Submitted:
28 May 2024
Posted:
28 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- A novel ResNet60 architecture is proposed for the classification of ovarian tumors as benign and malignant.
- The ResNet60 classifier performs the classification task with 97.5% accuracy on the test dataset, which is higher than other state-of-the-art architectures implemented - GoogLeNet (Inception-v1), Inception-v4, VGG16, VGG19, ResNet50, EfficientNetB0.
- The ResNet60 model results are then provided for interpretation to the explainable AI methods LIME, Saliency Map, Grad-CAM, Occlusion Analysis, SHAP and SmoothGrad.
- The above explainable AI methods highlighted the features and regions of interest that were given most importance by the ResNet60 model in obtaining the final classification output, such as the shape and neighbouring area of the tumor.
2. Related Work
2.1. Existing Work Explainability of Classification Models in the Medical Imaging Domain
3. Research Gap and Motivation
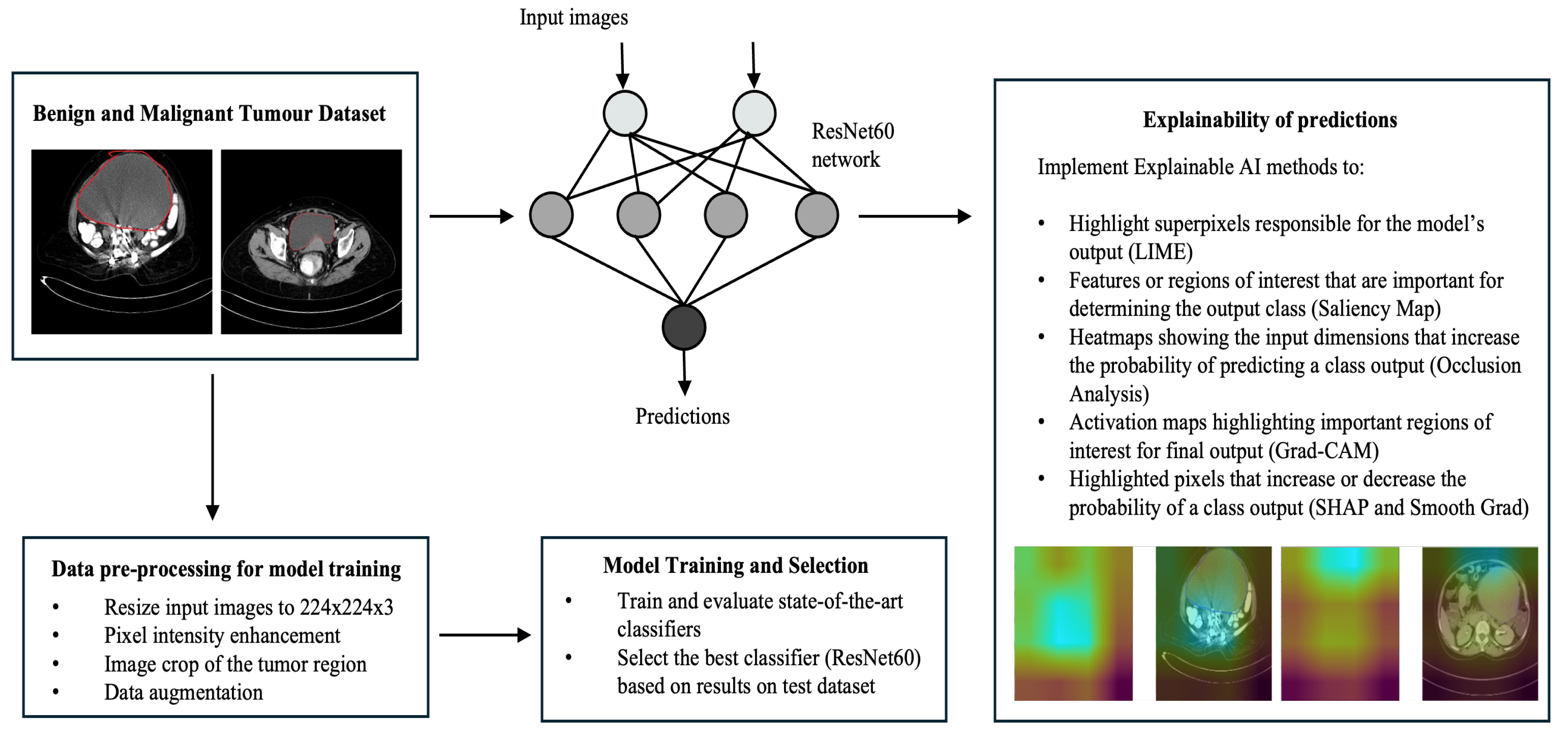
4. Methodology
4.1. Proposed ResNet60 Architecture
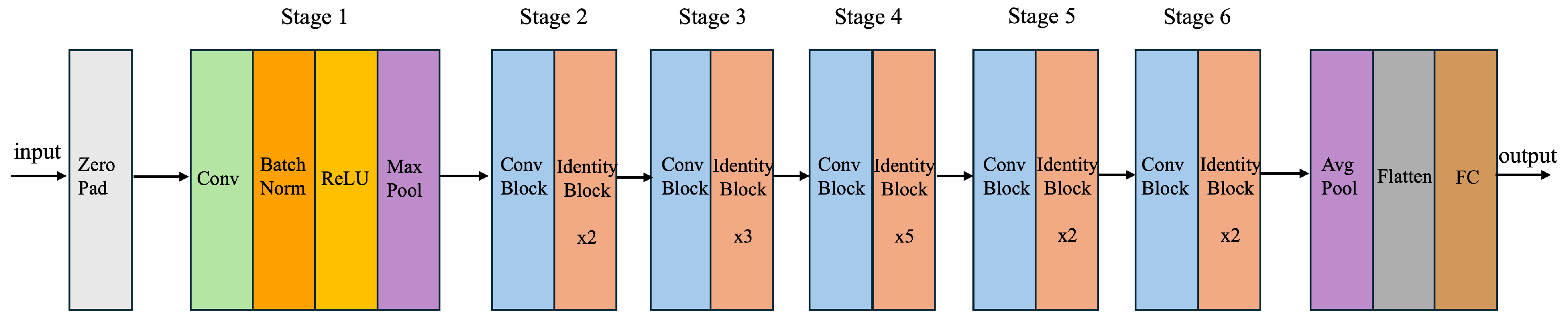
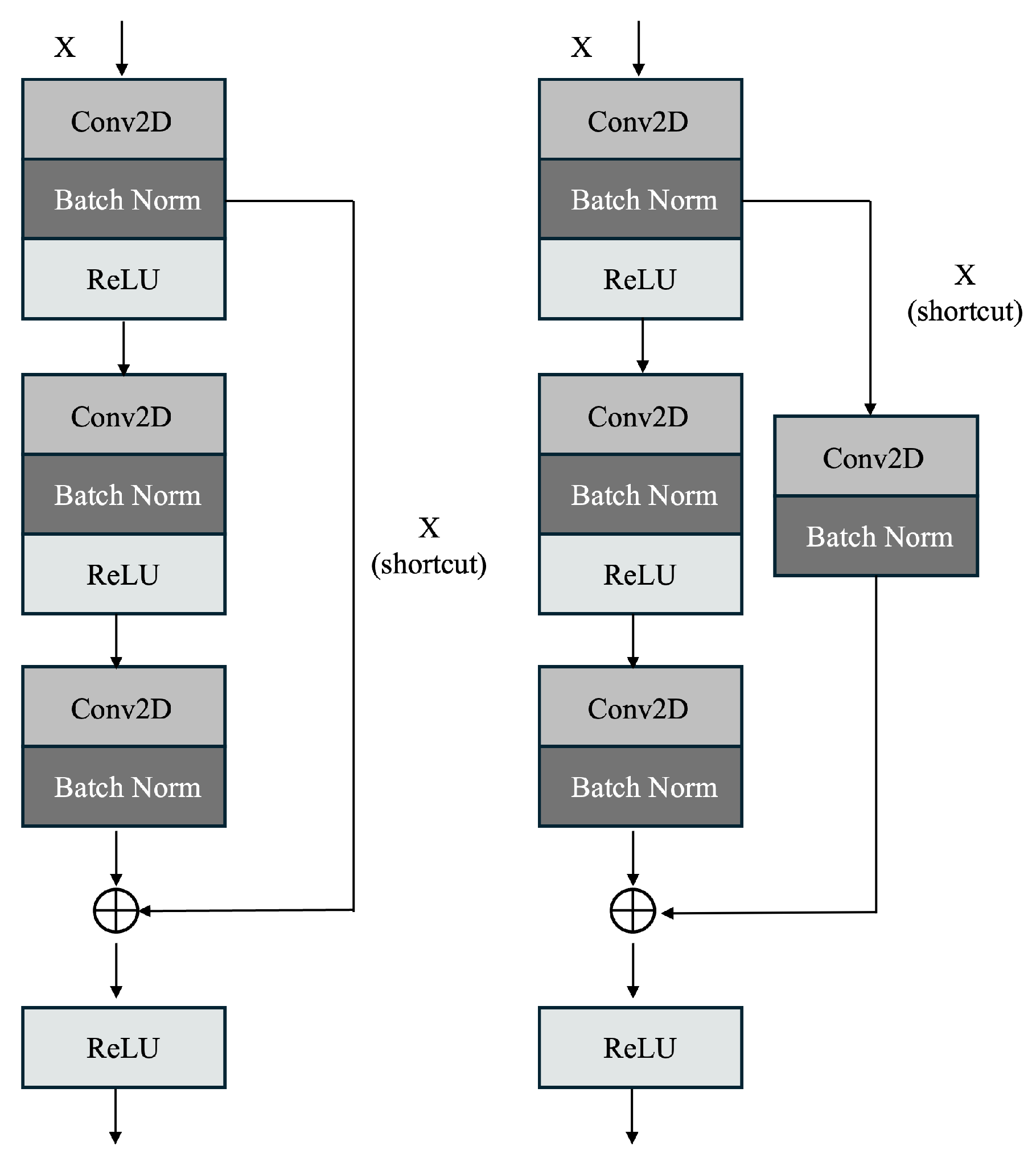
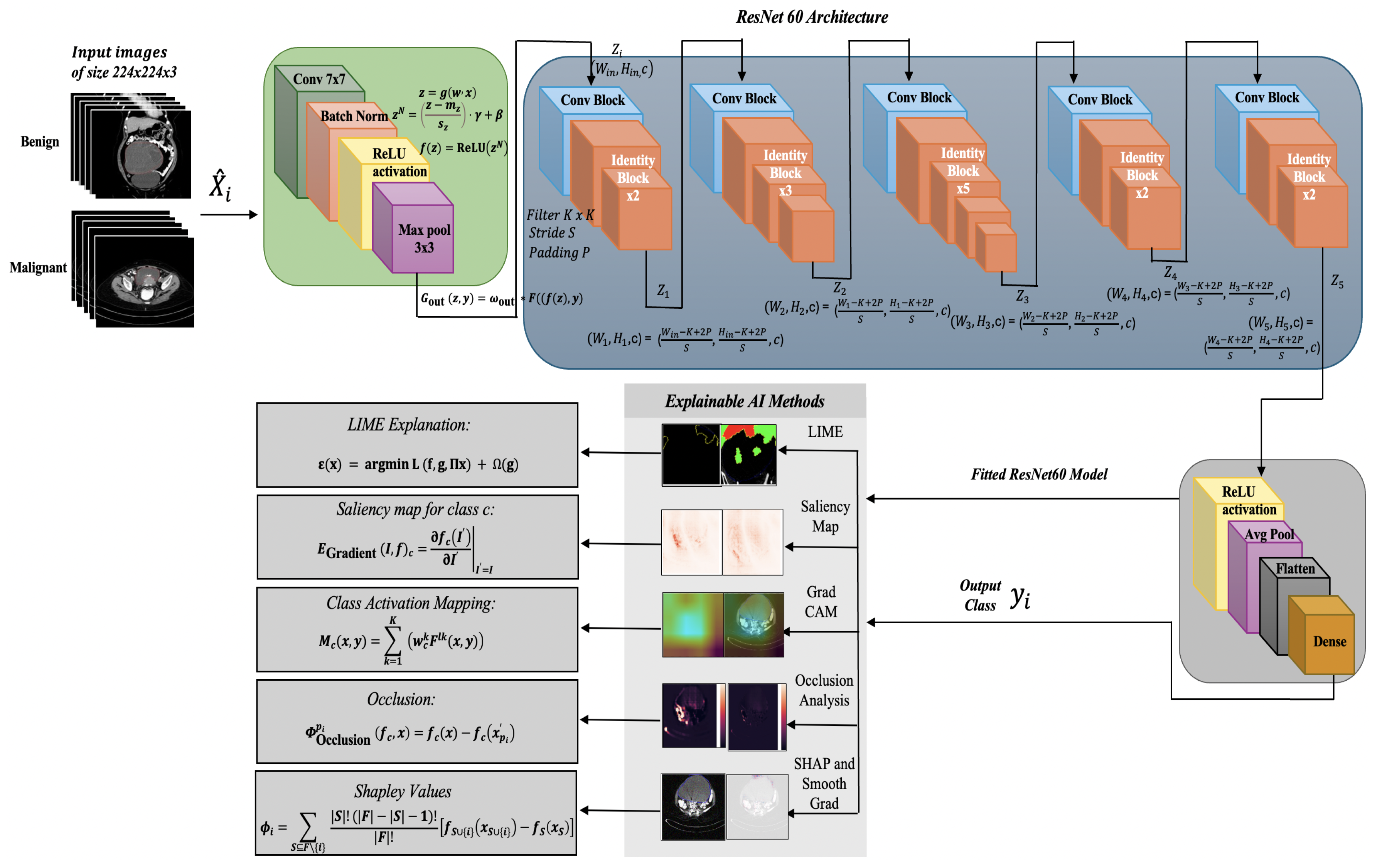
| Algorithm 1: Image Classification using custom ResNet60 Architecture |
 |
4.2. Explainable AI Methods
5. Results and Discussion
5.1. Data Source and Description
5.2. Data Preprocessing and Dataset Preparation for Training and Evaluation
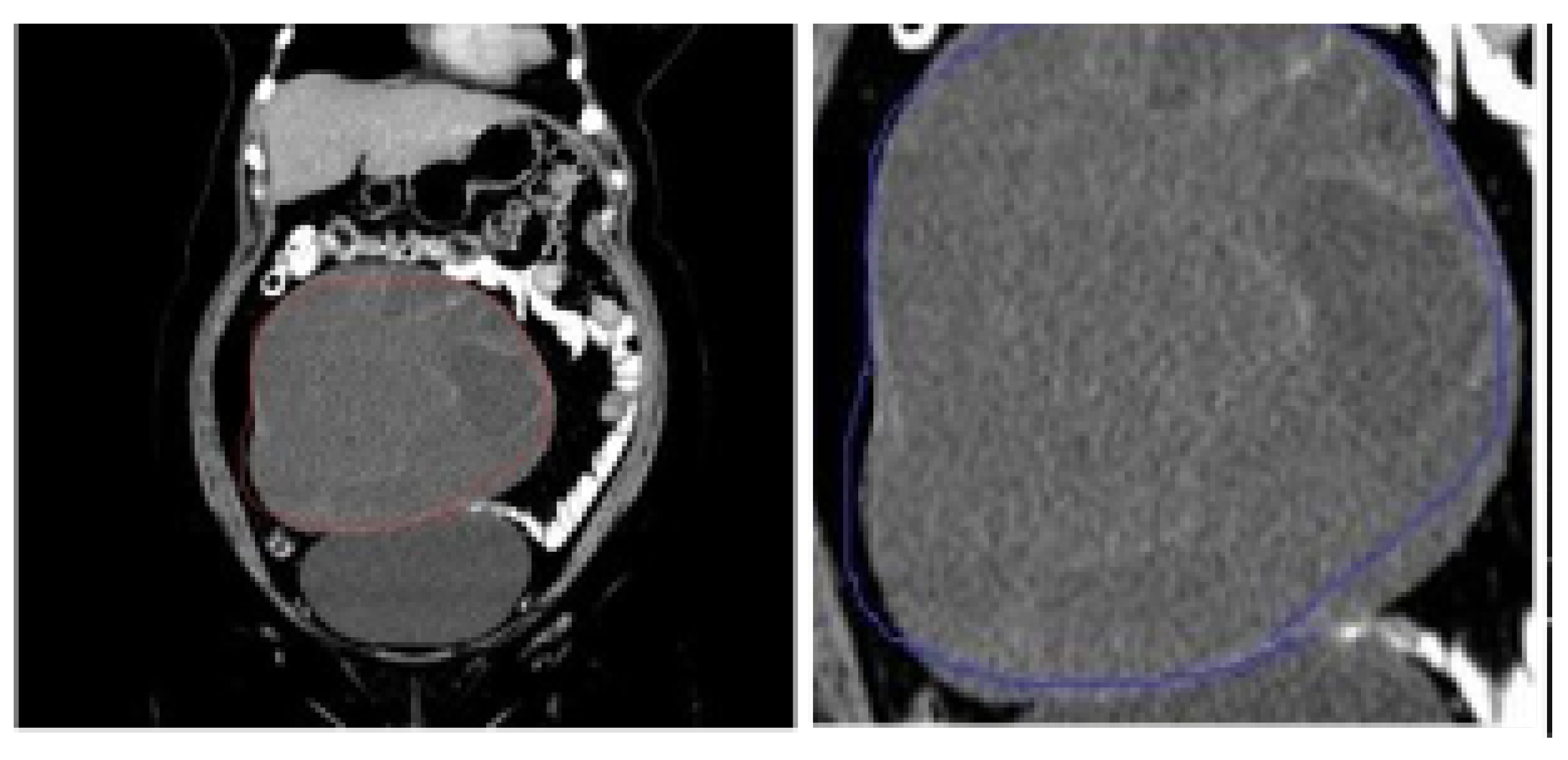
5.3. LIME Results
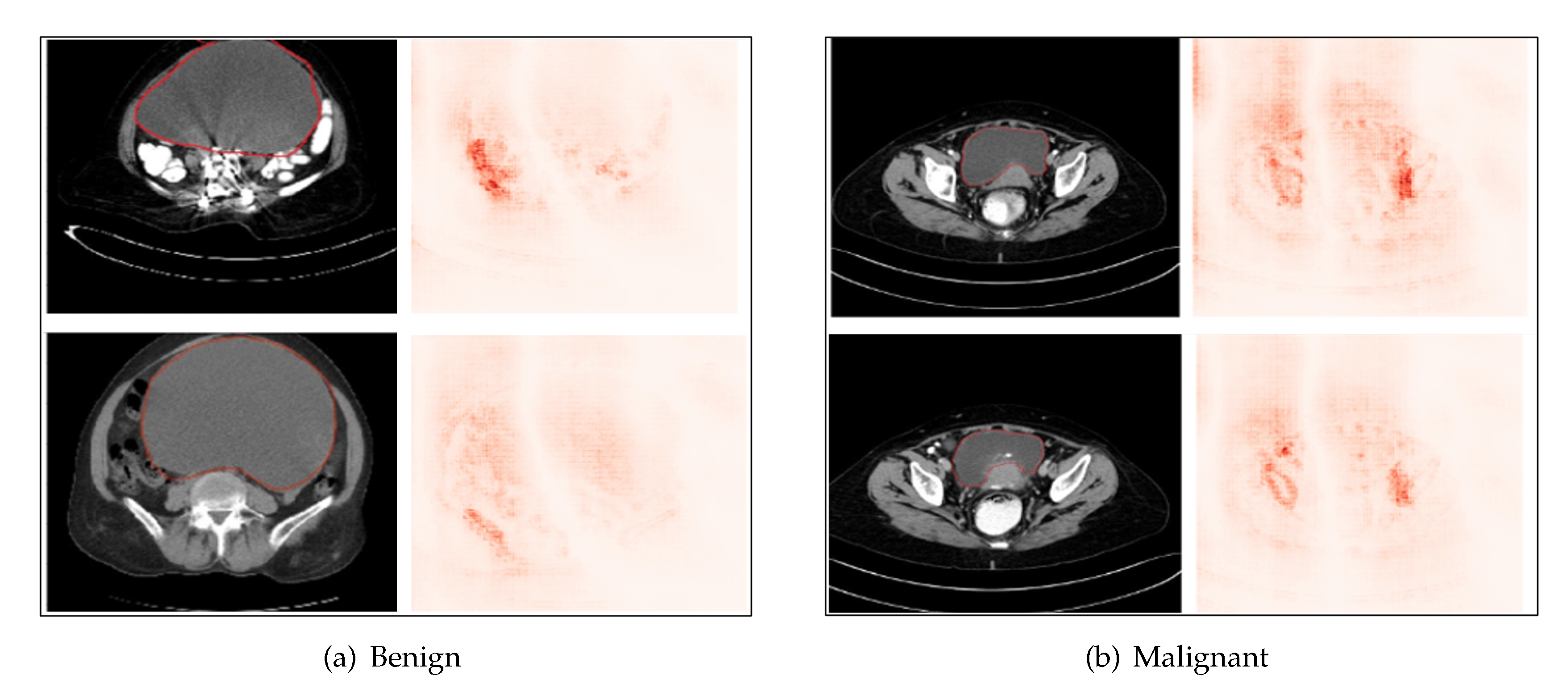
5.4. Saliency Map Results
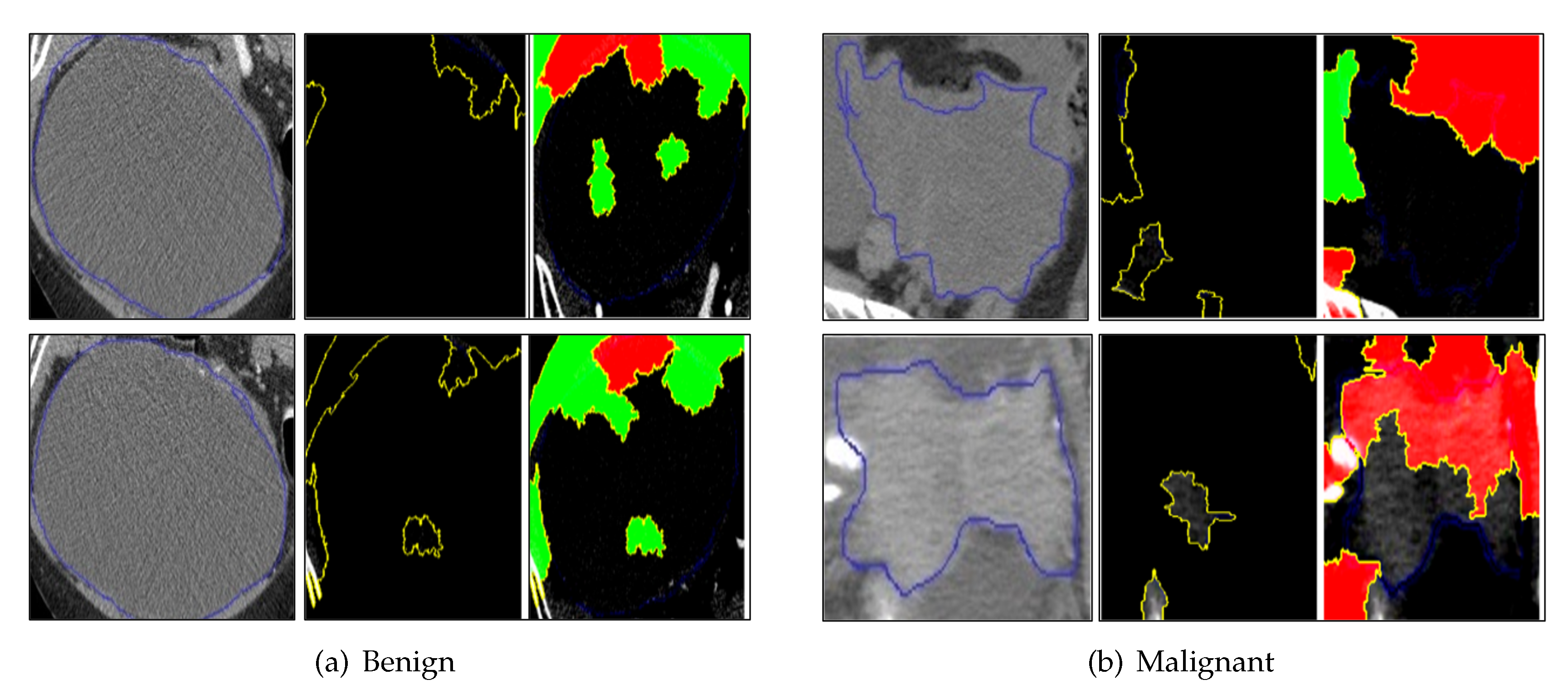
5.5. Occlusion Analysis Results
5.6. Grad-CAM Results
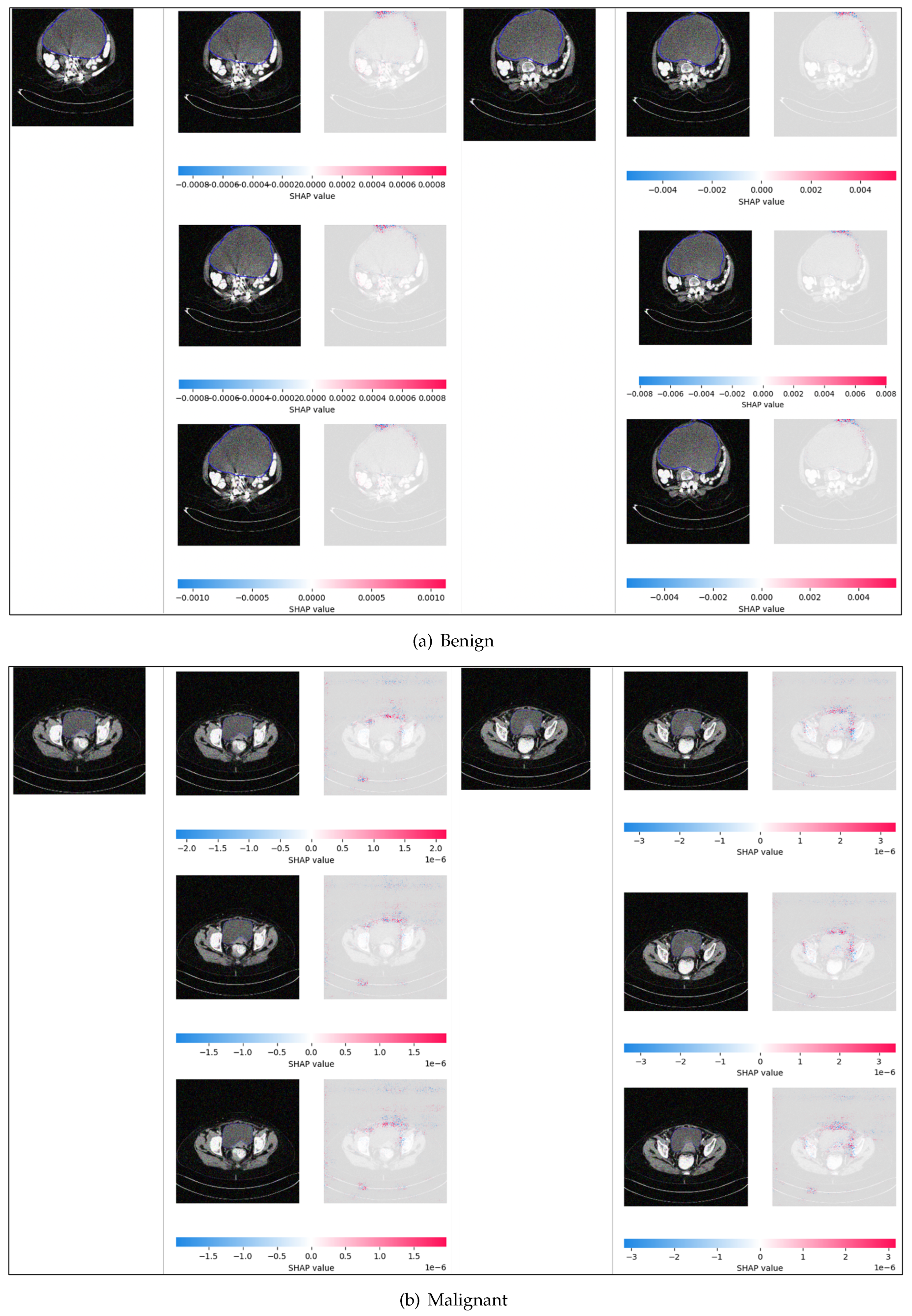
5.7. SHAP and SmoothGrad Results
5.8. Comparison of the Explainable AI Results


6. Conclusion and Scope of Future Work
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wojtyła, C., Bertuccio, P., Giermaziak, W., Santucci, C., Odone, A., Ciebiera, M., ... & La Vecchia, C. (2023). European trends in ovarian cancer mortality, 1990–2020 and predictions to 2025. European Journal of Cancer, 194, 113350.
- Asangba, A. E., Chen, J., Goergen, K. M., Larson, M. C., Oberg, A. L., Casarin, J., ... & Walther-Antonio, M. R. (2023). Diagnostic and prognostic potential of the microbiome in ovarian cancer treatment response. Scientific reports, 13(1), 730.
- Jan, Y. T., Tsai, P. S., Huang, W. H., Chou, L. Y., Huang, S. C., Wang, J. Z., ... & Wu, T. H. (2023). Machine learning combined with radiomics and deep learning features extracted from CT images: a novel AI model to distinguish benign from malignant ovarian tumors. Insights into Imaging, 14(1), 68.
- Vela-Vallespín, C., Medina-Perucha, L., Jacques-Aviñó, C., Codern-Bové, N., Harris, M., Borras, J. M., & Marzo-Castillejo, M. (2023). Women’s experiences along the ovarian cancer diagnostic pathway in Catalonia: A qualitative study. Health Expectations, 26(1), 476-487.
- Zacharias, J., von Zahn, M., Chen, J., & Hinz, O. (2022). Designing a feature selection method based on explainable artificial intelligence. Electronic Markets, 32(4), 2159-2184.
- Gupta, M., Sharma, S. K., & Sampada, G. C. (2023). Classification of Brain Tumor Images Using CNN. Computational Intelligence and Neuroscience, 2023.
- Yadav, S. S., & Jadhav, S. M. (2019). Deep convolutional neural network based medical image classification for disease diagnosis. Journal of Big data, 6(1), 1-18.
- Liang, J. (2020, September). Image classification based on RESNET. Journal of Physics Conference Series 1634(1).
- Saranya, A., & Subhashini, R. (2023). A systematic review of Explainable Artificial Intelligence models and applications: Recent developments and future trends. Decision analytics journal, 100230.
- Baehrens, D., Schroeter, T., Harmeling, S., Kawanabe, M., Hansen, K., & Müller, K. R. (2010). How to explain individual classification decisions. The Journal of Machine Learning Research, 11, 1803-1831.
- Xu, F., Uszkoreit, H., Du, Y., Fan, W., Zhao, D., & Zhu, J. (2019). Explainable AI: A brief survey on history, research areas, approaches and challenges. In Natural Language Processing and Chinese Computing: 8th CCF International Conference, NLPCC 2019, Dunhuang, China, October 9–14, 2019, Proceedings, Part II 8 (pp. 563-574). Springer International Publishing.
- Yang, W., Wei, Y., Wei, H., Chen, Y., Huang, G., Li, X., ... & Kang, B. (2023). Survey on explainable AI: From approaches, limitations and Applications aspects. Human-Centric Intelligent Systems, 3(3), 161-188.
- Samek, W., Wiegand, T., & Müller, K. R. (2017). Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv preprint arXiv:1708.08296.
- Singh, A., Sengupta, S., & Lakshminarayanan, V. (2020). Explainable deep learning models in medical image analysis. Journal of imaging, 6(6), 52.
- Van der Velden, B. H., Kuijf, H. J., Gilhuijs, K. G., & Viergever, M. A. (2022). Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Medical Image Analysis, 79, 102470.
- Linardatos, P., Papastefanopoulos, V., & Kotsiantis, S. (2020). Explainable ai: A review of machine learning interpretability methods. Entropy, 23(1), 18.
- Ribeiro, M. T., Singh, S., & Guestrin, C. (2016, August). " Why should i trust you?" Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining (pp. 1135-1144).
- An, J., Zhang, Y., & Joe, I. (2023). Specific-Input LIME Explanations for Tabular Data Based on Deep Learning Models. Applied Sciences, 13(15), 8782.
- Alqaraawi, A., Schuessler, M., Weiß, P., Costanza, E., & Berthouze, N. (2020, March). Evaluating saliency map explanations for convolutional neural networks: a user study. In Proceedings of the 25th international conference on intelligent user interfaces (pp. 275-285).
- Simonyan, K., Vedaldi, A., & Zisserman, A. (2013). Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034.
- Li, X. H., Shi, Y., Li, H., Bai, W., Song, Y., Cao, C. C., & Chen, L. (2020). Quantitative evaluations on saliency methods: An experimental study. arXiv preprint arXiv:2012.15616.
- Resta, M., Monreale, A., & Bacciu, D. (2021). Occlusion-based explanations in deep recurrent models for biomedical signals. Entropy, 23(8), 1064.
- Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., & Batra, D. (2017). Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision (pp. 618-626).
- Cao, Q. H., Nguyen, T. T. H., Nguyen, V. T. K., & Nguyen, X. P. (2023). A Novel Explainable Artificial Intelligence Model in Image Classification problem. arXiv preprint arXiv:2307.04137.
- Fu, R., Hu, Q., Dong, X., Guo, Y., Gao, Y., & Li, B. (2020). Axiom-based grad-cam: Towards accurate visualization and explanation of cnns. arXiv preprint arXiv:2008.02312.
- Kakogeorgiou, I., & Karantzalos, K. (2021). Evaluating explainable artificial intelligence methods for multi-label deep learning classification tasks in remote sensing. International Journal of Applied Earth Observation and Geoinformation, 103, 102520.
- Lundberg, S. M., & Lee, S. I. (2017). A unified approach to interpreting model predictions. Advances in neural information processing systems, 30.
- Bach, S., Binder, A., Montavon, G., Klauschen, F., Müller, K. R., & Samek, W. (2015). On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PloS one, 10(7), e0130140.
- Hooker, S., Erhan, D., Kindermans, P. J., & Kim, B. (2019). A benchmark for interpretability methods in deep neural networks. Advances in neural information processing systems, 32.
- Ishikawa, S. N., Todo, M., Taki, M., Uchiyama, Y., Matsunaga, K., Lin, P., ... & Yasui, M. (2023). Example-based explainable AI and its application for remote sensing image classification. International Journal of Applied Earth Observation and Geoinformation, 118, 103215.
- Shivhare, I., Jogani, V., Purohit, J., & Shrawne, S. C. (2023, January). Analysis of Explainable Artificial Intelligence Methods on Medical Image Classification. In 2023 Third International Conference on Advances in Electrical, Computing, Communication and Sustainable Technologies (ICAECT) (pp. 1-5). IEEE.
- Montavon, G., Lapuschkin, S., Binder, A., Samek, W., & Müller, K. R. (2017). Explaining nonlinear classification decisions with deep taylor decomposition. Pattern recognition, 65, 211-222.
- Shrikumar, A., Greenside, P., & Kundaje, A. (2017, July). Learning important features through propagating activation differences. In International conference on machine learning (pp. 3145-3153). PMLR.
- Smilkov, D., Thorat, N., Kim, B., Viégas, F., & Wattenberg, M. (2017). Smoothgrad: removing noise by adding noise. arXiv preprint arXiv:1706.03825.
- Soltani, S., Kaufman, R. A., & Pazzani, M. J. (2022). User-centric enhancements to explainable ai algorithms for image classification. In Proceedings of the Annual Meeting of the Cognitive Science Society (Vol. 44, No. 44).
- Springenberg, J. T., Dosovitskiy, A., Brox, T., & Riedmiller, M. (2015). Striving for simplicity: The all convolutional net. In arxiv: cs. arXiv preprint arXiv:1412.6806.
- Sundararajan, M., Taly, A., & Yan, Q. (2017, July). Axiomatic attribution for deep networks. In International conference on machine learning (pp. 3319-3328). PMLR.
- Vermeire, T., Brughmans, D., Goethals, S., de Oliveira, R. M. B., & Martens, D. (2022). Explainable image classification with evidence counterfactual. Pattern Analysis and Applications, 25(2), 315-335.
- Zeiler, M. D., & Fergus, R. (2014). Visualizing and understanding convolutional networks. In Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part I 13 (pp. 818-833). Springer International Publishing.
- Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., & Torralba, A. (2016). Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2921-2929).
- Wu, B., Fan, Y. and Mao, L. (2021) Large-scale image classification with explainable deep learning scheme, Research Square.


| Stage | Layers | Conv1 Filters | Conv2 Filters | Conv3 Filters | TotalConv Filters | Stride | Padding and ReLU | Batch Norm |
|---|---|---|---|---|---|---|---|---|
| 1 | Conv | 64 | 64 | 256 | 384 | (2, 2) | Valid | Yes |
| 1 | Identity (x2) | 64 | 64 | 256 | 384 | (1, 1) | Valid | Yes |
| 2 | Conv | 128 | 128 | 512 | 768 | (2, 2) | Valid | Yes |
| 2 | Identity (x3) | 128 | 128 | 512 | 768 | (1, 1) | Valid | Yes |
| 3 | Conv | 256 | 256 | 1024 | 1536 | (2. 2) | Valid | Yes |
| 3 | Identity (x5) | 256 | 256 | 1024 | 1536 | (1, 1) | Valid | Yes |
| 4 | Conv | 512 | 512 | 2048 | 3072 | (2, 2) | Valid | Yes |
| 4 | Identity (x2) | 512 | 512 | 2048 | 3072 | (1, 1) | Valid | Yes |
| 5 | Conv | 512 | 512 | 2048 | 3072 | (1, 1) | Valid | Yes |
| 5 | Identity (x2) | 512 | 512 | 2048 | 3072 | (1, 1) | Valid | Yes |
| 6 | Conv | 1024 | 1024 | 4096 | 6144 | (2, 2) | Valid | Yes |
| 6 | Identity (x2) | 1024 | 1024 | 4096 | 6144 | (1, 1) | Valid | Yes |
| Train | Validation | Test | |||
|---|---|---|---|---|---|
| Benign | Malignant | Benign | Malignant | Benign | Malignant |
| 2633 | 1011 | 1128 | 433 | 376 | 144 |
|
Model Name |
Variant Name |
Train Accuracy |
Train Loss |
Test Accuracy |
Test Loss |
|---|---|---|---|---|---|
| GoogLeNet (Inception v1) |
91.2% | 0.24 | 92.5% | 0.25 | |
| Inception | Inception v4 |
93.8% | 0.12 | 80% | 42.60 |
| VGG 16 |
71.8% | 2.46 | 72.5% | 2.42 | |
| VGG | VGG 19 |
74.4% | 0.56 | 74.37% | 0.57 |
| ResNet 50 |
96.2% | 0.16 | 90% | 0.52 | |
| ResNet | ResNet60 (proposed) |
95% | 0.19 | 97.5% | 0.14 |
| EfficientNet | EfficientNet B0 |
71.8% | 0.60 | 72.5% | 0.58 |
| DenseNet | Densenet 121 |
70% | 2.25 | 70.63% | 0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




