hepatocellular carcinoma |
| HCV | hepatitis C virus |
| HDV | hepatitis D virus |
| HGF | hepatocyte growth factor |
| HMGB1 | high mobility group box 1 protein |
| HNRNPA1 | heterogeneous nuclear ribonucleoprotein A1 |
| HSP47 | Heat shock protein 47 |
| ICAM-1 | intercellular adhesion molecule 1 |
| IL-6 | interleukin 6 |
| ILCs | innate lymphoid cells |
| KCs | Kupffer cells |
| lncRNAs | long noncoding RNAs |
| LPS | lipopolysaccharide |
| LRAT | lecithin retinol acyltransferase |
| LSEC | liver sinusoidal endothelial cells |
| MCP-1 | monocyte chemoattractant protein-1 |
| MDSCs | myeloid-derived suppressor cells |
| mito-DAMPs | mitochondria-derived danger signals |
| MMPs | matrix metalloproteinases |
| MOMP | mitochondrial outer membrane permeabilization |
| MSCs | mesenchymal stem cells |
| MSLN | mesothelin |
| MUCI 16 | mucin 16 |
| NAFLD | nonalcoholic fatty liver disease |
| NASH | nonalcoholic steatohepatitis |
| NETs | neutrophil extracellular traps |
| NKs | natural killer cells |
| NKTs | natural killer T cells |
| PAMPs | pathogen-associated molecular patterns |
| PBC | primary biliary cholangitis |
| PDGF | platelet-derived growth factor |
| PPARγ | peroxisome proliferator-activated receptor γ |
| PSC | primary sclerosing cholangitis |
| ROS | reactive oxygen species |
| SAMs | scar-associated macrophages |
| scRNA sequencing | single-cell RNA sequencing |
| SEREB-1c | sterol regulatory element-binding protein 1c |
| TET | methylcytosine dioxygenase |
| TGFβ | transforming growth factor β |
| TIMPs | tissue inhibitors of metalloproteinases |
| TLR-4 | toll-like receptor 4 |
| TRAIL | TNF-related apoptosis-inducing ligands |
| VCAM-1 | vascular cell adhesion molecule 1 |
References
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Wiering, L.; Subramanian, P.; Hammerich, L. Hepatic Stellate Cells: Dictating Outcome in Nonalcoholic Fatty Liver Disease. Cell Mol Gastroenterol Hepatol 2023, 15, 1277–1292. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Seki, E. In Vivo and In Vitro Models to Study Liver fibrosis: Mechanisms and limitations. Cell Mol Gastroenterol Hepatol 2023, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, T.; Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nature Reviews Gastroenterology & Hepatology 2021, 18, 151–166. [Google Scholar]
- Hammerich, L.; Tacke, F. Hepatic inflammatory responses in liver fibrosis. Nature Reviews Gastroenterology & Hepatology 2023, 20, 633–646. [Google Scholar]
- Xu, F.; Zhou, D.; Meng, X.; Wang, X.; Liu, C.; Huang, C.; Li, J.; Zhang, L. Smad2 increases the apoptosis of activated human hepatic stellate cells induced by TRAIL. Int Immunopharmacol. 2016, 32, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Marcellin, P.; et al. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a-5-year open-label follow-up study. Lancet 2013, 381, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Calvaruso, V.; Craxi, A. Hepatic benefits of HCV cure. J Hepatol 2020, 73, 1548–1556. [Google Scholar] [CrossRef]
- Villar-Gomez, E.; et al. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology 2015, 149, 367–378. [Google Scholar] [CrossRef]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Buonaguro, L.; Izzo, F.; Buonaguro, F.M. Molecular alterations in hepatocellular carcinoma associated with hepatitis B and hepatitis C infections. Oncotarget 2016, 7, 25087–25102. [Google Scholar] [CrossRef] [PubMed]
- Hirschfield, G.M.; Heathcote, E.J. Cholestasis and cholestatic syndromes. Curr. Opin. Gastroenterol. 2009, 25, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Mabire, M.; Hegde, P.; Hammoutene, A.; Wan, J.; Caör, C.; Sayegh, R.A.; Cadoux, M.; Allaire, M.; Weiss, E.; Thibaulth-Sogorb, T.; et al. MAITT cell inhibition promotes liver fibrosis regression via macrophage phenotype reprogramming. Nat Commun 2023, 14, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Gonzales-Rodriguez, A.; Garcia-Monzon, C.; et al. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis 2020, 11, 802. [Google Scholar] [CrossRef]
- Subramanian, P.; Hampe, J.; Tacke, F.; et al. Fibrogenic pathways in metabolic dysfunction associated fatty liver disease [MAFLD]. Int J Mol Sci 2022, 23, 6996. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Tabas, I.; Pajvani, U.B. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology 2020, 158, 1913–1928. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018, 24, 908–924. [Google Scholar] [CrossRef] [PubMed]
- Mederacke, I.; et al. Fate tracking reveals hepatic stellate cells as dominant contributors to live fibrosis independent of its etiology. Nat. Commun. 2013, 4, 2823. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Elfimova, N.; Müller, M.; Bachurski, D.; Koitzsch, U.; Drebber, U.; Mahabir, E.; Hansen, H.P.; Friedman, S.L.; Klein, S.; Dienes, H.P.; et al. Autophagy-Related Activation of Hepatic Stellate Cells Reduces Cellular miR-29a by promoting its Vesicular Secretion. Cell Mol Gastroenterol Hepatol 2022, 13, 1701–1716. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu Rev Pathol 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Kourtzelis, I.; Hajishengallis, G.; Chavakis, T. Phagocytosis of apoptotic cells in resolution of inflammation. Front Immunol 2020, 11, 553. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, F.; Matsuzaki, K.; Mori, S.T.; et al. p38 MAPK mediates fibrogenic signal through Smad3 phosphorylation in rat myofibroblasts. Hepatology 2003, 38, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Matsuzaki, K.; Mori, S.T.; et al. Transforming growth factor-beta and platelet-derived growth factor signal via c-jun N-terminal kinase-dependent Smad2/3 phosphorylation in rat hepatic stellate cells after acute liver injury. Am J Pathol 2005, 166, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Dewidar, B.; Meyer, C.; Dooley, S.; et al. TGF-beta in hepatic stellate cells activation and liver fibrogenesis-updated 2019. Cells 2019, 8, 1419. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.E.; McDonnel, M.A.; Law, B.K.; et al. Independent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J Biol Chem 1999, 274, 37413–37420. [Google Scholar] [CrossRef] [PubMed]
- Hanafusa, H.; Ninomiya-Tsuji, J.; Masuyama, N.; et al. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-beta-induced gene expression. J Biol Chem, 1999; 274, 27161–27167. [Google Scholar]
- Wipff, P.J.; Rifkin, D.B.; Meister, J.J.; et al. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J Cell Biol 2007, 179, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.M.; Nikolic-Peterson, D.J.; Lan, H.Y. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 2016, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.E. Non-Smad pathways in TGF-beta signaling. Cell Res 2009, 19, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.J.; Ren, J.J.; Zhang, Q.Q.; Kong, Y.Y.; Zhang, H.Y.; Guo, X.H.; Fan, H.Q.; Liu, L.X. IGFBPrP1 accelerates autophagy and activation of hepatic stellate cells via mutual regulation between H19 and P13K/AKT/mTOR pathway. Biomed Pharmacother 2019, 116, 109034. [Google Scholar] [CrossRef]
- Gabele, E.; Brenner, D.A.; Rippe, R.A. Liver fibrosis: signals leading to the amplification of the fibrogenic hepatic stellate cell. Front Biosci 2003, 8, 69–77. [Google Scholar]
- Pinzani, M.; Gesualdo, L.; Sabbah, G.M.; Abdoud, H.E. Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver-fat-storing cells. J Clin Invest 1989, 84, 1786–1793. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Arthur, M.J. Activation of cultured rat hepatic lipocytes by Kupffer cell conditioned medium: direct enhancement of matrix synthesis and stimulation of cell proliferation via induction of platelet-derived growth factor receptors. J Clin Invest 1989, 84, 1780–1785. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Matsuzaki, K. Differential regulation of TGF-beta/Smad signaling in hepatic stellate cells between acute and chronic liver injuries. Front Physiol 2012, 3, 53. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y.; Mao, H.; et al. Sonic hedgehog is an autocrine viability factor for myofibroblastic hepatic stellate cells. J Hepatol 2008, 48, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, A.; Pradhan-Sundd, T.; Singh, S.; et al. Platelet-derived growth factor receptor alpha contributes to human hepatic stellate cell proliferation and migration. Am J Pathol 2017, 187, 2273–2287. [Google Scholar] [CrossRef] [PubMed]
- Czochra, P.; Klopcic, B.; Meyer, E.; et al. Liver fibrosis induced by overexpression of PDGF-B in transgenic mice. J Hepatol 2006, 45, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Kocabayoğlu, P.; Lade, A.; Lee, Y.A.; et al. beta-PDGF receptor expressed by hepatic stellate cells regulates fibrosis in murine liver injury, but not carcinogenesis. J Hepatol 2015, 63, 141–147. [Google Scholar] [CrossRef]
- Lambrecht, J.; Verhuist, S.; Mannaerts, I.; et al. A PDGFβ-based score predicts significant liver fibrosis in patients with chronic alcohol abuse, NAFLD and viral liver disease. E BioMedicine 2019, 43, 501–512. [Google Scholar]
- Kisseleva, T.; Brenner, D.A. Mechanisms of fibrogenesis. Exp. Biol. Med 2008, 233, 109–122. [Google Scholar] [CrossRef]
- Ueno, T. Hepatic stellate cells and intrahepatic innervation in human liver cirrhosis. Hum Pathol 1997, 28, 953–959. [Google Scholar] [CrossRef]
- Goddard, C.; et al. Localisation and semiquantitative assessment of hepatic procollagen mRNA in primary biliary cirrhosis. Gut 1998, 43, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Iwaisako, K.; et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc. Natl. Acad. Sci. USA 2014, 111, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Nishio, T.; et al. Activated hepatic stellate cells and portal fibroblasts contribute to cholestatic liver fibrosis in MDR2 knockout mice. J Hepatol 2019, 71, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Dranoff, J.A.; Wells, R.G. Portal fibroblasts: underappreciated mediators of biliary fibrosis. Hepatology 2010, 51, 1438–1444. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc. Natl. Acad. Sci. USA 2001, 98, 6686–6691. [Google Scholar] [CrossRef] [PubMed]
- Taura, K.; Miura, K.; Iwaisako, K.; et al. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology 2010, 51, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Scolten, D.; et al. Genetic labelling does not detect epithelial-to-mesenchymal transition of cholangiocytes in liver fibrosis in mice. Gastroenterology 2010, 139, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.S.; et al. Lineage tracing demonstrates no evidence of cholangiocyte epithelial-to-mesenchymal transition in murine models of hepatic fibrosis. Hepatology 2011, 53, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.P.; et al. The bone marrow functionally contributes to liver fibrosis. Gastroenterology 2006, 130, 1807–1821. [Google Scholar] [CrossRef]
- Kallis, Y.N.; Forbes, S.J. The bone marrow and liver fibrosis: friend or foe? Gastroenterology 2009, 137, 1218–1221. [Google Scholar] [CrossRef]
- Short, B.J.; Brouard, N.; Simmonds, P.J. Prospective isolation of mesenchymal stem cells from mouse compact bone. Methods Mol Biol 2009, 482, 259–268. [Google Scholar] [PubMed]
- Kitto, L.I.; Henderson, N.C. Hepatic stellate cell regulation of liver regeneration and repair. Hepatology 2021, 5, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Canbay, A.; Friedman, S.; Gores, G.J. Apoptosis: the nexus liver injury and fibrosis. Hepatology 2004, 39, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Canbay, A.; Taimr, P.; Torok, N.; Higuchi, H.; Friedman, S.; Gores, G.J. Apoptotic body engulfment by a human stellate cell line is fibrogenic. Lab Invest 2003, 83, 665–663. [Google Scholar] [CrossRef] [PubMed]
- Casini, A.; Ceni, E.; Salzano, R.; Biondi, P.; Parola, M.; Galli, A.; et al. Neutrophil-derived superoxide anion induces lipid peroxidation and stimulates collagen synthesis in human hepatic stellate cells: role of nitric oxide. Hepatology 1997, 25, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Novo, E.; Povero, D.; Busletta, C.; Paternostro, C.; di Bonzo, L.V.; Cannito, S.; et al. The biphasic nature of hypoxia-induced directional migration of activated human hepatic stellate cells. J Pathol 2012, 226, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Pinzani, M. Unmet needs and blueprint for the future. Hepatology 2022, 75, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Campana, L.; Esser, H.; Huch, M.; Forbes, S. Liver regeneration and inflammation: from fundamental science to clinical applications. Nat Rev Mol Cell Biol 2021, 22, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Koyama, Y. Mesothelin/mucin 16 signaling in activated portal fibroblasts regulates cholestatic liver fibrosis. J Clin Invest 2017, 127, 1254–1270. [Google Scholar] [CrossRef]
- Mridha, A.R.; et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol 2017, 66, 1037–1046. [Google Scholar] [CrossRef]
- Brinkmann, V.; et al. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Moles, A.; et al. A TLR2/S100A9/CXCL-2 signaling network is necessary for neutrophil recruitment in acute and chronic liver injury in mouse. J Hepatol 2014, 60, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Bataller, R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011, 141, 1572–1585. [Google Scholar] [CrossRef]
- Mederacke, I.; et al. The purinergic P2Y14 receptor links hepatocyte death to hepatic stellate cell activation and fibrogenesis in the liver. Sci Trans Med 2022, 14, eebe5795. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.H.; et al. High-mobility group box 1 protein activates hepatic stellate cells in vitro. Transplant Proc 2008, 40, 2704–2705. [Google Scholar] [CrossRef]
- Wiesner, R.J.; Ruegg, J.C.; Morano, I. Counting target molecules by exponential polymerase chain reaction: copy number of mitochondrial DNA in rat tissues. Biochem Biophys Res Commun 1992, 183, 553–559. [Google Scholar] [CrossRef]
- An, P.; et al. Hepatocyte mitochondria-derived danger signals directly activate hepatic stellate cells and drive progression of liver fibrosis. Nat Commun 2020, 11, 2362. [Google Scholar] [CrossRef] [PubMed]
- Knor, J.; Wree, A.; Tacke, F.; Feldstein, A.E. The NLRP3 inflammasome in alcoholic and nonalcoholic steatohepatitis. Semin Liver Dis 2020, 40, 298–306. [Google Scholar] [CrossRef]
- Vonderlin, J.; Chavakis, T.; Sieweke, M.; and Tacke, F. The Multifaced Roles of Macrophages in NAFLD pathogenesis. Cell Mol Gastroenterol Hepatol 2023, 15, 1311–1324. [Google Scholar] [CrossRef]
- Krenkel, O.; et al. Myeloid cells in liver and bone marrow acquire a functionality distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 2020, 69, 551–563. [Google Scholar] [CrossRef]
- Pellicora, A.; Ramachandran, P.; Iredela, J.P.; and Fallowfield, J.A. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat Rev Immunol 2014, 14, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017, 17, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrate the regression of murine liver fibrosis. Proc Natl Acad Sci USA 2012, 109, 3186–3195. [Google Scholar] [CrossRef] [PubMed]
- Minutti, C.; Modak, R.V.; Macdonald, F.; et al. A macrophages-pericyte axis directs tissue restoration via amphiregulin-induced Transforming Growth Factor Beta Activation. Immunity 2019, 50, 645–654. [Google Scholar] [CrossRef] [PubMed]
- de Goiville, A.C.; et al. Inhibition of TGF-α signaling by an ALK5 inhibitor protects rats from dimethylnitrosamine-induced liver fibrosis. Br. J. Pharmacol. 2005, 145, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Bonniaud, P.; et al. TGF-α and Smad3 signaling link inflammation to chronic fibrogenesis. J Immunol 2005, 175, 5390–5395. [Google Scholar] [CrossRef] [PubMed]
- Kulkami, A.B.; Karisson, S. Inflammation and TGF-α: lessons from the TGF-β1 null mouse. Res Immunol 1997, 148, 453–456. [Google Scholar]
- Miura, K.; et al. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin 1-β in mice. Gastroenterology 2010, 139, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Sudo, K.; Yamada, Y.; Moriwaki, H.; Salto, K.; and Selshima, M. Lack of tumor necrosis factor receptor type 1 inhibits liver fibrosis induced by carbontetrachloride in mice. Cytokine 2005, 29, 236–244. [Google Scholar] [CrossRef]
- Seki, E.; et al. TLR4 enhances TGF-α signaling and hepatic fibrosis. Nat Med 2007, 13, 1324–1332. [Google Scholar] [CrossRef]
- Liu, C.; et al. Transcriptional repression of the transforming growth factor β [TGF-β] Pseudoreceptor BMP and activin membrane-bound inhibitor [BAMBI] by Nuclear Factor Kappa κB [NF-κB] p50 enhances TGF-β signaling in hepatic stellate cells. J Biol Chem 2014, 289, 7082–7091. [Google Scholar] [CrossRef] [PubMed]
- Kolls, J.K.; Linden, A. Interleukin-17 family members and inflammation. Immunity 2004, 21, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Remmerie, A.; et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity 2020, 53, 641–657. [Google Scholar] [CrossRef]
- Seidman, J.S.; et al. Niche-specific reprogramming of epigenetic landscape drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity 2020, 52, 1057–1074. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.; et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during nonalcoholic steatohepatitis. Immunity 2020, 53, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Daemen, S.; Gainullina, A.; Kalugatia, G.; et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep 2021, 34, 108626. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.A.; Adlung, L.; Thaiss, C.A.; et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 2019, 178, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Iredale, J.P. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest 2007, 117, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Baeck, C.; et al. Pharmacological inhibition of the chemokine CCL2 [MCP-1] diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut 2012, 61, 416–426. [Google Scholar] [CrossRef]
- Tacke, F.; Puengel, T.; Loomba, R.; Friedman, S.L. An integrated view of anti-inflammatory and antifibrotic targets for the treatment of NASH. J Hepatol. 2023, 79, 552–566. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; and Trauner, M. Gut-liver axis: pathophysiological concepts and clinical complications. Cell Metab 2022, 34, 1700–1718. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, A.; Hundermark, J.; Guillot, A.; and Tacke, F. Molecular and cellular mediators of the gut-liver axis in the progressive liver diseases. Front Med 2021, 8, 725390. [Google Scholar] [CrossRef] [PubMed]
- Chopyk, D.M.; Grakoul, A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology 2020, 159, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: current concepts and future challenges. Nature reviews Gastroenterology & hepatology 2019, 16, 411–428. [Google Scholar]
- Parthasarathy, G.; Revelo, X.; Malhi, H. Pathogenesis of nonalcoholic steatohepatitis an overview. Hepatol Commun 2020, 4, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G. ; Hill C The interaction between bacteria and bile. FEMS Microbiol Rev 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- Mouries, J.; et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol 2019, 71, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Adolph, T.E.; Trouner, M. Gut-liver axis: Pathophysiological concepts and clinical implications. Cell Metab 2022, 34, 1700–1718. [Google Scholar] [CrossRef]
- Ramachandran, P.; Mathchett, K.P.; Dobie, R.; Wilson-Kanamori, J.R.; and Henderson, N.C. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat Rev Gastoenterol Hepatol 2020, 17, 457–472. [Google Scholar] [CrossRef]
- Islam, K.B.; et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; et al. Farnesoid x receptor antagonizes nuclear factor κB in hepatic inflammatory response. Hepatology 2008, 48, 1632–1643. [Google Scholar] [CrossRef]
- Azzu, V.; Vacca, M.; Virtue, S.; Allison, M.; and Vidal-Puig, A. Adipose tissue-liver crosstalk in the control of whole-body metabolism: implications in nonalcoholic fatty liver disease. Gastroenterology 2020, 158, 1899–1912. [Google Scholar] [CrossRef]
- Imajo, K.; et al. Hyperresponsivity to low-dose endotoxin during progression to nonalcoholic steatohepatitis is regulated by leptin-mediated signaling. Cell Metab 2012, 16, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; et al. Leptin is key to peroxynitrite-mediated oxidative stress and Kupffer cell activation in experimental non-alcoholic steatohepatitis. J Hepatol 2013, 58, 778–784. [Google Scholar] [CrossRef]
- Gong, J.; Tu, W.; Liu, J.; Tian, D. Hepatocytes: A key role in liver inflammation. Front Immunol 2022, 13, 1–22. [Google Scholar] [CrossRef]
- Povero, D.; Panera, N.; Japtok, L.; et al. Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cells via microRNA targeting PPAR-gamma. Cell Mol Gastroenterol Hepatol 2015, 1, 646–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-D.; Zhou, J.; Chen, E.-Q. Molecular Mechanisms and Potential New Therapeutic Drugs for Liver Fibrosis. Front Pharmacol, 2022; 13, 1–14. [Google Scholar]
- Wang, X.; Cal, B.; Yang, X.; et al. Cholesterol stabilizes TAZ in hepatocytes to promote experimental non-alcoholic steatohepatitis. Cell Metab 2020, 31, 969–986. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; Caparros, E.; Fernandez-Iglesias, A.; Ruben Frances. Role of liver endothelial cells in liver diseases. Nat Rev Gastroenterol Hepatol 2021, 16, 411–432. [Google Scholar] [CrossRef]
- Poisson, J.; Lemoinne, S.; Boulanger, C.; Durand, F.; Moreau, R.; Valla, D.; Rautou, P.-E. Liver sinusoidal endothelial cells: Physiology and role in liver disease. J Hepatol 2017, 66, 212–227. [Google Scholar] [CrossRef]
- DeLeve, L.D. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology 2015, 61, 1740–1746. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.; Haddad, P.G.; Garcia-Cardena, G.; Frangos, J.A.; Mennone, A.; Grozzman, R.J.; et al. Liver sinusoidal endothelial cells are responsible for nitric oxide modulation of resistance in the hepatic sinusoids. J Clin Invest 1997, 100, 2923. [Google Scholar] [CrossRef] [PubMed]
- Wohileber, D.; Knolle, P.A. The role of liver sinusoidal cells in local hepatic immune surveillance. Clin Trans Immunol 2016, 5, e117. [Google Scholar] [CrossRef]
- Shetty, S.; Lalor, P.F.; Adams, D.H. Liver sinusoidal endothelial cells – gatekeepers of hepatic immunity. Nat Rev Gastroenterol. Hepatol 2018, 15, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.; et al. Platelet interactions with liver sinusoidal endothelial cells and hepatic stellate cells lead to hepatocyte proliferation. Cell 2020, 9, 1243. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; et al. Increased oxidative stress in cirrhotic rat livers: a potential mechanism contributing to reduced nitric oxide bioavailability. Hepatology 2008, 47, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Le Guillou, D.; Chen, J.Y. Cellular stress in the pathogenesis of nonalcoholic steatohepatitis and liver fibrosis. Nature Reviews Gastroenterology & Hepatology 2023, 20, 662–678. [Google Scholar]
- DeLeve, L.D.; Wang, X.; Guo, Y. Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescent. Hepatology 2008, 48, 920–930. [Google Scholar] [CrossRef]
- Xie, G.; Wang, I.; Wang, I.; Atkinson, R.D.; Kanel, G.C.; et al. Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology 2012, 142, 918–927. [Google Scholar] [CrossRef]
- Gracia-Sancho, J.; et al. Enhanced vasoconstrictor prostanoid production by sinusoidal endothelial cells increases portal perfusion pressure in cirrhotic rat livers. J Hepatol 2007, 47, 220–227. [Google Scholar] [CrossRef]
- Rockey, D.C.; Weisiger, R.A. Endothelin induced contractility of stellate cells from normal and cirrhotic rat liver: implications for regulation of portal pressure and resistance. Hepatology 1996, 24, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; et al. Evidence against a role for NADPH oxidase modulating hepatic vascular tone in cirrhosis. Gastroenterology 2007, 133, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; et al. Endothelial expression of transcription factor Kruppel-like factor 2 and its vasoprotective target genes in the normal and cirrhotic rat liver. Gut 2011, 60, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Lisman, T.; Luyendyk, J.P. Platelets as modulators of liver diseases. Semin Thromb Hemost 2018, 44, 114–128. [Google Scholar] [PubMed]
- Tripodi, A.; Primignani, M.; Mannucci, P.M.; and Caldwell, S.H. Changing concepts of cirrhotic coagulopathy. Am J Gastroenterol 2017, 112, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Ehling, I.; Bartneck, M.; Wei, X.; Gremse, F.; Fech, V.; Möckel, D.; et al. CCL2-dependent infiltrating macrophages promote angiogenesis in progressive liver fibrosis. Gut 2014, 63, 1960–1971. [Google Scholar] [CrossRef] [PubMed]
- Thabut, D.; Shah, V. Intrahepatic angiogenesis and sinusoidal remodeling in chronic liver disease: new targets for the treatment of portal hypertension? J Hepatol 2010, 53, 976–980. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Tripathi, D.; Dongre, K.; Garg, V.; Rooge, S.; Mukopadyay, A.; et al. Increased number and function of endothelial progenitor cells stimulate angiogenesis by resident liver sinusoidal endothelial cells [LSECs] in cirrhosis through paracrine factors. J Hepatol 2012, 57, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Rautou, P.-E. Endothelial progenitor cells in cirrhosis: the more, the merrier? J Hepatol 2012, 57, 1163–1165. [Google Scholar] [CrossRef]
- Ding, B.-S.; Cao, Z.; Lis, R.; Nolan, D.J.; Guo, P.; Simons, M.; et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 2013, 505, 97–102. [Google Scholar] [CrossRef]
- Huang, E.; Peng, N.; Xiao, F.; et al. The Roles of Immune Cells in the Pathogenesis of Fibrosis. Int J Mol Sci 2020, 21, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Liedke, C.; Nevzorova, Y.A.; Luedde, T.; et al. Liver Fibrosis: From Mechanisms of Injury to Modulation of Disease. Front Med, 2022; 8, 1–32. [Google Scholar]
- Ebbo, M.; Crinier, A.; Vely, F.; et al. Innate lymphoid cells: Major players in inflammatory diseases. Nat Rev Immunol 2017, 17, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Mikami, Y.; Takada, Y.; Hagihari, Y.; et al. Innate lymphoid cells in organ fibrosis. Cytokine Growth Factor Rev 2018, 42, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shen, L.; Sun, X.; et al. Adipose group 1 innate lymphoid cells promote adipose tissue fibrosis and diabetes in obesity. Nat Commun 2019, 10, 3254. [Google Scholar] [CrossRef] [PubMed]
- Forkel, M.; Berglin, L.; Kekalainen, E.; et al. Composition and functionality of the intrahepatic innate lymphoid cell-component in human nonfibrotic and fibrotic livers. Eur J Immunol 2017, 47, 1280–1294. [Google Scholar] [CrossRef] [PubMed]
- Calvalheiro, T.; Zimmermann, M.; Radtake TRDJ, et al. Novel insights into dendritic cells in pathogenesis of systemic sclerosis. Clin Exp Immunol 2020, 201, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Sun, R.; Wei, H.; et al. Accelerated liver fibrosis in hepatitis B virus transgenic mice: Involvement of natural killer T cells. Hepatology 2011, 53, 219–229. [Google Scholar] [CrossRef]
- Böttcher, K.; Rambouts, K.; Saffioti, F.; et al. MAIT cells are chronically activated in patients with autoimmune liver disease and promote profibrogenic hepatic stellate cell activation. Hepatology 2011, 68, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Galati, D.; De Martino, M.; Trotta, A.; et al. Peripheral depletion of, N.K.; cells and imbalance of the Treg/Th17 axis in idiopathic pulmonary fibrosis. Cytokine, 2014; 66, 119–126. [Google Scholar]
- Deldago, M.E.; Candenas, B.L.; Farcan, N.; et al. Metabolic reprogramming of liver fibrosis. Cells 2021, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Khanam, A.; Saleeb, P.G.; and Koltilil, S. Pathophysiology and Treatment Options for Hepatic Fibrosis. Can it be Completely Cured? Cells 2021, 10, 1–22. [Google Scholar] [CrossRef]
- Yan, Y.; Zeng, J.; Xing, L.; et al. Extra- and Intra-Cellular Mechanisms of Hepatic Stellate Cell Activation. Biomedicines 2021, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Hyun, J.; Premont, R.T.; et al. Hedgehog Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells. Gastroenterology 2018, 154, 1465–1479. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Chitneni, S.K.; Suzuki, A.; et al. Increased glutaminolysis Marks Active Scarring in Nonalcoholic Steatohepatitis Progression. Cell Mol Gastroenterol Hepatol 2020, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Lian, N.; Jiang, J.; Zhang, F.; et al. Curcumin regulates cell fate and metabolism by inhibiting hedgehog signaling in hepatic stellate cells. Lab Invest 2015, 95, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Le, C.T.; Sung, K.Y.; et al. Succinate induces hepatic fibrogenesis by promoting activation, proliferation, and migration, and inhibiting apoptosis of hepatic stellate cells. Biochem Biophys Res Commun 2018, 496, 673–678. [Google Scholar] [CrossRef]
- Ge, J.; Cui, H.; Xe, N.; et al. Glutaminolysis promotes collagen translation and stability via α-ketoglutarate-mediated mTOR activation and proline hydroxylation. Am J Respir Cell Mol Biol 2018, 58, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z. Integrin αvβ6 critically regulates hepatic progenitor cell function and promote ductular reaction, fibrosis, and tumorigenesis. Hepatology 2016, 63, 217–232. [Google Scholar] [CrossRef]
- Smith-Cortinez, N.; van Eunen, K.; Heegsma, J.; et al. Simultaneous induction of glycolysis and oxidative phosphorylation during activation of hepatic stellate cells novel mitochondrial targets to treat liver fibrosis. Cells 2020, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, J.; Ping, J.; et al. TGF beta1-induced autophagy activates hepatic stellate cells via the ERK and JNK signaling pathways. Int J Mol Med 2021, 47, 256–266. [Google Scholar] [CrossRef]
- Chen, M.; Liu, J.; Yang, W.; et al. lipopolysaccharide mediates hepatic stellate cell activation by regulating autophagy and retinoic acid signaling. Autophagy 2017, 13, 1813–1827. [Google Scholar] [CrossRef]
- Cao, Y.; Mai, W.; Li, R.; et al. Macrophages evoke autophagy of hepatic stellate cells to promote liver fibrosis in NAFLD mice via the PGE2/EP4 pathway. Cell Mol Life Sci 2022, 79, 303. [Google Scholar] [CrossRef] [PubMed]
- Mridha, A.R.; Wree, A.; Robertson AAB, et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol 2017, 66, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Gea, v.; Ghiassi-Nejad, Z.; Rozenfeld, R.; et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 2012, 142, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Thoen, L.F.R.; Guimoraes, F.L.M.; Dolle, L.; et al. a role of autophagy during hepatic stellate cell activation. J Hepatol 2011, 55, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Gea, V.; Hilscher, M.; Rozenfeld, R.; et al. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol 2013, 59, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Kluwe, J.; Pradere, J.-P.; Gwak, G.-Y.; et al. Modulation of hepatic fibrosis by c-jun-N-terminal kinase inhibition. Gastroenterology, 2010; 138, 347–359. [Google Scholar]
- Liu, Y.; Wen, D.; Ho, C.; Yu, L.; Zheng, D.; O’Relly, S.; Gao, Y.; Li, Q.; Zhang, Y. Epigenetics as a versatile regulator of fibrosis. J Trans Med 2023, 21, 1–20. [Google Scholar] [CrossRef]
- Shi, C.-X.; Wang, Y.; Jiao, F.-Z.; Chen, Q.; Cao, P.; Pei, M.-H.; Zhang, L.-Y.; Guo, J.; Deng, W.; Wang, L.-W.; Gong, Z.-J. Epigenetic regulation of Hepatic Stellate Cell Activation and Macrophage in Chronic liver Inflammation. Front Physiol 2021, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Claveria-Cabello, A.; Colyn, L.; Arechederra, M.; et al. Epigenetics in Liver Fibrosis: Could HDACs be Therapeutic Target? Cells 2020, 9, 1–22. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Korutidis, A. LNCcation: IncRNA location and function. J Cell Biol 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Lin, Z.-J.; Li, C.-C.; Lin, X.; Shan, S.-K.; Guo, B.; Zheng, M.-H.; Li, F.; Yuan, L.-Q.; Li, Z.-H. Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study. Signal Transduct Target Ther, 2023; 8, 98. [Google Scholar]
- Li, X.-J.; Zhou, F.; Li, Y.-J.; Xue, X.-Y.; Qu, J.-R.; Fan, G.-F.; Liu, J.; Sun, R.; Wu, J.-Z.; Zheng, Q.; and Liu, R.-P. LncRNA H19-EZH2 interaction promotes liver fibrosis via reprogramming H3K27me3 profiles. Act Pharma Sinica, 2023; 44, 2479–2491. [Google Scholar]
- Wen, Y.; Hou, Y.; Yi, X.; Sun, S.; He, X.; et al. EZH2 activates CHK1 signaling to promote ovarian cancer chemoresistance by maintaining the properties of cancer stem cells. Theranostics 2021, 11, 1795–1813. [Google Scholar] [CrossRef]
- Liu, X.; Brenner, D.A. DNA methylation controls liver fibrogenesis. Nat Rev Gastroenterol Hepatol, 2016; 13, 126–128. [Google Scholar]
- Page, A.; Paoli, P.; Salvador, E.M.; White, S.; French, J.; Mann, J. Hepatic stellate cells transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J Hepatol 2016, 64, 661–673. [Google Scholar] [CrossRef]
- Murphy, S.K.; et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 2013, 145, 1076–1087. [Google Scholar] [CrossRef]
- Komatsu, Y.; Waku, T.; Iwasaki, N.; Ono, W.; Yamaguchi, C.; Yanagisawa, J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med. Genomics 2012, 5, 1–12. [Google Scholar]
- Götze, S.; Schumacher, E.C.; Kordes, C.; Houssinger, D. Epigenetic changes during hepatic stellate cell activation. PLoS ONE, 2015; 10, e0128745. [Google Scholar]
- El Toghdouini, A.; Serensen, A.I.; Reiner, A.H.; Coll, M.; Verhulst, S.; Mannaerst, I.; Qie, C.I.; Semedsrod, B.; Nojimi, M.; Sohal, E.; et al. Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells. Oncotarget 2015, 6, 26729–26745. [Google Scholar] [CrossRef]
- Bian, E.B.; Huang, C.; Wang, H.; Chen, X.X.; Zhang, Y.; Lv, X.W.; Li, J. Repression at Smad7 mediated by DNMT1 determines hepatic stellate cell activation and liver fibrosis in rats. Toxicol Lett 2014, 224, 175–185. [Google Scholar] [CrossRef]
- Bian, E.B.; Huang, C.; Wang, H.; Chen, X.X.; Zhang, Y.; Lv, X.W.; Li, J. DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell activation and liver fibrogenesis in rats. Toxicol Appl Pharmacol 2012, 264, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Maron-Salvador, E.; Mann, J. Epigenetics and liver fibrosis. Cell Moll Gastroenterol Hepatol 2017, 4, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Page, A.; Mann, D.A.; Mann, J. The Mechanisms of Hepatic Stellate Cell Activation and Epigenetic Regulation of Hepatic Stellate Cell Phenotypes. Curr Pathobiol Rep 2014, 2, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.; Chu, D.C.K.; Maxwell, A.; Oakley, F.; Zhu, N.L.; Tsukomato, H.; Mann, D.A. MsCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 2010, 138, 705–714. [Google Scholar] [CrossRef]
- Hazra, S.; Xiang, S.; Wang, J.; Rippe, R.A.; Chatterjee, V.K.K.; Tsukamoto, H. Peroxisome Proliferator-activated Receptor gamma Induces a Phenotypic Switch from Activated to Quiescent Hepatic Stellate Cells. J Biol Chem 2004, 279, 11392–11401. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Wilson, C.L.; Zeybel, M.; Wash, M.; Amin, S.; Robinson, S.; White, S.A.; Burt, A.D.; Odelay, F.; Tsukamoto, H.; et al. Histone methyltransferases ASH1 orchestrates fibrogenic gene transcription during myofibroblast differentiation. Hepatology 2012, 56, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Martin-Mateos, R.; De Assuncao, T.M.; Arab, J.P.; Jalon-Sakrikar, N.; Yaqoob, M.; Greuter, T.; Verma, V.K.; Mathison, A.J.; Cao, S.; Lomberk, G.; et al. Enhancer of Zeste Homologue 2 Inhibition Attenuates TGF-α Dependent Hepatic Stellate Cell Activation and Liver Fibrosis. CMGH 2019, 7, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Zeybel, M.; Luli, S.; SAbater, L.; Hardy, T.; Oakley, F.; Leslie, J.; Page, A.; Maron Savador, E.; Sharkey, V.; Tsukamoto, H.; et al. A Proof-of-Concept for Epigenetic Therapy of Tissue Fibrosis: Inhibition of Liver Fibrosis Progression by 3-Deazaneplacocin, A. Mol Ther 2017, 25, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, X.X.; Li, W.X.; Wu, X.Q.; Huang, C.; Xie, J.; Zhao, X.X.; Merg, X.M.; Li, J. EZH2-mediated repression of Dkk1 promotes hepatic stellate cell activation and hepatic fibrosis. J Cell Mol Med 2017, 21, 2317–2328. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; She, H.; Han, Y.P.; Wang, J.; Xiong, S.; Asahina, K.; Tsukamoto, H. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol 2007, 294, G39–49. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, T.; Cong, M.; Palk, Y.; Scholten, D.; Jiang, C.; Benner, C.; Iwaisako, K.; Moore-Morris, T.; Scott, B.; Tsukamoto, H.; Evans, S.M.; Dillman, W.; Glass, C.K.; and Brenner, D.A. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl. Acad. Sci. USA 2012, 109, 9448–9453. [Google Scholar] [CrossRef] [PubMed]
- Issa, R.; Willams, E.; Trim, N.; et al. Apoptosis of hepatic stellate cells: involvement in resolution of biliary fibrosis, and regulation by soluble growth factors. Gut 2001, 34, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Xie, X.-L.; Wang, M.-M.; et al. The role of the apoptosis-related BCL-B in the regulation of mitophagy in hepatic stellate cells during the regression of liver fibrosis. Exp Mol Med 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guicciardi, M.E.; and Gores, G. Paving the TRAIL to Anti-Fibrotic Therapy. Hepatology 2016, 64, 29–31. [Google Scholar] [CrossRef]
- Oh, Y.; Park, O.; Swierczewska, M.; Hamilton, J.P.; et al. Systemic PEGylated TRAIL Treatment Ameliorates Liver Cirrhosis in Rats by Eliminating Activated Hepatic Stellate Cells. Hepatology 2016, 64, 209–223. [Google Scholar] [CrossRef]
- Trivedi, P.; Wang, S.; and Friedman, S.L. The power of plasticity – metabolic regulation of Hepatic Stellate Cells. Cell Metab, 2021; 33, 242–247. [Google Scholar]
- Kendall, T.J.; Hennedige, S.; Aucott, R.L.; et al. p75 Neurotrophin receptor signaling regulates hepatic myofibroblast proliferation and apoptosis in recovery from rodent liver fibrosis. Hepatology 2009, 49, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Serna-Salas, S.; Damba, T.; et al. Hepatic stellate cell senescence in liver fibrosis. Characteristics, mechanisms and perpective. Mech Ageing Dev, 2021; 199, 1–14. [Google Scholar]
- Collado, M.; Serrano, M. Senescence in tumors: evidence from mice and humans. Nat Rev Cancer 2010, 10, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Troeger, A.S.; Mederacke, I.; Gwak, G.-Y.; et al. Deactivation of Hepatic Stellate Cells During Liver Fibrosis Resolution in Mice. Gastroenterology 2012, 143, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Coligiuri, A.; Gentilini, A.; Pastore, M.; et al. cellular and Molecular Mechanisms Underlying Liver Fibrosis Regression. Cells 2021, 10, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Radaeva, S.; Park, O. Liver natural killer and natural killer T cells: immunobiology and emerging roles in liver disease. J Leukoc Biol 2009, 86, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Baglieri, J.; Kisseleva, T.; et al. Mechanisms of liver fibrosis and its role in liver cancer. Exp Biol Med 2020, 245, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Iredale, J.P.; Benyan, R.C.; Pickering, J.; et al. Mechanisms of spontaneous resolution of rat liver fibrosis: Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J Clin Invest 1998, 102, 538–549. [Google Scholar] [CrossRef]
- Lo, R.; Kim, H. Histopathological evaluation of liver fibrosis and cirrhosis regression. Clin Mol Hepatol 2017, 23, 302–307. [Google Scholar] [CrossRef]
- Harris, E.N. Will inhibition of cellular crosstalk resolve liver fibrosis. Hepatology 2022, 76, 558–560. [Google Scholar] [CrossRef]
- Wu, H.; Chen, C.; Ziani, S.; et al. Fibrotic events in the progression of cholestatic liver disease. Cells 2021, 10, 1107–1121. [Google Scholar] [CrossRef]
- Scnable, B.; Purbeck, C.A.; Choi, Y.H.; et al. Replicative senescence of activated human hepatic stellate cells is accompanied by pronounced inflammatory but less fibrogenic phenotype. Hepatology 2003, 37, 653–664. [Google Scholar]
- Van der Heide, D.; Weiskirchen, R.; Ransal, R. Therapeutic targeting of hepatic macrophages for the treatment of liver diseases. Front Immunol 2019, 10, 2852. [Google Scholar] [CrossRef]
- Jeong, W.I.; Park, O.; Radaeva, S.; et al. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating, N.K.; cell cytotoxicity. Hepatology, 2006; 44, 1441–1451. [Google Scholar]
- Zhang, J.; Liu, Q.; He, J.; et al. Novel Targets in liver Fibrosis. Front Mol Biosci 2021, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P.N.; Elsharkawy, A.M.; Kendall, T.J.; Loomba, R.; Mann, D.A.; Fallowfield, J.A. Antifibrotic therapy in nonalcoholic steatohepatitis: time for a human-centric approach. Nat Rev Gastroenterol Hepatol, 2023; 20, 679–688. [Google Scholar]
- Duffield, J.S.; Forbes, S.J.; Constandinou, C.M.; et al. Selective depletion of macrophages reveals distinct opposing roles during liver injury and repair. J Clin Invest 2005, 115, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Ding, J.; Wang, M.; et al. Kupffer-derived matrix metalloproteinase-9 contributes to liver fibrosis resolution. Int J Biol Sci 2018, 14, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Piaz, V.D.; Giovanni, M.P. Phosphodiesterase 4 inhibitors structurally unrelated to rolipram, as promising agents for the treatment of asthma and other pathologies. Eur J Med Chem 2000, 35, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; She, H.; Han, Y.P.; et al. Wnt antagonism inhibits hepatic stellate cells activation and liver fibrosis. Am J Physiol Liver Physiol 2008, 294, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, H.; Xiao, D.; et al. Forsenoid X receptor [FXR]: Structures and ligands. Comput Struct Biotechnol 2021, 19, 2148–2159. [Google Scholar] [CrossRef]
- Wang, Y.D.; Chen, W.D.; Wang, M.; et al. Farnesoid X receptor antagonizes nuclear factor kappa B in hepatic inflammatory response. Hepatology 2008, 48, 1632–1643. [Google Scholar] [CrossRef]
- Gracia-Sancho, J.; Manicardi, N.; Ortega-Ribera, M.; et al. Emricasan ameliorates portal hypertension and liver fibrosis in cirrhotic rats through a hepatocyte-mediated paracrine mechanism. Hepatol Commun 2019, 3, 987–1000. [Google Scholar] [CrossRef]
- Chen, L.; Brenner, D.A.; Kisseleva, T. Combatting fibrosis: Exome-based therapies in the regression of liver fibrosis. Hepatol Commun 2018, 3, 180–192. [Google Scholar] [CrossRef] [PubMed]
- De Jong, O.G.; Van Balkom, B.W.; Gremmels, H.; et al. Exosomes from hypoxic endothelial cells have increased collagen crosslinking activity through up-regulation of lysyl oxidase-like 2. J Cell Mol Med 2016, 20, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhao, L.; Duan, J.; et al. strategies to improve efficiency of mesenchymal stem cell transplantation for reversal of liver fibrosis. J Cell Mol Med 2019, 23, 1657–1670. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Dong, M.Q.; Liu, Z.M.; et al. A strategy of vascular-targeted therapy for liver fibrosis. Hepatology 2022, 76, 660–675. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author[s] and contributor[s] and not of MDPI and/or the editor[s]. MDPI and/or the editor[s] disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




