1. Introduction
Human louse
Pediculus humanus is an obligatory blood feeding ectoparasite. Two known ecotypes of human louse exist: the head louse
Pediculus humanus capitis and the body louse
Pediculus humanus humanus [
1,
2]. Currently, pediculosis due to head louse infestations remains one of the most prevalent parasitic infestation in humans essentially due to the misuse of non-chemical products (silicon-based formulations) and development of resistance to the commonly used pediculicides [
3]. Human body louse is a known vector of three bacterial pathogens:
Rickettsia prawazakii responsible for epidemic typhus[
4,
5],
Borrelia recurrentis responsible for relapsing fever and
Bartonella quintana that causes trench fever [
6]. In the past, massive epidemics of relapsing fever have affected Africa and Eurasia, and recently many cases of louse-borne relapsing fever were diagnosed in Gremany [
7], Netherlands [
8], Switzerland [
9], Finland [
10] and Italy [
11].
GABA (γ-aminobutyic acid) receptors, members of the cys-loop ligand gated ion channels (cys-loop LGICs) superfamily, are among the main pharmacological targets for insecticides [
12,
13]. They share common molecular features: the cys-loop located in the extra cellular N-terminal domain, four transmembrane domains (TM1-TM4) architecting the body of the channel and large highly variable intracellular domain regulating the channel functions and containing most of the protein activation sites. In insects, four subunits of GABA receptors are well described. At phylogenetic level, these subunits form four distinct clades revealing orthologous relationships: resistant to dieldrin (RDL), glycin like receptor of drosophila (GRD), ligand gated chloride channel homologue3 (LCCH3), and [
14,
15,
16,
17,
18,
19].
Our previous exploration of GABA receptor subunits in human body louse
Pediculus humanus humanus revealed that the genome encodes for three GABA receptor subunits Phh-RDL, Phh-GRD, and Phh-LCCH3 that are expressed throughout the developmental stages and in different tissues [
20]. Moreover, Phh-RDL was able to reconstitute anion selective functional homomeric receptor while Phh-GRD and Phh-LCCH3 combined to form cation selective hetero-pentameric receptors [
20,
21].
Interestingly, besides the four well described GABA receptor subunits, an additional gene closely related to GRD and 8916 was described in
Blattella germanica (Bg-8916_2) and
Periplaneta americana (Pa-8916_2) [
16] as well as in the Chelicerata species,
Ixodes ricinus (Ir-GABA3),
Parasteatoda tepidarorium (Pt-GABA3), and
Galendromus occidentalis (Go-GABA3) [
22].
In the present study, our analysis in the genome of human body louse allowed to identify a homolog of these genes annotated as gamma-aminobutyric-acid receptor alpha-2 subunit precursor (PHUM507160; XM_002430955.1). This gene was characterized, the phylogenetic relationships among other GABA receptor subunits and the spatiotemporal expression in different parts and developmental stages of human body louse were investigated.
2. Materials and Methods
2.1. Insects
Strains of human body louse Pediculus humanus humanus were reared at BioMAP laboratory (University of Tours, France), maintained at standard conditions (temperature of 30˚C, relative humidity of 60-70%) and fed on rabbit blood five times per week. These strains of human louse were never exposed to chemical compounds.
2.2. RNA Extraction and cDNA Synthesis
Total RNA was extracted from 30 mg of young adult louse, nits, larva (L1, L2, L3), heads, thoraxes and abdomens using RNA plus extraction kit (Machery Nagel) as per manufacturer’s instructions. The first strand cDNA was synthesized by reverse transcription using superscript III reverse transcriptase (InvitrogenTM) and oligo dT primer.
2.3. Sequence Analysis and Phylogeny
By using BLASTN, we used the genomic sequences of
Blattella germanica (Bg-8916_2),
Periplaneta americana (Pa-8916_2),
Ixodes ricinus (Ir-GABA3),
Parasteatoda tepidarorium (Pt-GABA3), and
Galendromus occidentalis (Go-GABA3) searching for homologues in the genome of human body louse
Pediculus humanus humanus from Vector Base
® [
23]. For sequence analysis, all cloned transcripts were compared with the putative sequences of
P. humanus deposited in Vector Base
® by using Geneious software (Biomatters) and Basic Local Alignment Search Tool (BLAST
®, U.S. National Library of Medicine,
https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences deduced from full-length transcripts were obtained from ExPASy translate (Swiss Institute of Bioinformatics,
https://web.expasy.org/translate/), signal peptide cleavage sites were predicted using the SignalP-5.0 server (Center for Biological Sequence Analysis,
http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011), and the transmembrane domains were identified using the TMHMM program (
http://www.cbs.dtu.dk/services/TMHMM/).
Multiple sequence alignments were done by Clustal omega algorithm [
24], then viewed and annotated by Jalview software with the sequences of
Laodelphax striatellus Ls-alphalike (RZF33096.1),
Blatella germanica Bg-8916_2 (QQH14694.1
), Periplaneta americana Pa-8916_2 (QQH14658.1),
Ixodes ricinus Ir-GABA3 (UOV21278.1),
Galendromus occidentalis Go-GABA-3 (XP_028968418.1
), Parasteatoda tepidarorium Pt-GABA3 (XP_015925419.1).
The phylogenetic trees were constructed by Molecular Evolutionary Genetics Analysis (MEGAXI) software [
25] using the Neighbour-Joining (Poisson model) or Maximum likelihood methods based on the Whelan And Goldman, the best model being assessed with MEGAXI, and branch support being assessed with 1000 bootstrap replication.
The trees were constructed with the sequences of RDL, GRD, LCCH3, 8916, alpha-like, and GABA3 of insects, Collembola, Crustacea and Chelicerata, obtained from the National Center for Biotechnology Information (NCBI) database (
https://www.ncbi.nlm.nih.gov), accession numbers of sequences were indicated in
Figure 2 and
Figure 3.
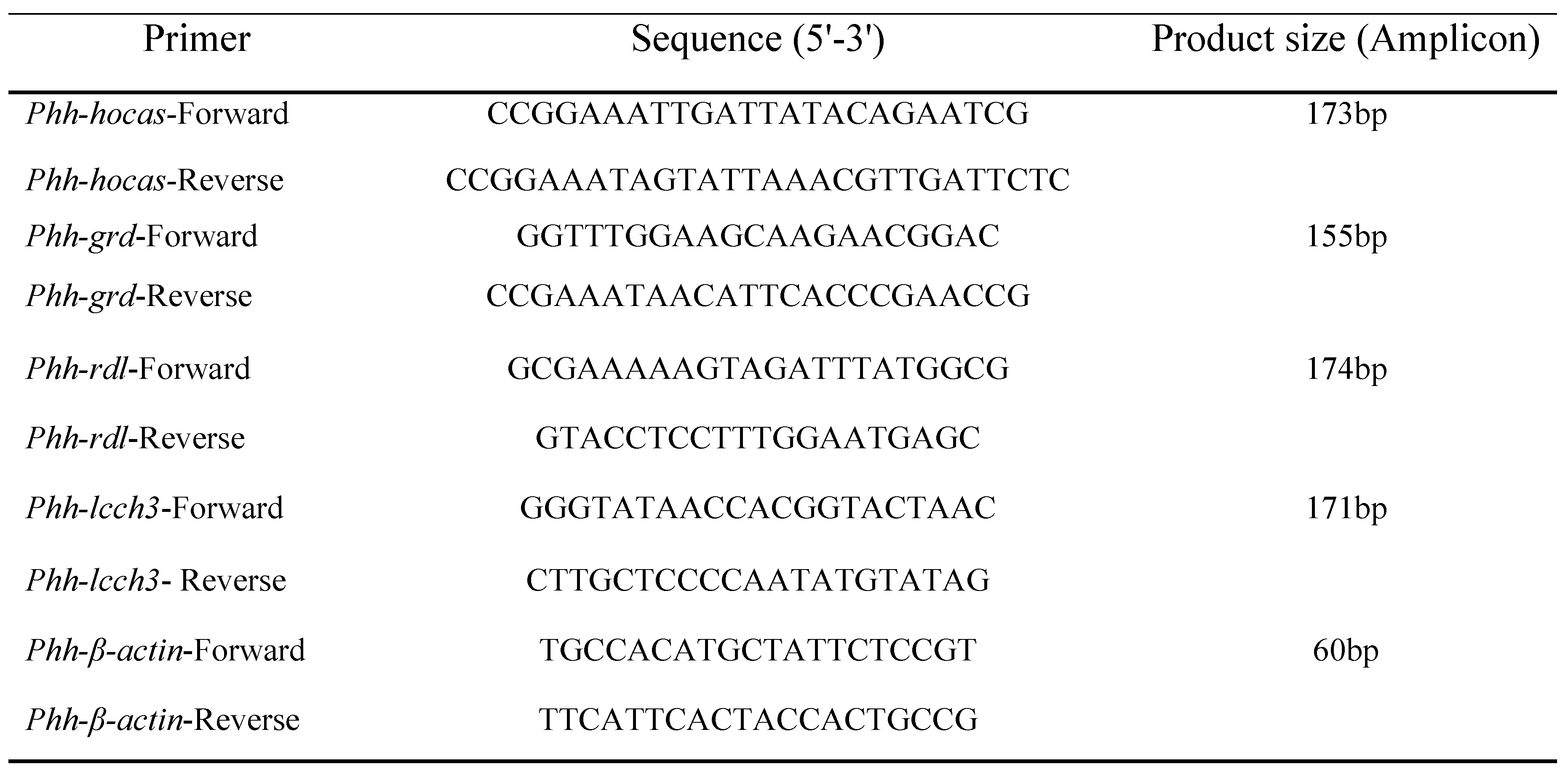
2.4. RACE-PCR and Cloning of Full Length Transcripts
The 5' ends and 3'ends of
gamma-aminobutyric-acid receptor alpha-2 subunit precursor (PHUM507160; XM_002430955.1) were characterized by rapid amplification of cDNA ends (RACE-PCR) using Gene Racer
® kit (Invitrogen) as previously described [
20] using gene-specific and nested specific primers (
Figure 1,
Table 1). Cycles for first and second nested PCR was 94°C for 5 min then 35 cycles of 94°C for 30 sec, 58°C for 30 sec, 72°C for 1 min and final extension 72°C for 5 min. Full length
gamma-aminobutyric-acid receptor alpha-2 subunit precursor (PHUM507160; XM_002430955.1) was amplified using 1 µl of cDNA and 10 pmol of two gene specific primers (
Table 1) using GoTaq DNA Polymerase (Promega, Charbonnières les Bains, France) according to the manufacturer’s instructions. RACE-PCR, and RT-PCR products were cloned in PGEM-T Easy vector (Promega) and sequenced by Eurofins genomics.
2.5. Spatial and Temporal Expression
Quantitative PCR (qPCR) analysis was performed on 100 ng of RNA extracted from adult lice, nits, L1, L2, L3 larvae stages, as well from heads, thoraxes and abdomens of adult louse using the primers listed in (
Table 2) and by applying comparative cycle threshold experiment 2
-(ΔCt) with
β-actin as endogenous control as described [
20]. Data of resulting relative expressions were analyzed by ANOVA test followed by Tukey's multiple comparisons test and plotted as median using GraphPad Prism 7.
3. Results
3.1. Phylogenetic Analysis and Sequence Identity
Using the mRNA sequences of
Blattella germanica (Bg-8916_2),
Periplaneta americana (Pa-8916_2),
I. ricinus (Ir-GABA3),
P. tepidarorium (Pt-GABA3), and
G. occidentalis (Go-GABA3) in BLASTN analysis against the genome of human body louse
Pediculus humanus humanus (Vector Base
®) we retrieve a putative
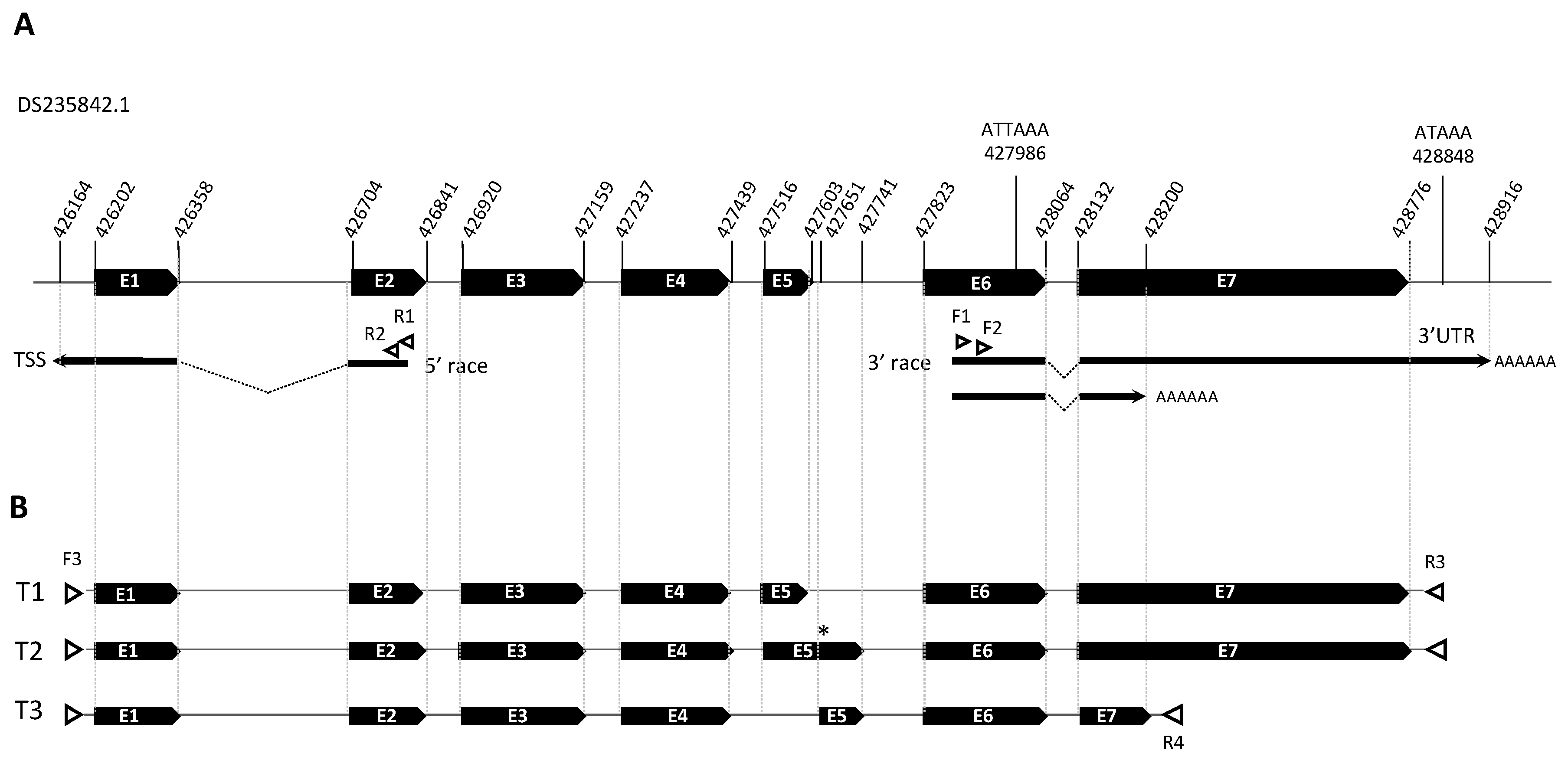
gamma-aminobutyric-acid receptor alpha-2 subunit precursor gene (PHUM507160; XM_002430955.1). The genomic sequence of PHUM507160 contains 2,575 bp organized in 7 exons located in the super contig DS235842.1 (
Figure 1a).
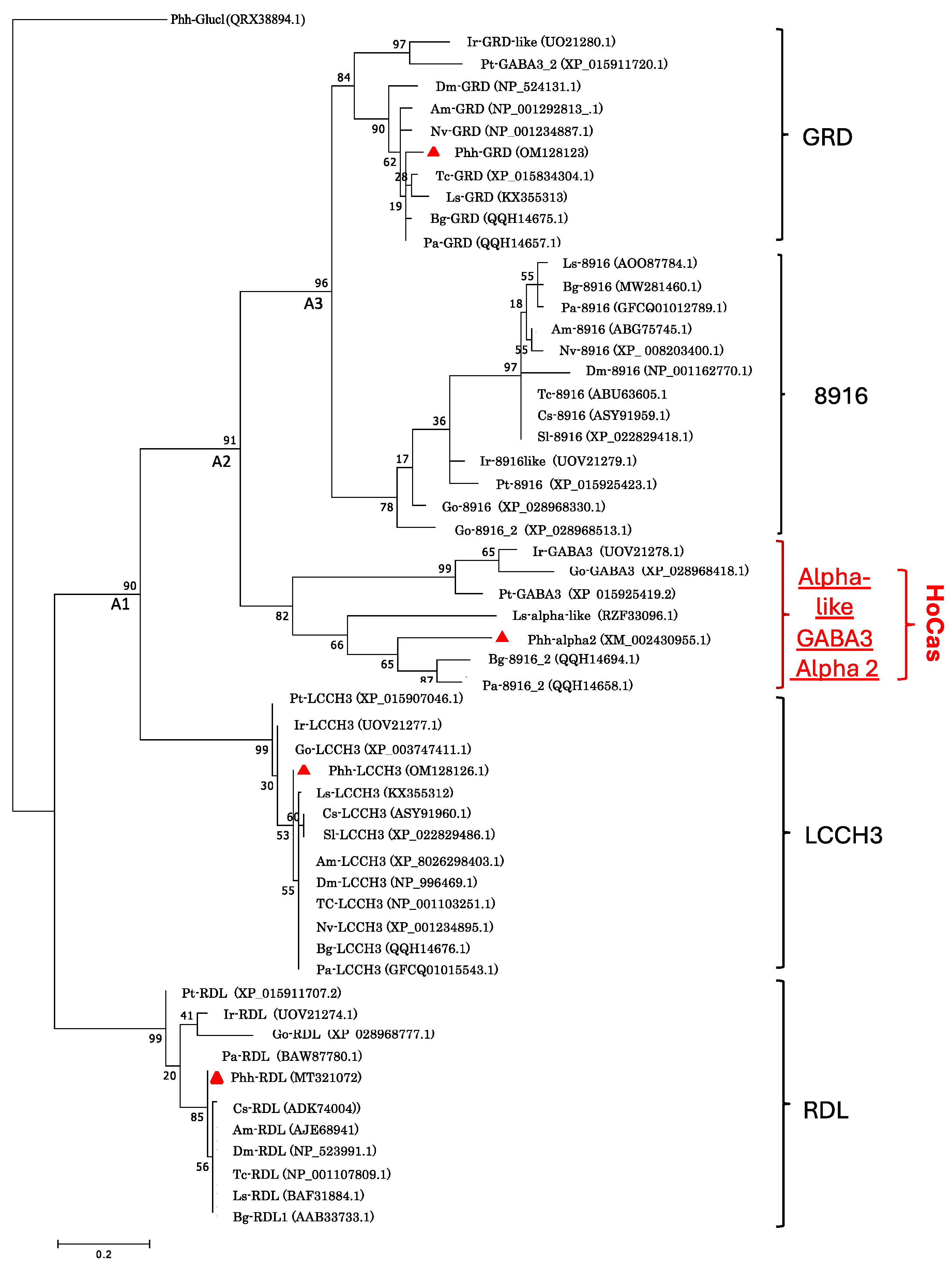
Phylogenetic tree constructed with the protein sequences of GRD, LCCH3, RDL, 8916, alpha-like, and GABA3 subunits of selected organisms revealed that GABA receptor proteins are segregated in five clades : the well-known four clades (RDL, LCCH3, GRD and 8916) and a fifth clade containing the sequences of gamma-aminobutyric-acid receptor Phh-alpha-2, Pa-8916_2, Bg-8916_2, Ls-alpha-like, Go-GABA3, Ir-GABA3 and Pt-GABA3 (
Figure 2).
Figure 2.
Phylogenic tree of Phh-GABA receptor subunits compared with those of other arthropod species. The phylogenic tree was constructed by Maximum likelihood methods based on the Whelan And Goldman, the best model being assessed with MEGAXI, and branch support being assessed with 1000 bootstrap replications. P. humanus sequences are labeled by red triangle. A1: Common ancestor of LCCH3, HoCas, 8916 and GRD. A2: Common ancestor of HoCas, 8916 and GRD. A3: Common ancestor of 8916 and GRD.
Figure 2.
Phylogenic tree of Phh-GABA receptor subunits compared with those of other arthropod species. The phylogenic tree was constructed by Maximum likelihood methods based on the Whelan And Goldman, the best model being assessed with MEGAXI, and branch support being assessed with 1000 bootstrap replications. P. humanus sequences are labeled by red triangle. A1: Common ancestor of LCCH3, HoCas, 8916 and GRD. A2: Common ancestor of HoCas, 8916 and GRD. A3: Common ancestor of 8916 and GRD.
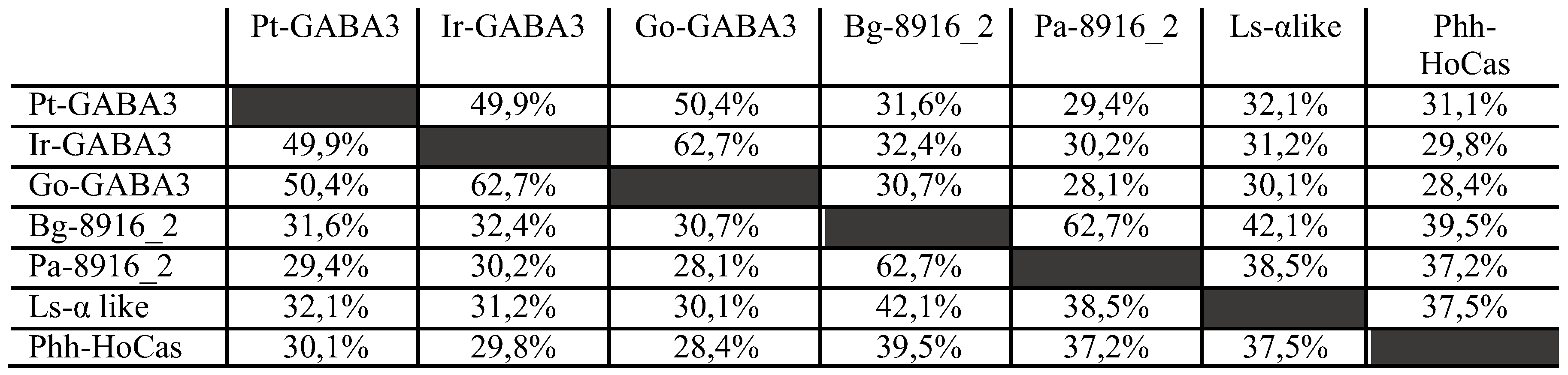
Moreover, arthropods 8916, GRD and the new (fifth) clade showed close phylogenetic relationship, suggesting the possibility of having a common ancestor. Considering these results, we hypothesized the existence of a new GABA receptor subunit in arthropods that we named HoCas for
Homologous to
Cys-loop
alpha-like
subunit. At the sequence identity level, insect HoCas share identity from 37,3 to 62,7% whereas Chelicerata-HoCas share identity from 49,9 to 62,7% (
Table 3). Inside insects, Phh-HoCas is more closed to Bg-8916_2 with 39,5% sequence identities.
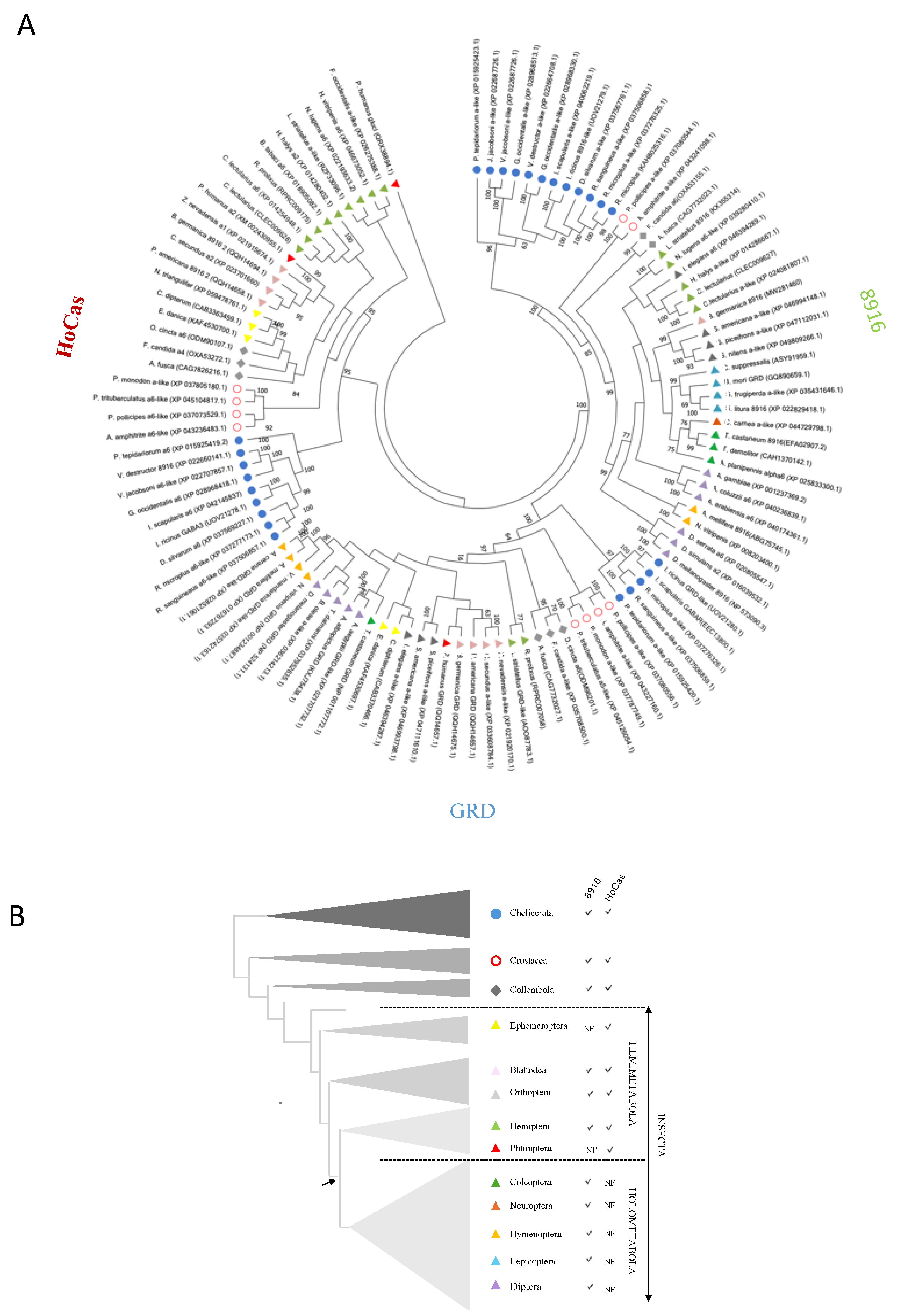
In order to confirm the existence of this new clade/subunit, using BLASTP we compared the protein sequences of Phh-HoCas, Am-8916 and Phh-GRD as templates against different insect orders, Crustacea, Collembola and Chelicerata. Firstly, our results confirmed the existence of the three separate clades (GRD, 8916, and HoCas) (
Figure 3A). Secondly, 8916 is conserved among arthropods as sequences were present in Chelicerata, Collembola, Crustacea and most of the tested insect orders: Blattodae, Orthoptera, Hemiptera, Coleoptera, Neuroptera, Hymenoptera, Lepidoptera and Diptera (
Figure 3B). Systematic search for 8916 encoding gene in the genome of
Pediculus humanus (Phthiraptera) ended up with no result, suggesting that human body louse possesses the four GABA subunit genes:
Phh-grd,
Phh-rdl,
Phh-lcch3 and
Phh-hocas, but possibly misses
Phh-8916. (
Figure 3A,B). Finally, the sequences of the newly described clade “HoCas” were found in Chelicerata, Crustacea, Collembola, and Hemimetabola, but not in Holometabola (
Figure 3B). Inside each clade (GRD, 8916, HoCas), the sequences of Insecta, Collembola, Crustacea and Chelicerata are well segregated (
Figure 3A). Interestingly,
Bombyx mori Bm-GRD clustered with other insect 8916 genes (
Figure 3A), confirming that, as previously described, Bm-GRD sequence deposited in NCBI (Accession numbers: GQ890659.1 or NP_001182633.1) may be Bm-8916 gene [
26].
Figure 3.
A. Phylogenic analysis of GRD, HoCas and 8916 subunits in different arthropods. Neighbour-joining phylogenic tree of GRD, 8916 and HoCas subunits. The phylogeny includes sequences from Chelicerata, Crustacea, Collembola and representative insects. Phh-GluCl was used to root the tree. B. Schematic representation showing the presence of 8916 and HoCas in Arthropodes. The tree adapted from Thomas et al., 2020. NF not found.
Figure 3.
A. Phylogenic analysis of GRD, HoCas and 8916 subunits in different arthropods. Neighbour-joining phylogenic tree of GRD, 8916 and HoCas subunits. The phylogeny includes sequences from Chelicerata, Crustacea, Collembola and representative insects. Phh-GluCl was used to root the tree. B. Schematic representation showing the presence of 8916 and HoCas in Arthropodes. The tree adapted from Thomas et al., 2020. NF not found.
3.2. Cloning and Sequence Analysis of Phh-hocas
We characterized the
Phh-hocas mRNA ends by 5’ and 3’ RACE-PCR on total RNA extracted from nits by using set of primers binding in exon 2 and in exon 6, respectively (
Table 1;
Figure 1a). For 5’ RACE PCR, we identified one TSS 38 nt upstream from the starting codon, and for 3’RACE two 3’ end sites located respectively 146 nt and 68 nt downstream from an ATTAAA/ AT polyA signal (
Figure 1A). Reverse transcription PCR (RT-PCR) on total RNA extracted from lice with the couple of primer F3-R3 and F3-R4 allowed to amplify 3 transcripts (
Figure 1B). The sequences of
Phh-hocas transcripts were deposited in gene bank with the corresponding accession numbers: the transcript
Phh-hocas-1 (OQ831857) confirmed the
in-silico annotation, the second transcript
Phh-hocas-2 (OQ851502) resulted from a partial retention of intron 5, and the third transcript
Phh-hocas-3 (OQ851503) resulted from alternative splicing of exon 5 and ends early at the beginning of E7. Translation and analysis of the encoded proteins revealed that both
Phh-hocas-2 (with a premature stop codon in the retained intron) and
Phh-hocas-3 are coding for truncated proteins missing TM4. Finally, the full-length cDNA of
Phh-hocas-1 is 1,819 bp, with an ORF of 1,713 bp encoding a protein of 570 amino acids.
The multiple sequence alignment of HoCas in Insecta and Chelicerata revealed that they possess all the typical features of cys-loop LGICs: the two cysteines: Phh-HoCas-C
161 and Phh-HoCas-C
175 characteristic of the cys loop motif, 4 TM domains: TM1 (Phh-HoCas-Y
242-S
261), TM2 (Phh-HoCas-R
272-S
292), TM3 (Phh-HoCas-V
307-L
334) and TM4 (Phh-HoCas-S
533-F
552), the six loops (A-F) involved in binding to the natural ligand, and a signal peptide at position 1 to 23 (
Figure 4).
3.3. Spatial and Temporal Expression of GABA Receptor Subunits
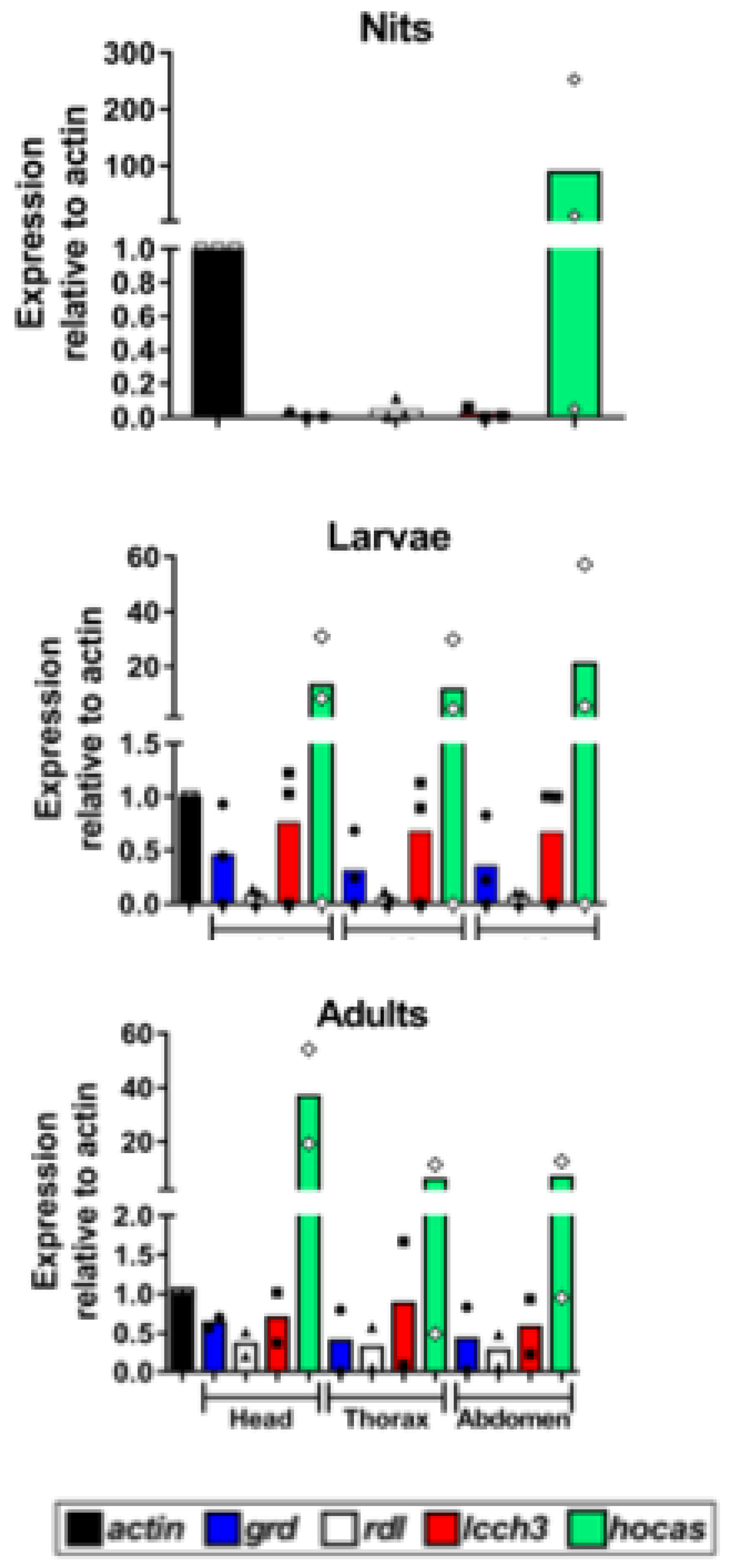
In order to verify the expression of
Phh-hocas in the human louse, we performed qPCR experiments with RNA extracted throughout the development stages (nits, L1, L2, L3 larval stages and adults) and in different parts (head, thorax and abdomen) (
Figure 5).
Phh-hocas was found to be expressed in all parts of adult body louse (
Figure 5C) and throughout the development stages (
Figure 5A, B). Relative to
actin, the expression of
Phh-hocas is highest in nits (about 100-fold) (
Figure 5A) followed by larval stages (20-fold) (
Figure 5). In adults, relative to
actin, the highest expression of
Phh-hocas was in head (40-fold) then in thorax and abdomen (about 10-fold) (
Figure 5C). Furthermore,
Phh-hocas was always much more expressed than the other subunits.
4. Discussion
GABA is the main inhibitory neurotransmitter in insects targeted by insecticides. In insects, four GABA subunits segregated in 4 separate clades are well described: RDL, GRD, LCCH3 and 8916. Depending on their association to formulate functional receptors, they could have distinct sensitivities to the natural ligand and different pharmacological profiles [
15,
18,
27,
28,
29,
30,
31].
In the present work, in addition to our previous studies describing three GABA subunits in human body louse,
Phh-rdl [
21] ,
Phh-grd and
Phh-lcch3 [
20] we have now identified another gene encoding for a putative GABA receptor subunit that we named
Phh-hocas for Homologous to Cys-loop alpha like subunit. Phh-HoCas formed a separate clade distinct from the well-known GRD, LCCH3, RDL and 8916 subunits. In this clade, Phh-HoCas clustered with the non-classified Bg-8916_2, Pa-8916_2, Ir-GABA-3, Go-GABA3, Pt-GABA3, and Ls-alphalike sequences, suggesting the existence of a new (fifth) GABA receptor clade in arthropods.
Our systematic search revealed that HoCas and 8916 subunits are present in Insecta, Collembola, Crustacean and Chelicerata, and share a common ancestor with GRD subunits who lived about 600 million years ago [
32,
33]. It has been shown that, in the genome of
A. mellifera, D. melanogaster and
T. castaneum, the
8916 gene is located close to
lcch3, while in the genome of
Pediculus humanus, Phh-hocas is close to
Phh-lcch3. Interestingly, our search revealed that in the genomes of Hemiptera, Chelicerata and Blattodae
8916,
hocas and
lcch3 genes are located close to each other (data not shown), suggesting that these genes originated from three duplication events: the first duplication event from the ancestral gene A1 leading to LCCH3 and the ancestral gene A2, A2 was further duplicated in HoCas and the ancestral gene A3 that finally duplicated to give 8916 and GRD (
Figure 2). However, the absence of
hocas in all Holometabola (
Figure 3B) is intriguing. If a common ancestor of
hocas existed as suggested previously, one hypothesis would be the loss of this gene during speciation event of Holometabola sub-order [
33].
In the genome of
P. humanus humanus, our analysis failed to identify the gene encoding for 8916. Assembly of insect genome is challenging due to the large stretch of AT, we cannot exclude defaults in assembly of the human louse genome, as revealed by the missed sequences found in the
Phh-GRD genomic annotation and the strikingly very long
Phh-GRD gene (> 7 kb) encompassing 3 contigs [
20]. Confirmation of the genomic annotation by comparing with the transcriptomics data is a good method to have a complete view of cys-loop LGICs as observed in
I. ricinus and
I. scapularis [
22]. In human body louse, the transcriptomics analysis [
34] validated the genomic annotation [
23] confirming the absence of
Phh-8916. The obligatory parasitism status of
P. humanus could explain the loss of some genes like
8916 in favor of others, similar to what described in
Acyrtosyphon pisum lacking
grd and
lcch3 but having a second
GluCl subunit [
35]
In this study, we cloned the complete cDNA sequence of
Phh-hocas of
Pediculus humanus humanus (Phthiraptera) by using qPCR and RACE-PCR techniques. Moreover, besides the full-length transcript, we identified two transcript variants of
Phh-hocas resulted from a partial intron 5 retention/alternative splicing and premature polyA signal, both encoding proteins missing TM4. The possible role of these truncated variants is not clear and requires more investigations. Alternative splicing and retention of introns in the genes encoding cys-loop LGICs have been described in many insects and resulting in proteins with variable functionalities and sensitivities to insecticides. For example, retentions located at the intracellular domain between TM3 and TM4 were observed in
varroa destructor (Vd-GRD), leading to marked changes in GABA sensitivities compared to variant without retentionsm [
30].
Results of the relative expressions revealed a remarkable higher expression of Phh-HoCas compared to other subunits in all tested parts of human body louse and throughout the development stage, while the expression of Phh-grd, Phh-lcch3 and Phh-rdl is almost the same, raising a question about the possible physiological role of Phh-hocas. It would be interesting to see if similar expression profiles obtained in other species and to test the effect of inhibition of gene expression in the louse physiology using RNAi. Since Phh-hocas was found to be expressed at higher levels compared to other GABA subunits of human louse especially in nits, it remains to determine whether Phh-HoCas is able to reconstitute homo or hetero pentameric functional GABA receptor. If so, and hence most of the commonly used pediculicides are not active against nits, Phh-HoCas could be a potential target to design a novel pediculicides against adult louse and nits.
Funding
This work was supported by Campus France (N° 934152H to O.H.), the government of Sudan (to O.H. and A.A.E.A.), the University of Tours (to B.T., I.D.P., F.D.G. and C.D.), the INRAE (to C.L.C. and C.N.) and the Fédération de Recherche en Infectiologie of the Region Centre Val-de-Loire (to C.L.C. and C.D.).
CRediT authors contribution statement
Omar Hashim: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - original draft. Claude L. Charvet: Formal analysis, Writing - review & editing. Berthine Toubaté: Methodology. Aimun A. E. Ahmed: Validation, Writing - review & editing, Supervision, Funding acquisition. Cédric Neveu: Validation, Writing - review & editing, Funding acquisition. Isabelle Dimier-Poisson: Validation, Writing - review & editing, Funding acquisition. Françoise Debierre-Grockiego: Validation, Formal analysis, Data curation, Supervision. Catherine Dupuy: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Supervision, Funding acquisition.
Informed Consent Statement
Declaration of competing interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Fouad Lahmer for his participation in the cloning of Phh-HoCas during his master internship. We sincerely thank Dr. Franck Dedeine (IRBI, University of Tours) for helpful discussion and advice on phylogenetic analysis.
References
- Sangaré AK, Doumbo OK, Raoult D. Management and Treatment of Human Lice. Biomed Res Int 2016;2016.
- Amanzougaghene N, Fenollar F, Raoult D, Mediannikov O. Where Are We With Human Lice? A Review of the Current State of Knowledge. Front Cell Infect Microbiol 2020;9.
- Amanzougaghene N, Fenollar F, Davoust B, Djossou F, Ashfaq M, Bitam I, et al. Mitochondrial diversity and phylogeographic analysis of Pediculus humanus reveals a new Amazonian clade “F.” Infection, Genetics and Evolution 2019;70:1–8.
- Bechah Y, Capo C, Mege JL, Raoult D. NCBI genome https://www.ncbi.nlm.nih.gov/genome/?term=rickettsia. Lancet Infect Dis 2008;8:417–26.
- Straub MH, Roy AN, Martin A, Sholty KE, Stephenson N, Foley JE. Distribution and prevalence of vector-borne diseases in California chipmunks (Tamias spp.). PLoS One 2017;12:1–19.
- Raoult D, Roux V. The Body Louse as a Vector of Reemerging Human Diseases [Internet]. 1999. Available from: https://academic.oup.com/cid/article/29/4/888/451618.
- Hoch M, Wieser A, Löscher T, Margos G, Pürner F, Zühl J, et al. Louse-borne relapsing fever (Borrelia recurrentis) diagnosed in 15 refugees from northeast Africa: Epidemiology and preventive control measures, Bavaria, Germany, July to October 2015. Eurosurveillance 2015;20.
- Wilting KR, Stienstra Y, Sinha B, Braks M, Cornish D, Grundmann H. Louse-borne relapsing fever (Borrelia recurrentis) in asylum seekers from Eritrea, The Netherlands, July 2015. Eurosurveillance 2015;20.
- Osthoff M, Schibli A, Fadini D, Lardelli P, Goldenberger D. Louse-borne relapsing fever - report of four cases in Switzerland, June-December 2015. BMC Infect Dis 2016;16.
- Hytönen J, Khawaja T, Grönroos JO, Jalava A, Meri S, Oksi J. Louse-borne relapsing fever in Finland in two asylum seekers from Somalia. Apmis 2017;125:59–62.
- Antinori S, Mediannikov O, Corbellino M, Grande R, Parravicini C, Bestetti G, et al. Louse-Borne Relapsing Fever (Borrelia recurrentis) in a Somali Refugee Arriving in Italy: A Re-emerging Infection in Europe? PLoS Negl Trop Dis 2016;10:0–5.
- Knipple DC, Soderlund DM. The ligand-gated chloride channel gene family of Drosophila melanogaster. Pestic Biochem Physiol 2010;97:140–8.
- Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel superfamily of the honeybee, Apis mellifera. Invertebrate Neuroscience 2006;6:123–32.
- Ffrench-Constant RH. Corrections and Retraction: Molecular Cloning and Transformation of Cyclodiene Resistance in Drosophila: An Invertebrate -Aminobutyric Acid Subtype A Receptor Locus. Proceedings of the National Academy of Sciences 1992;89:7849b–7849.
- Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the red flour beetle, tribolium castaneum. BMC Genomics 2007;8:1–16.
- Jones AK, Goven D, Froger JA, Bantz A, Raymond V. The cys-loop ligand-gated ion channel gene superfamilies of the cockroaches Blattella germanica and Periplaneta americana. Pest Manag Sci 2020.
- Wei Q, Wu SF, Gao CF. Molecular characterization and expression pattern of three GABA receptor-like subunits in the small brown planthopper Laodelphax striatellus (Hemiptera: Delphacidae). Pestic Biochem Physiol 2017;136:34–40.
- Sheng CW, Jia ZQ, Ozoe Y, Huang QT, Han ZJ, Zhao CQ. Molecular cloning, spatiotemporal and functional expression of GABA receptor subunits RDL1 and RDL2 of the rice stem borer Chilo suppressalis. Insect Biochem Mol Biol [Internet] 2018;94:18–27. [CrossRef]
- Wang J da, Chen L fei, Lin D jiang, Zhang J song, Zhao J han, Xiao D, et al. Molecular cloning, characterization and functional analysis of GluCl from the oriental armyworm, Mythimna separata Walker. Pestic Biochem Physiol [Internet] 2019;156:56–62. [CrossRef]
- Hashim O, Charvet CL, Toubaté B, Ahmed AAE, Lamassiaude N, Neveu C, et al. Molecular and functional characterization of GABA receptor subunits GRD and LCCH3 from human louse Pediculus humanus humanus S. 2022;
- Lamassiaude Id N, Toubate Id B, Dric Neveu C, Charnet Id P, Dupuy C, Oise Debierre-Grockiego Id F, et al. The molecular targets of ivermectin and lotilaner in the human louse Pediculus humanus humanus: New prospects for the treatment of pediculosis. 2021. [CrossRef]
- Rispe C, Hervet C, de la Cotte N, Daveu R, Labadie K, Noel B, et al. Transcriptome of the synganglion in the tick Ixodes ricinus and evolution of the cys-loop ligand-gated ion channel family in ticks. BMC Genomics 2022;23:1–21.
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A 2010;107:12168–73.
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 2019;47:W636–41.
- Kumar S, Stecher G, Tamura K, Dudley J. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol [Internet] 2016;33:1870–4. Available from: http://www.ncbi.nlm.nih.gov/guide/taxonomy/.
- Jia ZQ, Sheng CW, Tang T, Liu D, Leviticus K, Zhao CQ, et al. Identification of the ionotropic GABA receptor-like subunits from the striped stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). Pestic Biochem Physiol 2019;155:36–44.
- Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel superfamily of the honeybee, Apis mellifera. Invertebrate Neuroscience 2006;6:123–32.
- Jiang F, Zhang Y, Sun H, Meng X, Bao H, Fang J, et al. Identification of polymorphisms in Cyrtorhinus lividipennis RDL subunit contributing to fipronil sensitivity. Pestic Biochem Physiol [Internet] 2015;117:62–7. [CrossRef]
- Taylor-Wells J, Brooke BD, Bermudez I, Jones AK. The neonicotinoid imidacloprid, and the pyrethroid deltamethrin, are antagonists of the insect Rdl GABA receptor. J Neurochem 2015;135:705–13.
- Ménard C, Folacci M, Brunello L, Charreton M, Collet C, Mary R, et al. Multiple combinations of RDL subunits diversify the repertoire of GABA receptors in the honey bee parasite Varroa destructor. Journal of Biological Chemistry 2018;293:19012–24.
- Henry C, Cens T, Charnet P, Cohen-Solal C, Collet C, van-Dijk J, et al. Heterogeneous expression of GABA receptor-like subunits LCCH3 and GRD reveals functional diversity of GABA receptors in the honeybee Apis mellifera. Br J Pharmacol 2020;177:3924–40.
- Thomas GWC, Dohmen E, Hughes DST, Murali SC, Poelchau M, Glastad K, et al. Gene content evolution in the arthropods. Genome Biol 2020;21.
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science (1979) 2014;346:763–7.
- Previte D, Olds BP, Yoon K, Sun W, Muir W, Paige KN, et al. Differential gene expression in laboratory strains of human head and body lice when challenged with Bartonella quintana, a pathogenic bacterium. Insect Mol Biol 2014.
- Del Villar SG, Jones AK. Cloning and functional characterisation of the duplicated RDL subunits from the pea aphid, acyrthosiphon pisum. Int J Mol Sci 2018;19.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).